Abstract
BACKGROUND:
Cardiomyopathy (CM) is a life-threatening disease with progressive degeneration of cardiac muscle. From a representative animal model of CM, the BIO14.6 hamster, arose the TO-2 strain manifesting a severe, dilated form of CM. Previous studies demonstrated that both strains have an identical genomic deletion disrupting the delta-sarcoglycan gene to cause CM.
OBJECTIVE:
To elucidate an additional pathogenesis for cardiac dilation in the TO-2 hamster.
METHODS:
Reverse transcription-polymerase chain reaction (RT-PCR) was used to assess the expression levels of genes for cardiac hypertrophy, such as beta-myosin heavy chain and preproendothelin-1. The involvement of apoptosis in CM was tested using terminal deoxynucleotidyl transferase-mediated dUTP-biotin end-labelling (TUNEL) and DNA ladder assays. The progression of cardiac degeneration was visualized using specific histological staining techniques. Echocardiography was performed to estimate the wall thickness and the movement of a living hamster heart.
RESULTS:
RT-PCR showed the expression of the genes involved in cardiac hypertrophy to be higher in TO-2 hamsters than in BIO14.6 hamsters, contary to what the thin myocardium may suggest. DNA ladder and TUNEL assays showed no significant difference in apoptosis in the hearts of either strain. In contrast, the infiltrate of inflammatory cells was prominent in the myocardium of TO-2 hamsters. In addition to dilation of the cardiac chamber, echocardiography also revealed disharmonized contraction in TO-2 hearts without apparent arrhythmias.
CONCLUSIONS:
Impairment of compensatory cardiac hypertrophy or involvement of apoptosis is less likely to be an additional pathogenesis in the TO-2 hamster. The present data suggest that augmented necrosis is the principal cause of severe CM in the TO-2 hamster. Further analysis of the molecular pathogenesis of TO-2 would help to disclose the final common pathways for the manifestation of CM.
Keywords: Apoptosis, Cardiac hypertrophy, Cardiomyopathy, Genetic modifier, Necrosis, TO-2 hamster
Cardiomyopathy (CM) leads to serious congestive heart failure (CHF), which is a principal cause of mortality and morbidity in children and adults (1). This disease often requires cardiac transplantation, which costs US$200 million annually in the United States (2). Thus, new therapeutic alternatives for CM are eagerly awaited.
CM is classified mainly into hypertrophic and dilated cardiomyopathies (HCM and DCM, respectively) according to the gross appearance of the heart. HCM is characterized by marked ventricular wall thickening and hypercontractile systolic function with diastolic dysfunction (3). It constitutes a principal cause of sudden death in young people and athletes, and also leads to CHF (3). In the case of DCM, the myocardial wall is thin and, together with its increased ventricular chamber size, the pumping ability is greatly reduced. It is the most common cause of CHF, affecting 40 people in every 100,000 of the population (4).
Recent advancements in molecular genetics identified a number of mutated genes responsible for both HCM and DCM (5). However, the pathogenetic relationship between the causative genes is not well understood. Deep understanding of the pathogenetic interplay between each causative gene is crucial to devise a novel therapeutic strategy for CM. For this purpose, an animal model of hereditary CM is extremely useful (6,7).
The cardiomyopathic hamster is a representative animal model for hereditary CM (8). From the most famous animal model, the BIO14.6 hamster, arose several sublines with various severities of CM, outstanding among which is the TO-2 hamster (9). The BIO14.6 hamster exhibits marked cardiac hypertrophy in its early stage. The TO-2 hamster, on the other hand, does not manifest macroscopic cardiac hypertrophy and has a shorter life expectancy than the BIO14.6 hamster. The present author and another group of investigators demonstrated that the genetic cause for the BIO14.6 hamster is a mutation of the gene encoding delta-sarcoglycan (δ-SG) (10,11), one of the dystrophin-associated proteins (12–15). The authors also demonstrated that the TO-2 hamster shares with the BIO14.6 hamster the same deletion of the genomic interval that spans about 30 kilobases and includes the two promoters of the δ-SG gene. This deletion causes the loss of its protein product, δ-SG (11,16).
These findings indicate the presence of a genetic modifier in the TO-2 hamster (11,17) that might be involved in compensatory cardiac hypertrophy (18). The present study was aimed at the preliminary pathogenetic characterization of severe CM in the TO-2 hamster.
METHODS
Animals
The normal golden hamster was purchased from SLC (Shizuoka, Japan). The BIO14.6 and TO-2 hamsters were from BIO Breeders, USA. All the animals were housed in 12 h light/12 h dark with free access to water and nutrition. Before the experiments, the animals were anesthetized with pentobarbital (50 mg/kg of body weight, intraperitoneally) and, if needed, sacrificed according to the guidelines of the institutional animal committee.
Relative expression levels of genes involved in cardiac hypertrophy
Total RNA was extracted from the hamster’s left ventricle with ISOGEN reagent (Nippon Gene, Japan). Complementary DNA was synthesized using random primer and subjected to reverse transcription-polymerase chain reaction (RT-PCR) as described previously (11). A series of amplification cycles (25, 30, 35 and 40 cycles) for RT-PCR was employed to compare the relative expression levels of the genes of interest among golden, BIO14.6 and TO-2 hamsters, aged 26 weeks. RT-PCR was carried out with the following conditions: preheated at 94°C for 2 min, followed by various cycles of 94°C for 20 s, 55°C for 30 s and 72°C for 30 s. Primer sets used in this study were as follows: c-myc 5′-ACAGCAAACCTCCGCACAG-3′/5′-TGGT-CACGCAGGGCAAAAA-3′;c-ras 5′-CACCATAGAGGT-GAGCTCTG-3′/5′-TCCTCTTGGCCTGCTGTGTC-3′; preproendothelin-1 5′-CCAAGGAGCTCCAGAAACAG-3′/5′-TTGACCCAGATGATGTCCAG-3′; brain natriuretic peptide (BNP) 5′-ATCCAAGATGCAGACTCTGC-3′/5′-GTGTCTGCAGCTGGGAGCTC-3′; GATA-4 5′-TCCTGT-GCCAACTGCCAGAC-3′/5′-CCAAGAGTCCTGCTTG GAGC-3′; beta-myosin heavy chain 5′-CGCATGGACCTG-GAGCGAGC-3′/5′-CGCCGCATCTTCCGGAACTC-3′; alpha-skeletal muscle actin 5′-TGTGCGACGAAGACGA-GACC-3′/5′-TCTCAAACATGATCTGGGTC-3′; and glycer-aldehyde 3-phosphate dehydrogenase 5′-ACCACAGTC-CATGCCATCAC-3′/5′-TCCACCACCCTGTTGCTGTA-3′.
PCR was carried out using GeneAmp PCR system 9700 (PE Applied Biosystems, USA), and Ex Taq (TaKaRa, Shiga, Japan) was chosen as a thermo-stable polymerase, otherwise noted, throughout this study.
Pathogenetic analyses
Three millimetre sections of formaldehyde-fixed/paraffin-embedded left ventricular samples were stained with Masson’s trichrome or toluidine blue dye. Terminal deoxynucleotidyl transferase-mediated dUTP-biotin end-labelling (TUNEL)-positive cells (19) at the midline section of the left ventricle were detected using an Apoptosis in situ Detection Kit (Wako, Japan). For the detection of genomic DNA laddering indicative of apoptosis (19), genomic DNA of the left ventricle was extracted using DNeasy (Qiagen, Germany), subjected to acrylamide gel electrophoresis and stained with SYBR Green I (Molecular Probes, USA).
Echocardiographic and electrocardiographic analyses
An echocardiographic study of the hamsters was carried out at various stages (six weeks, 12 weeks and 26 weeks, n=6) using an ASPEN ultrasound system with an 11 MHz microprobe (Acuson, USA). The thickness of the interventricular septum at diastole and percent of fractional shortening were measured in the movement mode. Statistical analyses (two-way analysis of variance [ANOVA], one-way ANOVA and Student’s t test) were performed using StatView software (SAS Institute Inc, USA). P<0.05 was considered significant. The electrocardiographic study was done as previously described (11).
RESULTS
Shorter life expectancy of the TO-2 hamster
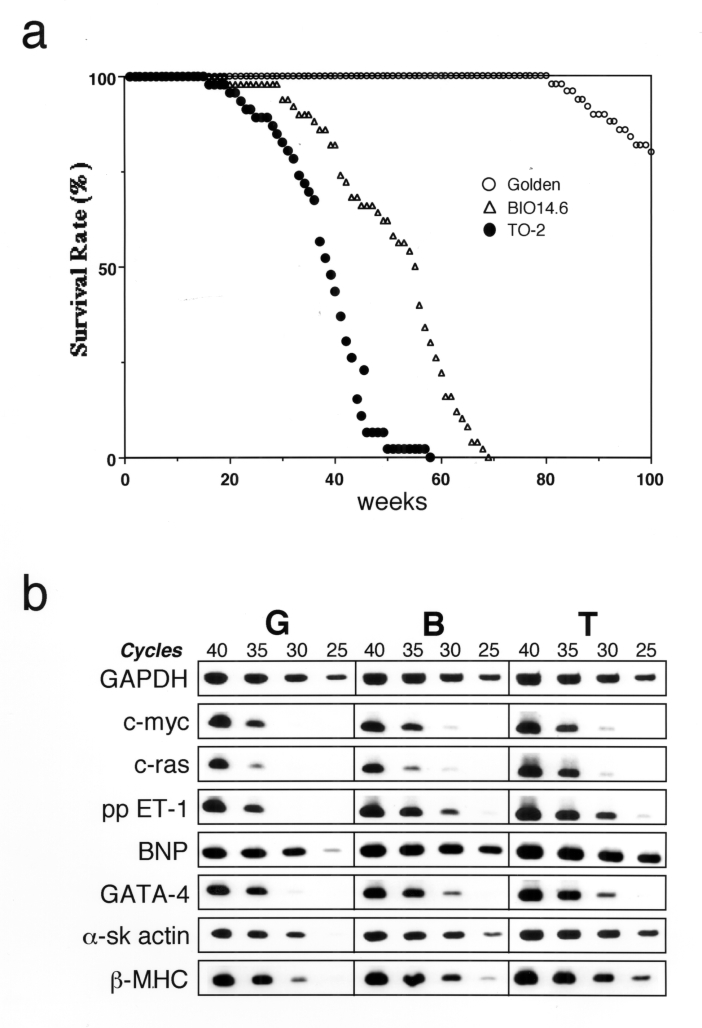
To begin, the natural life course of golden, BIO14.6 and TO-2 hamsters (n ≈ 50 for each strain) was delineated. The 50% life expectancy of TO-2 hamsters (38 weeks) was significantly shorter than that of BIO14.6 hamsters (55 weeks) (Figure 1a). Combined with the fact that TO-2 hamsters do not exhibit macroscopic cardiac hypertrophy (11), the TO-2 hamster was assumed to have another genetic defect in the compensatory machinery to survive CHF or, more specifically, cardiac hypertrophy (18).
Figure 1).
Initial characterization of TO-2 hamsters. a Survival rate. The 50% life expectancies for the golden (n=50), BIO14.6 (n=50) and TO-2 (n=46) hamsters were 114 weeks, 55 weeks and 38 weeks, respectively. b Semiquantitative reverse transcriptase polymerase chain reaction shows the relative expression levels of the genes involved in cardiac hypertrophy. The fewer the number of cycles needed for amplification, the more target complementary DNA (cDNA) was considered to be present. For example, 35 cycles were necessary for the detection of preproendothelin-1 (ppET-1) in golden hamsters (G), whereas 30 cycles and 25 cycles were enough in BIO14.6 (B) and TO-2 (T) hamsters, respectively. Thus, the relative expression level of ppET-1 was ordered from TO-2 to BIO14.6 to golden hamsters. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal standard for each template cDNA. α-sk Alpha-skeletal muscle actin; β-MHC Beta-myosin heavy chain; BNP Brain natriuretic peptide
Marked activation of the signalling pathway for cardiac hypertrophy in the TO-2 hamster
Expression levels of genes involved in cardiac hypertrophy (20) in the left ventricles of hamsters were compared using semiquantitative RT-PCR. Contrary to a previous expectation, the signalling pathway for cardiac hypertrophy was more activated in TO-2 hamsters than in BIO14.6 hamsters. Genes constituting a hypertrophied heart, such as alpha-skeletal muscle actin and beta-myosin heavy chain, in addition to genes inducing cardiac hypertrophy, such as preproendothelin-1, were upregulated in the left ventricle of TO-2 hamsters compared with those of BIO14.6 hamsters (Figure 1b).
Severe necrosis as the primary pathology in the TO-2 hamster
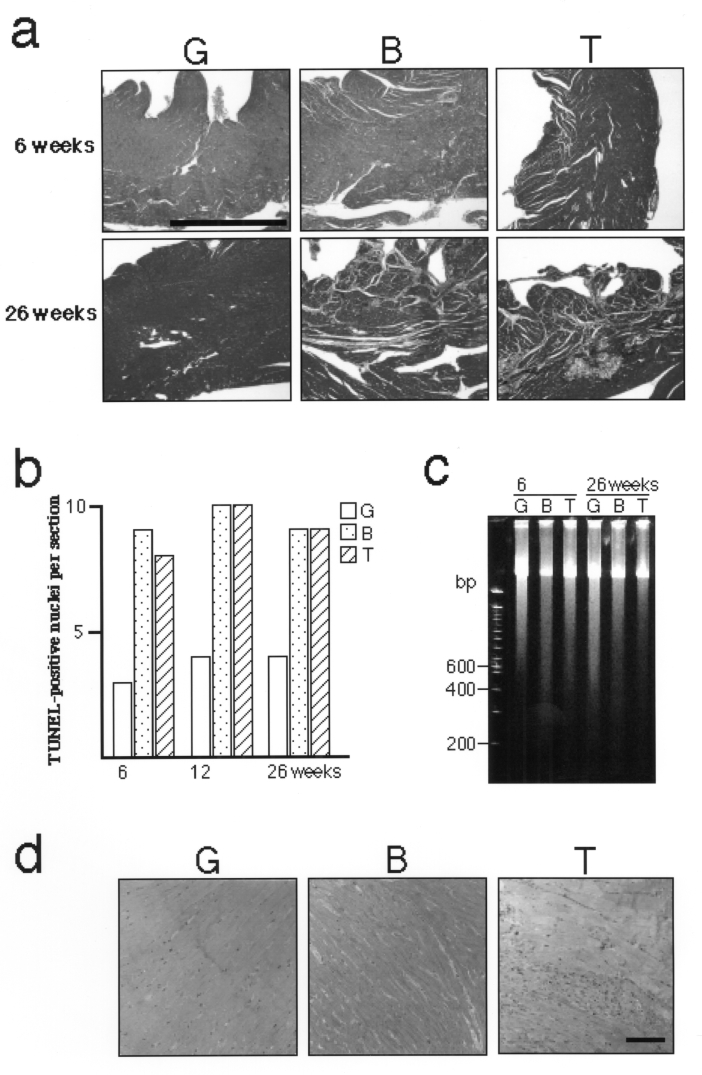
The absence of macroscopic cardiac hypertrophy, despite the upregulation of the relevant genes, suggested that the TO-2 hamster experiences further myocardial degeneration that could not be compensated by cardiac hypertrophy. Masson’s trichrome staining indeed revealed greater fibrosis in the TO-2 hamster than in the BIO14.6 hamster (Figure 2a).
Figure 2).
Pathogenetic investigation of TO-2 hamsters. a Masson’s trichrome staining. Note marked fibrosis in the TO-2 hamster (bar = 1 mm). b The number of terminal deoxynucleotidyl transferase-mediated dUTP-biotin end-labelling (TUNEL)-positive nuclei per mid-line section of the hamster ventricle. No significant difference was observed between BIO14.6 and TO-2 hamsters. c Electrophoresis of genomic DNA from the left ventricle. No significant DNA ladder indicative of apoptosis was observed in BIO14.6 or TO-2 hamsters. d Toluidine blue staining of the left ventricle. Note the marked infiltrate of inflammatory cells in the TO-2 hamster (bar = 100 μm)
First, a possible involvement of augmented apoptosis (21) in cardiac degeneration (22) in TO-2 hamsters was assessed. The number of TUNEL-positive nuclei (19) per midline cross-section of heart was not significantly different between TO-2 and BIO14.6 hamsters, although it was slightly increased compared with that of golden hamsters (Figure 2b), which was consistent with a recent report (23). Second, DNA fragmentation (19) in the left ventricle was not revealed by electrophoresis in either BIO14.6 or TO-2 hamsters (Figure 2c). Taken together, apoptosis was not considered to be the principal cause of the severe cardiac degeneration in the TO-2 hamster. Conversely, the infiltrate of inflammatory cells, a hallmark of necrosis (21), was more marked in the ventricles of TO-2 hamsters than in other strains (Figure 2d).
These findings strongly suggested that augmented necrosis is the leading pathogenetic factor for the severe CM in the TO-2 hamster.
Irregular chamber wall movement in the TO-2 hamster
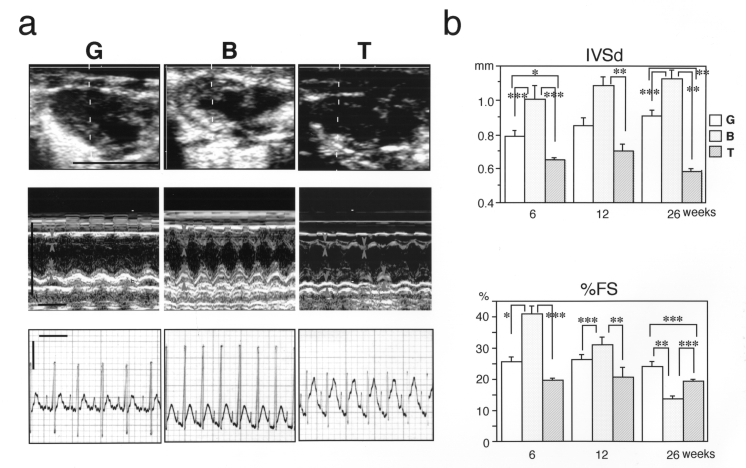
An echocardiographic study was performed to further investigate the functional abnormalities of CM in the TO-2 hamster. Dilation of the left ventricle and its thin wall were visualized, and the results were consistent with the previous histological study (11) (Figure 3b, upper panel). In addition, the percent of fractional shortening was significantly reduced in the TO-2 hamster (Figure 3a and b, lower panels). The most striking finding was discordant contraction of the ventricular chamber in the TO-2 hamster, which was disclosed in a two-dimensional movie (data not shown). The irregular movement of the ventricular wall was also detected easily in the M-mode recording (Figure 3a, middle panel). This irregularity in the movement of the ventricular wall was apparently not due to arrhythmias because no arrhythmias were detected in any of the three strains in the electrocardiogram recordings (Figure 3a, lower panel).
Figure 3).
Echocardiographic and electrocardiographic analyses of hamster hearts. a) Upper panel Two-dimensional view of diastolic hearts of 12-week-old hamsters. Representative figures are shown (n=6). The dotted lines indicate the scanning section by M-mode as shown in the middle panel (bar = 1 cm). Middle panel M-mode view of 12-week-old hamster hearts. Representative figures are shown (n=6). Margins of the interventricular septum (IVS) or left ventricular posterior wall (LVPW) are marked by arrows or arrowheads, respectively. Irregular movements of the IVS and LVPW in TO-2 hamsters are marked by asterisks or double asterisks, respectively (horizontal bar = 0.2 s; vertical bar = 1 cm). Lower panel Lead II of electrocardiogram. Note that no apparent arrhythmia was present in TO-2 hamsters (horizontal bar = 0.2 s, vertical bar = 0.25 V). b) Upper panel Measurement of the thickness of the interventricular septum at diastole (IVSd). Lower panel Measurement of percent of fractional shortening (%FS). Note that in the BIO14.6 animals, the IVSd became larger but %FS became smaller during development, indicating that the cardiac function of the BIO14.6 hamsters was gradually deteriorating or failing. In the TO-2 hamsters, both the IVSd and the %FS were consistently smaller than those of the BIO14.6 hamsters. The mean ± SEM was presented for all data. *P<0.05; **P<0.01; ***P<0.001
DISCUSSION
Observation of the natural life course of golden, BIO14.6 and TO-2 hamsters (n ≈ 50 for each strain) elucidated that the 50% life expectancy of the TO-2 hamster (38 weeks) was less than three-fourths that of the BIO14.6 hamster (55 weeks) (Figure 1a). Thus, cardiac hypertrophy was closely related to life expectancy in the cardiomyopathic hamsters.
Greater activation of the signalling pathway for cardiac hypertrophy in the TO-2 hamster than in the BIO14.6 hamster seemed paradoxical at first because the TO-2 hamster has dilated ventricles with thin walls (11). I suspected that some degeneration mechanisms might surpass the compensatory hypertrophy in the TO-2 hamster and set out to determine which type of degeneration, apoptosis or necrosis, prevails in the heart of the TO-2 hamster. A series of experiments strongly suggested that severe necrosis mainly aggravates the clinicopathological course of CM in the TO-2 hamster. Discordant contraction of the ventricular chamber, as revealed by echocardiography, was one of the most striking features of CM in the TO-2 hamster.
These observations suggested that a putative genetic modifier influences the structural and functional integrities of cardiac muscle in the TO-2 hamster. Identification of a modifier gene for the CM in the TO-2 hamster will elucidate a novel pathogenetic paradigm for CM and cardiac failure (24).
Acknowledgments
The author thanks Ms A Kamata and Mr H Kirai for helping with the echocardiographic and electrocardiographic recordings. The author also thanks Drs T Masaki and K Ono for continuous encouragement and critical reading of the manuscript, respectively. This work was supported in part by grants from the Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research of Japan.
REFERENCES
- 1.Kelly DP, Strauss AW. Inherited cardiomyopathies. N Engl J Med. 1994;330:913–9. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell JB, Bristow MR. Economic impact of heart failure in the United States: Time for a different approach. J Heart Lung Transplant. 1994;13:S107–12. [PubMed] [Google Scholar]
- 3.Maron BJ. Hypertrophic cardiomyopathy. Lancet. 1997;350:127–33. doi: 10.1016/S0140-6736(97)01282-8. [DOI] [PubMed] [Google Scholar]
- 4.Codd MB, Sugrue DD, Gersh BJ, Melton LJ., III Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation. 1989;80:564–72. doi: 10.1161/01.cir.80.3.564. [DOI] [PubMed] [Google Scholar]
- 5.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: From mutation identification to mechanistic paradigms. Cell. 2001;104:557–67. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 6.Maass A, Leinwand LA. Animal models of hypertrophic cardiomyopathy. Curr Opin Cardiol. 2000;15:189–96. doi: 10.1097/00001573-200005000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda Y, Ross J., Jr Models of dilated cardiomyopathy in the mouse and the hamster. Curr Opin Cardiol. 2000;15:197–201. doi: 10.1097/00001573-200005000-00013. [DOI] [PubMed] [Google Scholar]
- 8.Homburger F, Baker JR, Nixon CW, Whitney R. Primary, generalized polymyopathy and cardiac necrosis in an inbred line of Syrian hamsters. Med Exp. 1962;6:339–45. doi: 10.1001/archinte.1962.03620230106015. [DOI] [PubMed] [Google Scholar]
- 9.Sole MJ. Hamster cardiomyopathy: Understanding the pathogenesis of heart failure. Hamster Information Service. 1986;8:3–6. [Google Scholar]
- 10.Nigro V, Okazaki Y, Belsito A, et al. Identification of the Syrian hamster cardiomyopathy gene. Hum Mol Genet. 1997;6:601–7. doi: 10.1093/hmg/6.4.601. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto A, Ono K, Abe M, et al. Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, delta-sarcoglycan, in hamster: An animal model of disrupted dystrophin-associated glycoprotein complex. Proc Natl Acad Sci USA. 1997;94:13873–8. doi: 10.1073/pnas.94.25.13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox GF, Kunkel LM. Dystrophies and heart disease. Curr Opin Cardiol. 1997;12:329–43. [PubMed] [Google Scholar]
- 13.Straub V, Campbell KP. Muscular dystrophies and the dystrophinglycoprotein complex. Curr Opin Neurol. 1997;10:168–75. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 14.Ozawa E, Noguchi S, Mizuno Y, Hagiwara Y, Yoshida M. From dystrophinopathy to sarcoglycanopathy: Evolution of a concept of muscular dystrophy. Muscle Nerve. 1998;21:421–38. doi: 10.1002/(sici)1097-4598(199804)21:4<421::aid-mus1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 15.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto A, Abe M, Masaki T. Delineation of genomic deletion in cardiomyopathic hamster. FEBS Lett. 1999;447:124–8. doi: 10.1016/s0014-5793(99)00267-7. [DOI] [PubMed] [Google Scholar]
- 17.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–8. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 18.Zak R. Development and proliferative capacity of cardiac muscle cells. Circ Res. 1974;35(Suppl II):17–26. [PubMed] [Google Scholar]
- 19.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–83. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 20.Molkentin JD, Dorn II, GW II. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 21.Vermes I, Haanen C. Apoptosis and programmed cell death in health and disease. Adv Clin Chem. 1994;31:177–246. doi: 10.1016/s0065-2423(08)60336-4. [DOI] [PubMed] [Google Scholar]
- 22.Gill C, Mestril R, Samali A. Losing heart: The role of apoptosis in heart disease – A novel therapeutic target? FASEB J. 2002;16:135–46. doi: 10.1096/fj.01-0629com. [DOI] [PubMed] [Google Scholar]
- 23.Ryoke T, Gu Y, Ikeda Y, et al. Apoptosis and oncosis in the early progression of left ventricular dysfunction in the cardiomyopathic hamster. Basic Res Cardiol. 2002;97:65–75. doi: 10.1007/s395-002-8389-4. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto A.Electrical and ionic abnormalities in the heart of cardiomyopathic hamsters: In quest of a new paradigm for cardiac failure and lethal arrhythmia Mol Cell Biochem(In press) [DOI] [PubMed] [Google Scholar]