Abstract
Although peroxisome proliferator-activated receptor alpha (PPARα) is closely associated with myocardial fatty acid metabolism, the pathophysiological role of PPARα in myocardial infarction (MI) is not yet known. The aim of the present study was to clarify the relationship between cardiac energy metabolism and PPARα expression in the remodelling of myocardium after MI. We assayed the expression of PPARα and several metabolic genes in cultured cardiac cells (myocytes and nonmyocytes) and in MI hearts. PPARα was strongly expressed in cardiac myocytes but not in nonmyocytes (mainly fibroblasts). In MI rats, PPARα and PPARα-regulated genes (lipoprotein lipase, heart-type fatty acid binding protein, long-chain acyl-CoA dehydrogenase and uncoupling protein-3) were decreased concomitantly, whereas uncoupling protein-2 was not decreased in severely ischemic regions. Immunohistochemical staining for PPARα was less decreased in borderline myocardium than in sham-operated hearts. Furthermore, in electron microscopic study, there were no lipid droplet accumulations in surviving myocardium after MI. Our results suggest that the reduced expression of PPARα is closely related to that of fatty acid metabolism genes in infarcted myocardium, and PPARα may play an important role in cardiac energy metabolism during remodelling after MI.
Keywords: Energy metabolism, Fibroblast, Lipid metabolism, Myocardial infarction, Myocyte, PPARα, Remodelling
In response to a transmural myocardial infarction (MI), the left ventricle undergoes significant remodelling (1). Studies in the rat model of postinfarction remodelling have provided initial evidence that the expression of several genes may be altered in noninfarcted myocardium. Glucose oxidation is increased and fatty acid oxidation (FAO) is decreased in surviving rat myocardium (2). Atrial natriuretic factor (ANF) messenger RNA (mRNA) and protein levels are elevated in response to MI as a compensatory response to improve hemo-dynamics (3). Moreover, using complementary DNA (cDNA) microarray analysis, many post-MI changes in expression have been found in genes that encode proteins that have been implicated in cytoskeletal architecture, contractility and metabolism (4).
Long-chain fatty acids are important substrates for energy production in the heart. Intermediary metabolism in the heart changes markedly from fetal to adult life (5). In the adult heart, fatty acids are the preferred energy substrates. In contrast, the fetal heart appears to use lactate as its primary carbon substrate. Therefore, understanding long-chain fatty acid metabolism may help in elucidating the mechanism of various adult heart diseases. A key regulator of substrate switching in the heart is postulated to be peroxisome proliferator-activated receptor alpha (PPARα), a member of the ligand-activated nuclear receptor superfamily (6). PPARα has been shown to regulate the expression of several genes involved in FAO. These include the lipoprotein lipase (LPL), fatty acid translo-case/CD36, heart-type fatty acid binding protein (H-FABP), muscle-specific carnitine palmitoyltransferase I, as well as medium- and long-chain acyl-CoA dehydrogenase (MCAD and LCAD) (7–11). PPARα binds to peroxisome proliferator response elements in the promoter region of target genes as a heterodimeric partner with the retinoid X receptor (RXR) and is activated by a variety of ligands, including long-chain fatty acids (12). PPARα knockout mice exhibit low rates of FAO and abnormal accumulation of neutral lipids in both cardiac and hepatic tissues (13,14). In diabetes, plasma fatty acids are elevated and are able to activate PPARα with induction of PPARα-regulated genes (15). In contrast, pressure overload-induced left ventricular hypertrophy has decreased the expression of PPARα and FAO genes (16). Recent studies have demonstrated that PPARα reactivation in the hypertrophied heart or cardiac restricted overexpression of PPARα – signatures of diabetic cardiomyopathy – result in ventricular dysfunction (17,18). Moreover, another study in humans has suggested that PPARα regulates left ventricular growth in response to exercise and hypertension stimuli, illustrating the important role of cardiac fatty acid metabolism in cardiac growth (19). Although such evidence suggests that PPARα is a critical regulator of cardiac fatty acid uptake and utilization, the pathophysiological role of PPARα in MI in vivo is not yet known. Therefore, the aim of the present study was to clarify the relationship between cardiac energy metabolism and PPARα expression in myocardium undergoing remodelling after MI.
MATERIALS AND METHODS
Cell culture
Cultured neonatal rat cardiac myocytes were prepared as described previously, with minor modifications (20,21). Briefly, whole hearts removed from zero- to two-day-old Sprague-Dawley rats (Clea, Japan) were dispersed via agitation in balanced salt solution containing 0.03% collagenase type II and 0.06% pancreatin (SIGMA, USA). Cardiac myocytes were separated from cardiac nonmyocytes by the discontinuous Percoll (SIGMA, USA) gradient method. Cardiac myocytes and nonmyocytes were suspended in Dulbecco’s modified Eagle’s medium (DMEM; Nissui, Japan) supplemented with 10% fetal bovine serum (FBS; Invitrogen Corporation, USA). After incubation for 48 h, the cultures were incubated in serum-free DMEM for 24 h.
Animals and experimental MI
The investigation conformed with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication No. 85–23, revised 1996). Infarcts and sham operations were carried out in male Sprague-Dawley rats weighing 250 g to 300 g. Left anterior descending coronary artery (LAD) ligation was performed by a technique described previously (21,22). Briefly, the rats were anesthetized with ketamine hydrochloride (70 mg/kg, intramuscularly) and xylazine (4 mg/kg, intramuscularly). The LAD was ligated at approximately 2 mm from its origin. To standardize the nutritional state, rats were fasted for 24 h before sacrifice. Six weeks after the operation, the heart was excised. The heart excluding scar tissue was divided into the infarcted region (borderline myocardium, including adjacent noninfarcted myocardium) and the remote region (remote noninfarcted left ventricular free wall). Each tissue specimen was quickly frozen in liquid nitrogen and stored at −80°C.
RNA extraction and Northern blot analysis
Total RNA was isolated by the guanidine thiocyanate-phenol-chloroform extraction method of Chomczynski and Sacchi (23). Total RNA (15 μg) was subjected to electrophoresis in denaturing formaldehyde-agarose gel and transferred to a nylon membrane filter (Hybond-XL, Amersham Biosciences Corporation, USA). The filter was hybridized with cDNA fragments that were labelled with (α-32P) dCTP (Amersham Biosciences Corporation, USA) by the random priming procedure. The probes were obtained using reverse transcriptase-polymerase chain reaction (RT-PCR) from cDNAs encoding rat PPARα, glucose transporters 1 and 4 (GLUT1 and GLUT4), LPL, H-FABP, LCAD, hormone-sensitive lipase (HSL), succinate dehydrogenase (SDH), cytochrome oxidase (COX), uncoupling protein-2 and -3 (UCP-2 and UCP-3), and ANF. The PCR primers used in the present study (Table 1) were based on the sequences reported in GenBank. The sequences of the cDNA fragments were confirmed by the dideoxy sequence. To exclude the possibility that changes in gene expression after MI were due to changes in the myocyte-to-nonmyocyte ratio (eg, myocytes dropout, apoptosis and fibrosis), we also measured a muscle-specific marker, cardiac alpha-actin, in the rat hearts.
TABLE 1.
Polymerase chain reaction primers used for amplification of metabolic genes
| PPARα | Sense | AAGAGAATCCACGAAGCCTAC |
| (M88592, 662 bp) | Antisense | TGAAGGAGTTTTGGGAAGAGA |
| GLUT1 | Sense | CCCATCCACCACACTCACCAC |
| (M13979, 604 bp) | Antisense | GACCTTCTTCTCCCGCATCAT |
| GLUT4 | Sense | GATGACGGTGGCTCTGCTGCT |
| (D28561, 497 bp) | Antisense | CTAAAGTGCTGCGAGGAAAGG |
| LPL | Sense | ATGAAGAAAACCCCAGCAAG |
| (L03294, 413 bp) | Antisense | TGACCAGCGGAAGTAGGAGT |
| H-FABP | Sense | CATGACCAAGCCGACCACAAT |
| (M18034, 420 bp) | Nonsense | CCAGGAAAAGCCCAACCAAAG |
| LCAD | Sense | CTGCTCCTCCGCTCCCTTCGT |
| (J05029, 486 bp) | Nonsense | ACACTTGCCCGCCGTCATCTG |
| HSL | Sense | CTCGGCATTCTCACACAGCAT |
| (X51415, 442 bp) | Nonsense | GAAGAGCACTCCTGGTCGGTT |
| SDH | Sense | CCACACCAGCACCGGGGACG |
| (M60879, 535 bp) | Nonsense | ATGCACCGAGGCGCAGGCAG |
| COX | Sense | TAATAGAAGGCAACCGAAACC |
| (J01435, 630 bp) | Antisense | ATGGGAGGGGAAGTAGTAAGG |
| UCP-2 | Sense | GTCCGCGCAGCCTCTACAAT |
| (AB00613, 448 bp) | Antisense | CCGAAGGCAGAAGTGAAGTG |
| UCP-3 | Sense | CGAATTGGCCTCTACGACTCT |
| (AB00614, 508 bp) | Antisense | CCACCATCCTCAGCATACAGT |
| ANF | Sense | GTGAGCCGAGACAGCAAACAT |
| (M27498, 451 bp) | Antisense | TCAATCCTACCCCCGAAGCAG |
| Cardiac alpha-actin | Sense | CGCCCAAAGCACGCCTACAGA |
| (X80130, 483 bp) | Antisense | AGAAGCGTACAGGGACAGCAC |
Accession number and length of amplified fragments are shown in parentheses. ANF Atrial natriuretic factor; bp Base pairs; COX Cytochrome oxidase; GLUT Glucose transporter; H-FABP Heart-type fatty acid binding protein; HSL Hormone-sensitive lipase; LCAD Long-chain acyl-CoA dehydrogenase; LPL Lipoprotein lipase; PPARα Peroxisome proliferator-activated receptor alpha; SDH Succinate dehydrogenase; UCP Uncoupling protein
TaqMan RT-PCR
Real-time quantitative RT-PCR was performed on cDNA generated from 100 ng of total RNA. For the PCR, we used 200 nM of both sense and antisense primers, 100 nM of TAMRA-labelled primer probe and TaqMan Universal PCR Master Mix (Applied Biosystems, USA) in a final volume of 50 μL with an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, USA). Sense, antisense and probe primers were GCAGAGGTCCGATTCTTCCA, GCAAGGTAACCTG-GTCATTCAAG and CACGGAATTTGCCAAGGCTATC-CCA for rat PPARα; TTCCAGTATG- TTGCGGATGCT, GCCGAGATCTGGTCAAATGTC and TGGGTCCC-TACGTCTTCCTTCTATTTGCC for rat GLUT4; and GTGCATGGCCGTTCTTAGTTG, TGAACGCCACTT-GTCCCTCTA and CGATAACGAACGAGACTCTCG-GCATGC for 18S. Results were normalized to 18S to correct for differences in RNA quantity and quality and then expressed as a ratio of sham-operated hearts.
Immunohistochemical staining
Immunohistochemical staining was performed as previously described (21). Briefly, sections were deparaffinized and treated with 3% hydrogen peroxidase solution for 20 min. After incubation with normal rabbit serum for 20 min, sections were incubated overnight with goat polyclonal anti-PPARα antibody (Santa Cruz Biotechnology Inc, USA) at room temperature. Next, sections were incubated with biotinylated anti-goat immunoglobulin G (IgG; Vectastain ABC kit, Vector Laboratories Inc, USA) according to the manufacturer’s directions and stained with 3,3′-diaminobenzidine (DAKO, Denmark). For a negative control, the primary antibody was replaced by normal goat serum (DAKO, Denmark).
Electron microscopy
Tissues were prepared for histology by dissection in ice-cold fixative (2.5% glutaraldehyde in 0.1 mol/L of phosphate buffer, pH 7.4). After 2 h of fixation at 4°C, the tissues were washed with ice-cold phosphate buffer containing 8% sucrose (weight by volume) and postfixed in 2% osmium tetroxide at 4°C for 2 h. The tissues were dehydrated in an alcohol series and embedded in Epon resin. Ultrathin sections (70 nm) were stained with uranyl acetate and lead citrate, and viewed on a Hitachi H7500 electron microscope (Hitachi Ltd, Japan).
Statistical analysis
Results were expressed as the mean ± SEM. Differences between the groups were calculated by Student’s t test and one-way analysis of variance with Bonferroni adjustment. A value of P<0.05 was considered statistically significant.
RESULTS
Expression of PPARα mRNA in cardiac myocytes
To confirm the types of cells expressing PPARα mRNA, PPARα and other metabolic genes mRNA in total RNA obtained from cultured neonatal rat cardiac myocytes and non-myocytes (mainly fibroblasts) were measured by Northern blot analysis (Figure 1). PPARα mRNA, as well as mRNA of FAO genes (LPL, H-FABP and LCAD), was strongly expressed in cultured myocytes and was negligible in fibroblasts. The other metabolic gene mRNAs (HSL, SDH and COX) were mainly expressed in cardiac myocytes. GLUT1 and UCP-2 mRNA were expressed in both cultured cardiac myocytes and fibro-blasts. In contrast, the expressions of GLUT4 and UCP-3 mRNA were not found in either cells.
Figure 1).
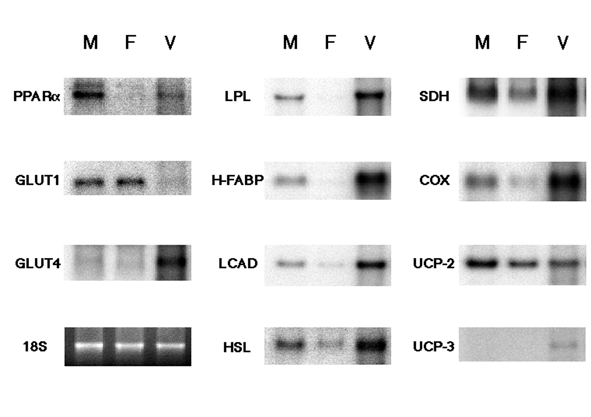
Northern blot analysis of messenger RNA expressions of peroxisome proliferator-activated receptor alpha (PPARα), glucose transporters 1 and 4 (GLUT1 and 4), lipoprotein lipase (LPL), heart-type fatty acid binding protein (H-FABP), long-chain acyl-CoA dehy-drogenase (LCAD), hormone-sensitive lipase (HSL), succinate dehydrogenase (SDH), cytochrome oxidase (COX), and uncoupling protein-2 and -3 (UCP-2 and UCP-3) in cultured rat cardiac cells. F Neonatal rat cardiac nonmyocytes; M Neonatal rat cardiac myocytes; V Adult rat heart tissues
General characteristics of animals and hearts
Rats with infarction sacrificed after six weeks did not exhibit symptoms of heart failure (lethargy, pericardial effusion and pleural effusion). Six weeks after surgery, systolic blood pressure was significantly lower and heart weight to body weight ratio was significantly increased in the infarcted rats (Table 2). There were no significant changes in the concentration of plasma cholesterol, triglycerides, free fatty acids, lactic acid or glucose between the two groups (data not shown).
TABLE 2.
Body weight, heart weight and hemodynamic parameters of sham-operated rats (Sham) and rats with coronary ligation (Infarcted)
| Experimental group | Sham (n=6) | Infarcted (n=6) |
|---|---|---|
| Heart rate (beats/min) | 343±11 | 336±6 |
| Systolic BP (mmHg) | 137±1.5 | 123±3.4* |
| Diastolic BP (mmHg) | 83±4.3 | 89±4.3 |
| Body weight (g) | 504±9 | 515±4 |
| Heart weight (mg) | 1151±36 | 1390±50* |
| Heart weight/body weight (mg/g) | 2.28±0.06 | 2.70±0.10* |
Results were expressed as the mean ± SEM.
P<0.05 compared with sham-operated rats. BP Blood pressure
Myocardial mRNA expressions of PPARα and other metabolic genes
To determine whether PPARα regulates the metabolism of myocardial fatty acids during remodelling after MI, the levels of PPARα mRNA were examined in the infarcted region, remote region and sham-operated rat hearts at six weeks after MI-induction. In Northern blot analysis, the expression of PPARα mRNA and GLUT4 mRNA was decreased in the infarcted region and only slightly increased in the remote region compared with in sham-operated hearts (Figure 2A). In contrast, ANF (a marker of hypertrophy) mRNA expression was increased in the remote region; this increase was much more pronounced in the infarcted region compared with in sham-operated hearts. The mRNA expression of GLUT1 in adult rat hearts was lower than that of GLUT4, and there were no significant changes of GLUT1 mRNA expression between the groups (data not shown).
Figure 2).
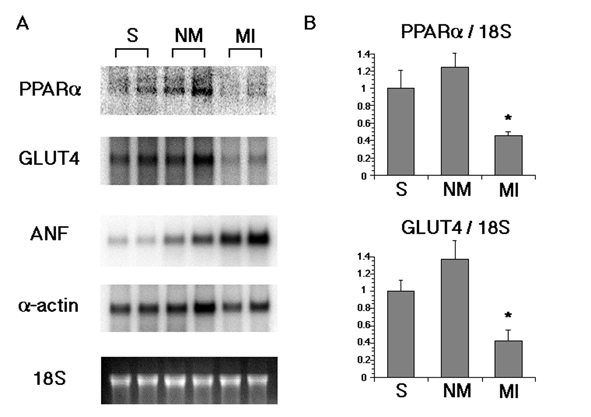
A Northern blot analysis of messenger RNA (mRNA) expressions of peroxisome proliferator-activated receptor alpha (PPARα), glucose transporter 4 (GLUT4), atrial natriuretic factor (ANF) and cardiac alpha-actin in rat hearts six weeks after myocardial infarct induction. B TaqMan reverse transcriptase-polymerase chain reaction analysis of mRNA expressions of PPARα and GLUT4 in rat hearts six weeks after myocardial infarct induction (n=4). Results were compared with values measured in sham-operated rat hearts and expressed as the mean ± SEM. All values are normalized against the expressions of 18S. *P<0.05 compared with sham-operated rat hearts. MI Infarcted myocardium including borderline region; NM Remote noninfarcted left ventricular free wall; S Sham-operated rat hearts
Moreover, to quantify the differences in PPARα and GLUT4 mRNA expression, TaqMan quantitative RT-PCR analysis was performed (Figure 2B). At six weeks after the operation, the mRNA level of PPARα was significantly decreased (−54±5%; P<0.05) and the mRNA level of GLUT4 was significantly decreased concomitantly (−57±12%; P<0.05) in the infarcted region, compared with in sham-operated hearts. Likely the result of Northern blot analysis, PPARα and GLUT4 mRNA tended to be slightly, although not significantly, higher in the remote region (25±16% and 38±21%, respectively).
Figure 3 depicts the myocardial mRNA expressions of FAO genes (LPL, H-FABP and LCAD), a lipolytic enzyme gene (HSL), oxidative phosphorylation enzyme genes (SDH and COX) and uncoupling proteins (UCP-2 and UCP-3) by Northern blot analysis. The mRNA expressions of these genes, except for UCP-2, were markedly decreased in the infarcted region compared with in the other groups. However, UCP-2 mRNA expression was only slightly increased in the remote and infarcted regions.
Figure 3).
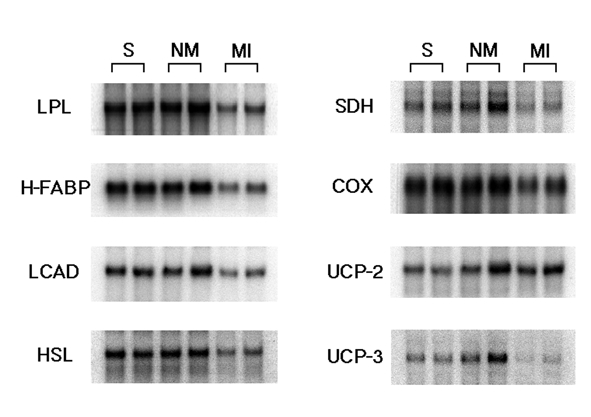
Northern blot analysis of messenger RNA expressions of lipoprotein lipase (LPL), heart-type fatty acid binding protein (H-FABP), long-chain acyl-CoA dehydrogenase (LCAD), hormone-sensitive lipase (HSL), succinate dehydrogenase (SDH), cytochrome oxidase (COX), and uncoupling protein-2 and -3 (UCP-2 and UCP-3) in rat hearts six weeks after myocardial infarct induction. MI Infarcted myocardium including borderline region; NM Remote noninfarcted left ventricular free wall; S Sham-operated rat hearts
Immunohistochemical staining for PPARα in MI hearts
To elucidate changes in the levels of PPARα, we immunohis-tochemically assayed rat heart tissue sections with antibody to the proteins (Figure 4A). Immunohistochemical staining for PPARα was less decreased in the borderline myocardium (fibroblast proliferation region) and slightly more increased in the remote region than in sham-operated hearts.
Figure 4).
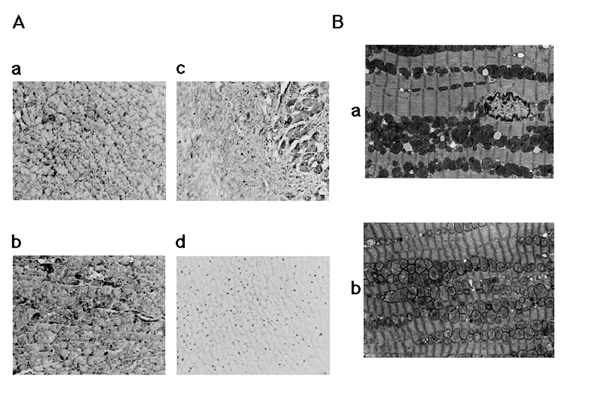
A Immunohistochemical localizations of peroxisome proliferator-activated receptor alpha (PPARα) in rat hearts six weeks after myocardial infarct induction. a PPARα immunoreactivity in sham-operated rat heart. b PPARα immunoreactivity in the remote noninfarcted left ventricular free wall. c PPARα immunoreactivity in borderline myocardium. d The reaction with normal goat serum. (Original magnification ×200) B Representative electron microscopic views of the left ventricular tissue from rats fasted for 24 h at six weeks after myocardial infarct induction. All experiments were carried out three times. a Tissue from the sham-operated rat heart. b Tissue from surviving myocardium in the borderline region. (Original magnification ×9200)
Electron microscopic analysis on lipid droplet accumulations in MI hearts
To explore the effect of ischemic status on lipid droplet accumulations in the rat heart, heart sections were studied using electron microscopy (Figure 4B). At six weeks after LAD ligation, infarcted myocardia exhibited coagulation necrosis and the ultrastructural alterations became even more marked, as characterized by a decrease in glycogen, swelling and fragmentation of mitochondria, and the presence of numerous amorphous electron-dense bodies. Comparing cross-sections of left ventricular heart tissues, surviving myocardium in the borderline region after MI exhibited no lipid droplet accumulations compared with sham-operated hearts after 24 h of fasting.
DISCUSSION
Myocardial hypertrophy after MI is accompanied by a shift from fatty acid to glucose metabolism and altered expression of regulatory proteins of energy metabolism (2,4,24). However, there are no previous studies on the role of PPARα in MI hearts. In the present study, we investigated the hypothesis that during postinfarction remodelling of the heart, PPARα plays an important role in myocardial fatty acid metabolism. Our findings provide further insight into the gene regulatory pathways involved in the energy substrate switch during cardiac remodelling after MI.
Because cardiac myocytes contract unremittingly, it is necessary to have a large energy production capacity. Cardiac myocytes prefer the oxidation of fatty acids, which provide more ATP than glucose per mole of substrate. In the present study, PPARα and FAO genes were mainly expressed in cultured myocytes. These results suggest that cardiac ATP production is greater in myocytes than in fibroblasts. The heart expresses two isoforms of glucose transporters: GLUT1 and GLUT4. GLUT4 predominates in adult myocardium, whereas GLUT1 predominates during prenatal life (25). In an in vitro study, we also observed the differences in GLUT1 and GLUT4 mRNA expression.
PPARα plays a key role in the transcriptional regulation of genes involved in fatty acid utilization pathways. In the present study, the changes in mRNA of FAO genes, a lipolytic enzyme gene and oxidative phosphorylation enzyme genes were accompanied by those in PPARα. A recent study reported that the expression of UCP-2 in the heart is 100-fold greater than that of UCP-3, and that the level of cardiac UCP-3 but not UCP-2 expression is severely reduced in PPARα knockout mice. Additionally, it showed that cardiac UCP-2 expression does not significantly change in response to high-fat feeding, fasting, diabetes or WY-14643 treatment, all of which lead to PPARα activation, in contrast to increasing cardiac UCP-3 expression under the same conditions. Taken together, these findings indicate that the expression of UCP-3, but not of UCP-2, is regulated by PPARα in the adult rodent heart (26). This report may be in accordance with our results that the expression of PPARα and UCP-3, not UCP-2, was decreased in the infarcted myocardium.
PPARα expression is downregulated in rats with left ventricular hypertrophy induced by pressure overload (16). However, in the remote noninfarcted region that showed myocyte hypertrophy and increased ANF mRNA, we did not observe downregulation of PPARα mRNA. Because we checked mRNA levels of PPARα at six weeks after LAD ligation – when the rats no longer demonstrated heart failure, having reached the compensation stage – there may have been no sufficient changes in the mRNA level of several metabolic genes, except for ANF in the remote region.
The present results imply that myocardial fatty acid metabolism genes are decreased in infarcted myocardium. It is possible that the downregulation of PPARα expression in the infarcted region is regulated by several factors. First, the upregulation of ANF expression brought on by hypertrophy of cardiac myocytes may downregulate PPARα expression. However, a recent study revealed that the expression of PPARα or RXR-alpha did not differ between Dahl salt-sensitive and Dahl salt-resistant rat hearts, and also that the PPARα/RXR-alpha complex was markedly downregulated in the hypertrophied heart, being mediated at the post-translational level (27). Second, although there were no significant changes in the concentration of plasma free fatty acids between sham-operated and MI rats, it seems that the concentration of cardiac circulated free fatty acids is decreased in the severe ischemic region. Lipid droplet accumulation in the borderline region after MI was not observed with electron microscopy. This was not due to upregulation of lipolysis by HSL, as shown in Figure 3, but was due to downregulation of the uptake of free fatty acids delivered from triglycerides by LPL. LPL plays a key role not only in the energy utilization and storage, but also in lipoprotein metabolism (28–30). Moreover, previous reports suggested that PPARα regulates the expression of the LPL gene (7) and that no obvious lipid storage could be detected in etomoxirtreated PPARα knockout mice (13). Thus, one of the reasons for decreased lipid droplet accumulation in the borderline region may be, in part, due to the downregulation of LPL expression regulated by PPARα. Third, in infarcted scar tissues, cardiac myocytes were replaced by fibroblasts, not expressed PPARα, in an in vitro study. In the present study, although the infarcted region was including the border zone, cardiac alpha-actin mRNA levels were comparable between the groups. Therefore, the downregulation of PPARα expression in the borderline myocardium can not be explained by the presence of contaminating fibroblasts.
We checked PPARα expression in cultured neonatal cardiac cells, not in adult cells, in the present study. A previous study showed that the rabbit heart switches from glucose to fatty acids as an energy substrate within the first week after birth (31). Future studies are required to investigate the expression of PPARα in adult cardiac myocytes and fibroblasts. Moreover, we only examined PPARα expression at six weeks after MI induction to avoid the influences of inflammatory changes of the early stage and congestive heart failure of the late stage. Recent reports have suggested that ligands of PPARα (clofibrate and WY-14643) reduce myocardial infarct size and may be useful in the therapy of conditions associated with ischemia-reperfusion of the heart (32). However, a mechanism of PPARα-dependent transcription in cardiac hypertrophy is still unclear at present (33,34). Our results suggest that myocardial fatty acid metabolism may be regulated by PPARα and PPARα-regulated genes in the MI heart. However, we checked only mRNA changes in several metabolic genes and found no direct relation between regional myocardial metabolism and expression of PPARα. Further studies of PPARα in relation to cardiac fatty acid metabolism should be performed in the future. In conclusion, the results of the present study suggest that the reduced expression of PPARα is closely related to that of fatty acid metabolic genes in infarcted myocardium, and PPARα may play an important role in cardiac energy metabolism during remodelling after MI.
Acknowledgments
We gratefully thank Dr Naomi Taniguchi and Ms Mari Kurata for their expert technical assistance. Part of the data was reported at the 2nd Annual Meeting of IACS (International Academy of Cardiovascular Sciences) Japan Section.
REFERENCES
- 1.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction: Experimental observations and clinical implications. Circulation. 1990;81:1161–72. doi: 10.1161/01.cir.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 2.Remondino A, Rosenblatt-Velin N, Montessuit C, et al. Altered expression of proteins of metabolic regulation during remodeling of the left ventricle after myocardial infarction. J Mol Cell Cardiol. 2000;32:2025–34. doi: 10.1006/jmcc.2000.1234. [DOI] [PubMed] [Google Scholar]
- 3.Mendez RE, Pfeffer JM, Ortola FV, et al. Atrial natriuretic peptide transcription, storage, and release in rats with myocardial infarction. Am J Physiol. 1987;253:H1449–55. doi: 10.1152/ajpheart.1987.253.6.H1449. [DOI] [PubMed] [Google Scholar]
- 4.Stanton LW, Garrard LJ, Damm D, et al. Altered patterns of gene expression in response to myocardial infarction. Circ Res. 2000;86:939–45. doi: 10.1161/01.res.86.9.939. [DOI] [PubMed] [Google Scholar]
- 5.Neely JR, Morgan HE. Relationship between carbohydrate and lipid metabolism and the energy balance of heart muscle. Annu Rev Physiol. 1974;36:413–59. doi: 10.1146/annurev.ph.36.030174.002213. [DOI] [PubMed] [Google Scholar]
- 6.Barger PM, Kelly DP. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc Med. 2000;10:238–45. doi: 10.1016/s1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- 7.Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, et al. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–48. [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Lee KA, Vork MM, De Vries JE, et al. Long-chain fatty acid-induced changes in gene expression in neonatal cardiac myocytes. J Lipid Res. 2000;41:41–7. [PubMed] [Google Scholar]
- 9.Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1998;273:23786–92. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 10.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA. 1994;91:11012–6. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyama T, Peters JM, Iritani N, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha) J Biol Chem. 1998;273:5678–84. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 12.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–7. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djouadi F, Weinheimer CJ, Saffitz JE, et al. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator-activated receptor alpha-deficient mice. J Clin Invest. 1998;102:1083–91. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–8. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley WC, Lopaschuk GD, McCormack JG. Regulation of energy substrate metabolism in the diabetic heart. Cardiovasc Res. 1997;34:25–33. doi: 10.1016/s0008-6363(97)00047-3. [DOI] [PubMed] [Google Scholar]
- 16.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. J Clin Invest. 2000;105:1723–30. doi: 10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young ME, Laws FA, Goodwin GW, Taegtmeyer H. Reactivation of peroxisome proliferator-activated receptor alpha is associated with contractile dysfunction in hypertrophied rat heart. J Biol Chem. 2001;276:44390–5. doi: 10.1074/jbc.M103826200. [DOI] [PubMed] [Google Scholar]
- 18.Finck BN, Lehman JJ, Leone TC, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–30. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamshidi Y, Montgomery HE, Hense HW, et al. Peroxisome proliferator-activated receptor alpha gene regulates left ventricular growth in response to exercise and hypertension. Circulation. 2002;105:950–5. doi: 10.1161/hc0802.104535. [DOI] [PubMed] [Google Scholar]
- 20.Harada M, Itoh H, Nakagawa O, et al. Significance of ventricular myocytes and nonmyocytes interaction during cardiocyte hypertrophy: Evidence for endothelin-1 as a paracrine hypertrophic factor from cardiac nonmyocytes. Circulation. 1997;96:3737–44. doi: 10.1161/01.cir.96.10.3737. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka N, Masamura K, Yoshida M, Kato M, Kawai Y, Miyamori I. A role of heparin-binding epidermal growth factor-like growth factor in cardiac remodeling after myocardial infarction. Biochem Biophys Res Commun. 2002;297:375–81. doi: 10.1016/s0006-291x(02)02197-6. [DOI] [PubMed] [Google Scholar]
- 22.Pfeffer MA, Pfeffer JM, Fishbein MC, et al. Myocardial infarct size and ventricular function in rats. Circ Res. 1979;44:503–12. doi: 10.1161/01.res.44.4.503. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Rosenblatt-Velin N, Montessuit C, Papageorgiou I, Terrand J, Lerch R. Postinfarction heart failure in rats is associated with upregulation of GLUT-1 and downregulation of genes of fatty acid metabolism. Cardiovasc Res. 2001;52:407–16. doi: 10.1016/s0008-6363(01)00393-5. [DOI] [PubMed] [Google Scholar]
- 25.Santalucia T, Camps M, Castello A, et al. Developmental regulation of GLUT-1 (erythroid/Hep G2) and GLUT-4 (muscle/fat) glucose transporter expression in rat heart, skeletal muscle, and brown adipose tissue. Endocrinology. 1992;130:837–46. doi: 10.1210/endo.130.2.1370797. [DOI] [PubMed] [Google Scholar]
- 26.Young ME, Patil S, Ying J, et al. Uncoupling protein 3 transcription is regulated by peroxisome proliferator-activated receptor (alpha) in the adult rodent heart. FASEB J. 2001;15:833–45. doi: 10.1096/fj.00-0351com. [DOI] [PubMed] [Google Scholar]
- 27.Kanda H, Nohara R, Hasegawa K, Kishimoto C, Sasayama S. A nuclear complex containing PPARalpha/RXRalpha is markedly downregulated in the hypertrophied rat left ventricular myocardium with normal systolic function. Heart Vessels. 2000;15:191–6. doi: 10.1007/s003800070022. [DOI] [PubMed] [Google Scholar]
- 28.Greenwood MR. The relationship of enzyme activity to feeding behavior in rats: Lipoprotein lipase as the metabolic gatekeeper. Int J Obes. 1985;9:67–70. [PubMed] [Google Scholar]
- 29.Eckel RH. Adipose tissue lipoprotein lipase. In: Borensztajn J, editor. Lipoprotein Lipase. Chicago: Evener Press; 1987. pp. 79–132. [Google Scholar]
- 30.Auwerx J, Leroy P, Schoonjans K. Lipoprotein lipase: Recent contributions from molecular biology. Crit Rev Clin Lab Sci. 1992;29:243–68. doi: 10.3109/10408369209114602. [DOI] [PubMed] [Google Scholar]
- 31.Lopaschuk GD, Spafford MA. Energy substrate utilization by isolated working hearts from newborn rabbits. Am J Physiol. 1990;258:H1274–80. doi: 10.1152/ajpheart.1990.258.5.H1274. [DOI] [PubMed] [Google Scholar]
- 32.Wayman NS, Hattori Y, McDonald MC, et al. Ligands of the peroxisome proliferator-activated receptors (PPAR-gamma and PPAR-alpha) reduce myocardial infarct size. FASEB J. 2002;16:1027–40. doi: 10.1096/fj.01-0793com. [DOI] [PubMed] [Google Scholar]
- 33.Kelly DP. Peroxisome proliferator-activated receptor alpha as a genetic determinant of cardiac hypertrophic growth: Culprit or innocent bystander? Circulation. 2002;105:1025–7. [PubMed] [Google Scholar]
- 34.Frey N, Olson EN. Modulating cardiac hypertrophy by manipulating myocardial lipid metabolism? Circulation. 2002;105:1152–4. [PubMed] [Google Scholar]


