Abstract
The area and volume of brain structural features, as assessed by high-resolution three-dimensional magnetic resonance imaging (MRI), are among the most heritable measures relating to the human CNS. We have conducted MRI scanning of all available monkeys >2 years of age (n = 357) from the extended multigenerational pedigree of the Vervet Research Colony (VRC). Using a combination of automated and manual segmentation we have quantified several correlated but distinct brain structural phenotypes. The estimated heritabilities (h2) for these measures in the VRC are higher than those reported previously for such features in humans or in other nonhuman primates: total brain volume (h2 = 0.99, SE = 0.06), cerebral volume (h2 = 0.98, SE = 0.06), cerebellar volume (h2 = 0.86, SE = 0.09), hippocampal volume (h2 = 0.95, SE = 0.07) and corpus callosum cross-sectional areas (h2 = 0.87, SE = 0.07). These findings indicate that, in the controlled environment and with the inbreeding structure of the VRC, additive genetic factors account for almost all of the observed variance in brain structure, and suggest the potential of the VRC for genetic mapping of quantitative trait loci underlying such variance.
Introduction
Brain structural variability is among the most promising phenotypes for genetic investigation of the CNS. Brain structural features correlate with functional measures such as cognitive performance (Hulshoff Pol et al., 2006; Narr et al., 2007) and variability in such features has been associated with behavioral abnormalities (Eckert, 2004; Honea et al., 2005; Kelly et al., 2007; Amaral et al., 2008). Therefore identifying genetic variants underlying interindividual differences in such features may provide insights into the biological basis of a wide range of phenomena including cognition and disease liability.
Many human brain structural phenotypes have heritabilities (h2) in the range of 0.5–0.9 (Bartley et al., 1997; Pennington et al., 2000; Pfefferbaum et al., 2000; Posthuma et al., 2000; Atwood et al., 2004; Peper et al., 2007), where h2 is defined as the proportion of total phenotypic variance due to additive genetic sources. The relatively high heritability of all of these features has generated excitement about the possibility of identifying quantitative trait loci (QTL) that contribute to their variance, but to date no well powered genetic mapping studies of brain structure have been reported; in part this may reflect practical issues that have so far made it not feasible to conduct magnetic resonance imaging (MRI) investigations of human pedigrees that would have sufficient power for either linkage analysis or association studies.
The development of genetic and genomic mapping tools in several nonhuman primate (NHP) species makes it feasible to mount QTL studies of brain structural features in extended NHP pedigrees. Such samples provide the opportunity to collect many phenotypic measurements, including behavioral, anatomical and biochemical measures from the same individuals at multiple time points. Relatively limited information is available regarding the heritability of brain structures in NHPs. Cheverud et al. (1990) estimated the heritability of intracranial volume at 0.60–0.75 in a free-ranging colony of rhesus macaques, Mahaney et al. (1993) estimated the heritability of postmortem brain weight at 0.41 in 601 members of a baboon pedigree, and Rogers et al. (2007) estimated the heritability of total brain volume at 0.52–0.82 using MRI in 109 members of the same baboon pedigree. The heritability of hippocampal volume was estimated at 0.54 by Lyons et al. (2001) using MRI in a group of 39 South American squirrel monkeys.
In this study, we use heritability estimates to characterize the potential for mapping brain structure QTLs in an extended vervet monkey pedigree (Chlorocebus aethiops sabaeus) of the Vervet Research Colony (VRC). We have expanded the phenotypic characterization of the VRC to include high-resolution MRI brain images from all available members of the colony over two years of age (n = 357). After MRI acquisition, we assembled enhanced signal-to-noise structural images, generated an affine-registered population atlas, and used a combination of automated and manual segmentation to produce high resolution, anatomically based measurements of total brain volume, cerebral volume, cerebellar volume, hippocampal volume and cross-sectional area of the corpus callosum for each animal in real world (nonaffine distorted) units. Heritability analysis of these phenotypes shows that almost all of the population variance is attributable to additive genetic sources.
Materials and Methods
Vervet research colony.
The vervet subjects scanned in this study are part of a pedigree [recently described by Freimer et al. (2007)] which has included >1000 individuals since its founding. Briefly, the Vervet Research Colony was established during the 1970s and 1980s from 58 founder animals captured from a wild population in St. Kitts. The animals were originally brought to St. Kitts from Africa on trading ships in the 1600s (McGuire et al., 1974). Breeding in the colony has been managed to promote diversity, to preserve each of the original matrilines, and to provide a species-typical social environment for developing offspring. Wild vervets live in matrilineal social groups that typically contain 15–45 members, with females remaining in the natal group for life and males emigrating at adolescence. At the VRC, animals are housed in large outdoor enclosures with their mothers and other matrilineal kin. Female offspring remain in the breeding groups, males are removed and housed in all-male groups at 4 years of age, and unrelated adult males are introduced into the breeding groups at 3–4 year intervals. Commercial laboratory chow is available ad libitum, supplemented by fresh fruits and produce. Under these circumstances, rates of aggression among breeding females are low, and there is no relationship between dominance rank and female body weight, or between rank and infant mortality (Fairbanks and McGuire, 1984) (L. A. Fairbanks, unpublished data).
Subjects for this study included all vervets >2 years of age from 16 matrilineal breeding groups and 3 all-male groups (N = 357). The matrilineal groups contained between 16 and 41 animals, including adult females, their immature offspring and 1–3 unrelated adult males (mean female/male ratio = 2.6). The all-male groups each contained 9 young adult males that had been removed from their natal groups at 4 years of age. All subjects were born and raised at the VRC. Information on the mother's dominance rank during infancy was available for all subjects born in 1999 and later (N = 207). Dominance rank, coded as high, middle or low, was calculated from the direction of aggressive encounters observed during ∼72 h per year of behavioral observations in each of the 16 matrilineal breeding groups.
MRI acquisition.
Animals were scanned during a three-month period starting in February 2007 using a mobile 1.5 Tesla Siemens Symphony unit. Studies were conducted as approved by the Animal Research Committee in the Office for Protection of Research Subjects at University of California, Los Angeles and the Institutional Animal Care and Use Committee at the Sepulveda Veterans Administration. Animals were fasted on the night before scanning. After initial intramuscular administration of 15 mg/kg ketamine, an intravenous line was established and used to initiate anesthesia using midazolam (0.25 mg/kg) and ketamine (5 mg/kg). Each animal was also given 0.027 mg/kg atropine and intubated to protect the airway and reduce risk of aspiration. After intubation, respiratory rate was monitored frequently and used to maintain adequate anesthesia using midazolam and ketamine. All animals tolerated the procedure well.
To generate enhanced images with good signal-to-noise properties, nine separate scans were acquired for each animal using an 8-channel high-resolution knee array coil (Invivo) as a receiver. The images were acquired as axial T1-weighted volumes with a three-dimensional magnetization prepared rapid acquisition gradient echo (MPRAGE). The repetition time was 1900 ms, the echo time 4.38 ms, and the inversion time was 1100 ms. A flip angle of 15° was used. Voxel resolution was 0.5 mm in all three planes. The total acquisition time for the nine T1 volumes was ∼1 h for each animal. All scans were examined at the time of acquisition. Any image with artifact was excluded in the subsequent image processing.
Generation of affine atlas.
To increase the reliability of the anatomic measurements, all images were resampled into an affine common space before anatomic segmentation. Even simple correction of head tilt has been shown to improve anatomic reliability (Bartzokis et al., 1998), and affine deformations provide the greatest degree of anatomic standardization that can be achieved by global (i.e., locally invariant) transformations (Woods et al., 1998; Woods, 2009). All volumes and cross-sectional areas measured in this affine space were transformed back to each animal's native space dimensions before quantitative genetic analysis. Relative to simply making measurements directly in native space, there is no loss of accuracy associated with making measurements in an image that has been resampled into an atlas space and transforming those measurements back to native space (Mazziotta et al., 1995). Before the generation of an affine atlas, the individual MPRAGE images for each animal were aligned to each other using the Automated Image Registration (AIR 5.2.5) software package (Woods et al., 1998) in pairwise rigid body registrations and averaged together to yield one high signal-to-noise image. This native-space image was then segmented to remove all nonbrain tissue using an iterative combination of manual and automated segmentation. All manual segmentation was performed using MultiTracer software (Woods, 2003b) by individuals who were blinded to age, sex and familial relationships. MultiTracer allows the simultaneous viewing of all three orthogonal planes enabling the tracer to use anatomical landmarks in two planes while editing in the third plane. In the initial phase of segmentation, examples of “brain” and “nonbrain” tissue were manually labeled on every three to eight axial slices depending on the complexity of the surrounding tissue. A three-dimensional implementation of the random-walker algorithm (Grady, 2006) was then used to generate a complete initial segmentation into brain and nonbrain. Each slice of the resultant brain images was then examined and manually corrected side by side with the corresponding original image in the axial plane using MultiTracer. One animal was chosen at random, and the corrected segmented images from each animal were registered to the corrected segmented images from this animal using an affine spatial transformation model and averaged to create an initial atlas using AIR. This initial atlas was then used as a new registration target in an iterative registration and averaging procedure that was repeated five times. With each iteration, the transformation matrices were adjusted so that each resulting atlas had the mean size, shape and orientation of the input images, using methods described by Woods (2003a). The atlas from the fifth iteration was adjusted with a rigid-body transformation to assure right-left symmetry and to orient the line connecting the anterior and posterior commissures along the y-axis of the images to create the final atlas. Transformation matrices were tracked across all registrations and were combined to compute a transformation matrix that would map each original image obtained from the MRI scanner into the final common atlas space. The original images obtained from the scanner were then resampled directly into this final affine atlas space and averaged for each animal. The brain segmentation process was then repeated on the affine-space images to avoid propagating any interpolation errors. The segmented images were bias field corrected (Shattuck et al., 2001) to generate the images used for phenotyping. All subsequent segmentation protocols used these affine-atlas registered images. The determinants of the three-dimensional rotation and scaling matrices used to transform each animal's brain from native space to affine space were computed to provide a scaling factor for converting the animal's affine space volumes back into native space volumes. Two-dimensional matrices for mapping between the affine space mid-sagittal plane and each animal's native space were derived from the corresponding three dimensional rotation and scaling matrices and the determinants of these two-dimensional matrices were computed to provide a scaling factor for converting affine space areas measured parallel to the mid-sagittal plane back into native space areas.
Structure segmentation.
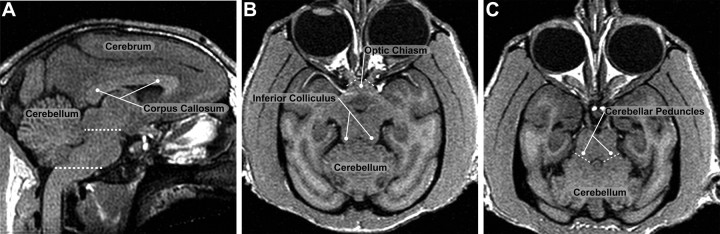
Total brain volume was defined to include the cerebrum, cerebellum and brainstem (Fig. 1). All CSF external to the cortex, cerebellum and brainstem was removed. Optic nerve tissue distal to the optic chiasm was removed (Fig. 1B). The caudal boundary of the brainstem was defined by the most inferior edge of the cerebellum (Fig. 1A). The cerebellum was segmented manually in the axial plane. Superiorly, the cerebellum was separated from the surrounding CSF and cerebrum. The cerebellar peduncles were transected orthogonally to the long axis of the peduncle midway between the cerebellum and brainstem (Fig. 1C). After removal of the cerebellum, the brainstem was removed from the cerebrum below the inferior boundary of the inferior colliculus as identified on the sagittal plane (Fig. 1A).
Figure 1.
Segmentation of total brain volume, cerebrum and cerebellum. A, Sagittal slice near the midline. The dashed lines indicate the rostral and caudal boundaries of the brainstem. The dense white matter of the corpus callosum is clearly distinguishable from the dorsal gray matter and the ventral CSF. B, An axial slice at the rostral end of the brainstem. Tissue distal to the optic chiasm (dashed lines) was removed during the brain/nonbrain segmentation process. C, An axial slice 2.5 mm caudal to the slice in B. The cerebellar peduncles were transected (dashed lines) orthogonal to the long axis to separate the brainstem and cerebellum.
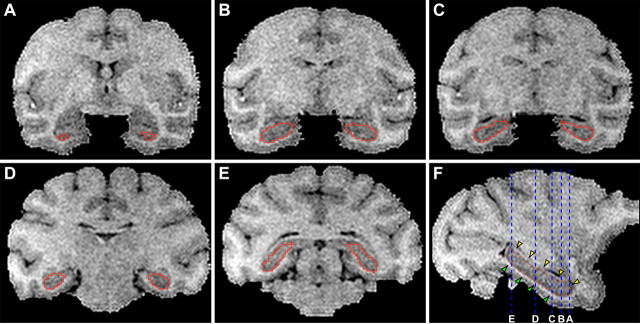
Hippocampi were segmented using a combination of manual and automated delineation. Specifically, 40 images were manually segmented in duplicate by 10 extensively trained research assistants using MultiTracer software. Before segmenting the final data set, the reliability of each tracer was assessed using a training-set of 20 expertly segmented gold standard hippocampi. The intraclass correlation coefficient on the set of 20 hippocampi for the 10 tracers exceeded 0.85. Each hippocampus was segmented in the coronal plane while concomitantly using the sagittal slices to verify anatomic details. The dorsal boundary of the hippocampus was defined by the alveus, which is clearly visible on sagittal slices (Fig. 2). Correspondingly, the anterior boundary of the hippocampus was defined by the ventral deflection of the alveus, also visible on sagittal sections (Fig. 2). The ventral boundary of the hippocampus was demarcated by the white matter of the parahippocampal gryus (Fig. 2). Reliability of the segmentations for each hippocampus was assessed using Dice's coefficient [(2*intersection)/(area1+area2)] to make pairwise comparisons for each of the ∼40 image slices containing hippocampal anatomy. Each hippocampus was manually segmented by at least four different research assistants. A research assistant's hippocampal segmentation was discarded if the average Dice coefficient relative to corresponding segmentations from other research assistants was <0.80. This procedure resulted in at least two reliable segmentations generated by different research assistants for each hippocampus. If more than two segmentations per hippocampus met the reliability criteria, two of the segmentations were randomly selected for subsequent steps of the analysis. The manual segmentations were used to train a hybrid discriminative/generative-learning algorithm (Tu et al., 2008), which, in turn, was used to segment the entire set of images to produce the final phenotypes. Visual inspection by a trained anatomical expert was done to verify the accuracy of segmentations generated by the trained algorithm (Fig. 2). Segmented volumes produced by the manual and learning algorithm methods were strongly correlated (r2 >0.75), and comparison by direct visual inspection showed greater anatomic consistency across animals in the automated segmentations. This was expected since the algorithm uniformly applies a set of rules determined during the learning phase, and the variance in hippocampal volumes was substantially smaller (0.08 vs 0.40) with the algorithmic segmentations. The automated segmentations were therefore used for the heritability analysis since the variance component method implemented by SOLAR (see below) would confound interindividual residual error due to manual segmentation with environmental sources of variance.
Figure 2.
Segmentation of the hippocampus. A–E, Coronal brain slices with hippocampal contours overlaid in red. F, A sagittal slice through the body of the hippocampus. Red dots indicate where the coronal traces transect the sagittal plane. The dorsal boundary of the hippocampus is defined by the alveus (yellow arrowheads) and ventral boundary is defined by the white matter of the parahippocampal gyrus (green arrowheads). Dashed blue lines indicate the location of coronal slices in A–E. A hybrid discriminative/generative-learning algorithm trained with manually delineated tracings generated the final contours.
Corpus callosum cross-sectional areas were measured by tracing the structure on the sagittal slices 1 mm to each side of the mid-sagittal plane (Fig. 1A). These slices were chosen to avoid the difficulty of distinguishing the fornix and septum pellucidum from the corpus callosum on mid-sagittal slices. The selected sagittal slices were separated from each image and randomly reassembled into composite image sets that were then traced by 10 trained research assistants. The inter-rater reliability of traces was assessed using Dice coefficients. Additional quality control slices were included on every tenth slice of the composite images to assess for tracer reliability. The intraclass correlation coefficient for the 10 tracers exceeded 0.85. The 10 independent measurements were averaged for each slice and then the cross-sectional areas from the two slices per brain were averaged to generate the final phenotype.
The R environment (The R Development Team, 2008) including the lme4 package [D. Bates (2007) lme4: Linear mixed-effects models using S4 classes. R package version 0.99875-9] was used for the basic statistical analysis of the phenotypes.
Heritability estimation.
Additive genetic heritability (narrow-sense heritability) for each phenotype was estimated using the software package SOLAR (Almasy and Blangero, 1998). SOLAR implements a variance components approach using maximum likelihood methods to estimate the additive polygenic component of variation considering the entire pedigree jointly. The pedigree was constructed using a panel of 14 highly polymorphic microsatellite markers to determine paternity and verify maternity (Newman et al., 2002). In addition, all individuals have been genotyped with >200 genome wide microsatellites, allowing clarification for the few ambiguous cases. The raw data for each phenotype were transformed to normality using the natural logarithmic function before analysis. Factors identified as having significant effect (p-value <0.05) on each trait using linear regression analysis were included as covariates in the heritability estimation. The heritability of weight was estimated including age, sex and an age-sex interaction as covariates.
Results
We scanned 357 individuals from the VRC (for sex, age, and weight characteristics of the study sample, see Table 1). We used a complete set of nine MPRAGE volumes to analyze the scans of 340 of these vervets. Due to image artifacts, one of the nine MPRAGE volumes was discarded in 17 animals, for which subsequent image processing steps used eight MPRAGE volumes.
Table 1.
Sample statistics
| n | Number abnormal | Age range, years (mean) | Weight range, kg (mean) | |
|---|---|---|---|---|
| Female | 256 | 4 | 2.3–24 (9.6) | 2.37–7.85 (4.78) |
| Male | 101 | 6 | 2.7–13 (5.8) | 2.59–10.52 (5.53) |
| Total | 357 | 10 | 2.3–24 (8.5) | 2.37–10.52 (4.99) |
We excluded from heritability analyses 10 animals (2.8%) in which the scans identified gross structural abnormalities, including six of 101 (5.9%) males and four of 256 females (1.6%). Three of these animals displayed unilateral enlargement of the lateral ventricles; an additional three animals demonstrated cavities extending caudally from one of the posterior horns of the lateral ventricles, and the remaining four animals showed large unilateral cavities in the parenchyma of the parietal-occipital lobes.
Phenotypes
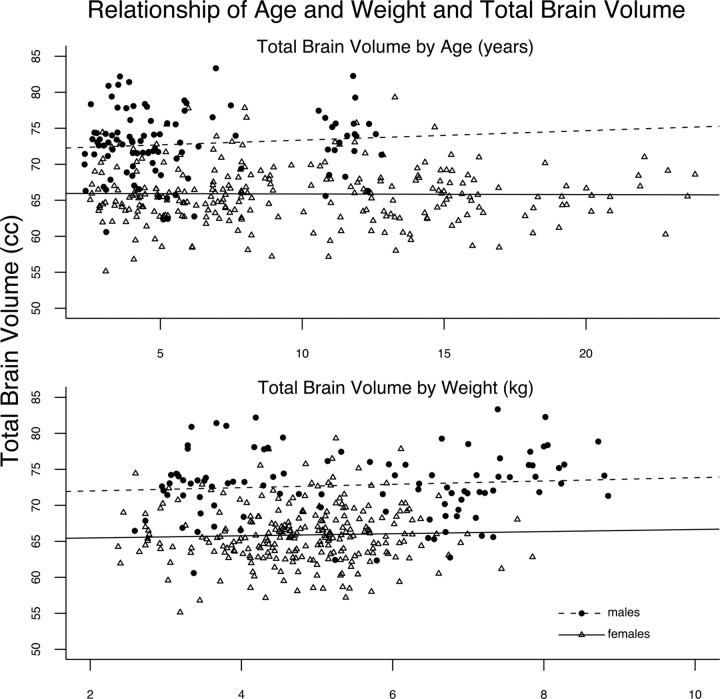
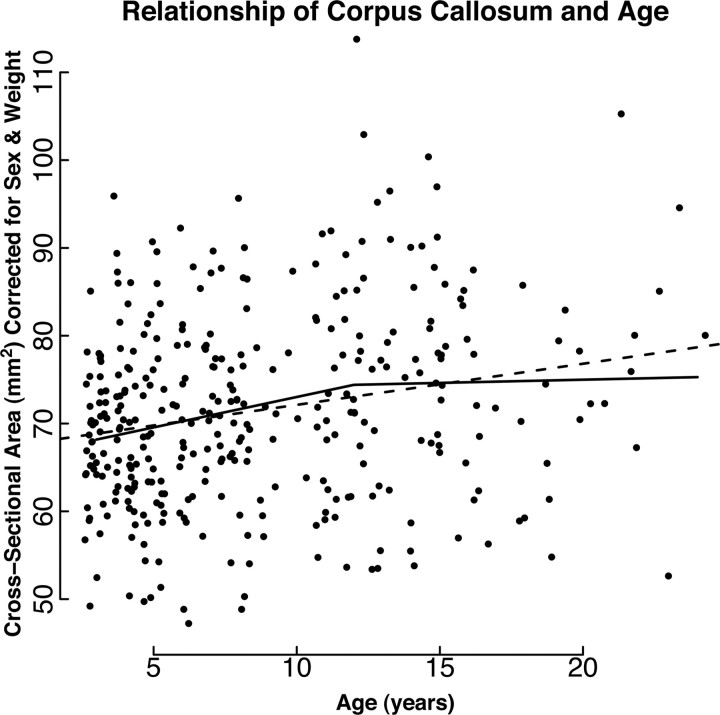
Summary statistics for each brain phenotype are presented in Table 2. Sex had a large effect on all phenotypes. For total brain volume and cerebral volume, sex was the only covariate that showed a significant effect (Fig. 3). Weight was a significant covariate for cerebellar volume, hippocampal volume and corpus callosal area, all of which increased in magnitude with increasing weight. Increasing age was positively associated with corpus callosal cross-sectional area (Fig. 4, dashed line). Modeling of the data using first-order splines suggests that the corpus callosal area increases until ∼12–15 years of age and then shows little change in later years (Fig. 4, solid line). Partial correlation, controlling age and sex, showed no significant relationship of mother's dominance rank during the year of birth with any of the five brain phenotypes (Table 3).
Table 2.
Summary statistics for phenotypes
| Mean (SD) |
Range | Significant covariates |
||||
|---|---|---|---|---|---|---|
| All | Male | Female | Factor | R2 | ||
| Total brain volume (cc) | 67.8 (5.2) | 72.8 (4.7) | 65.9 (3.9) | 55.1–83.3 | Sex | 0.36 |
| Cerebrum (cc) | 60.1 (4.7) | 64.4 (4.3) | 58.4 (3.7) | 48.8–73.7 | Sex | 0.32 |
| Cerebellum (cc) | 5.34 (0.50) | 5.81 (0.43) | 5.14 (0.36) | 4.27–6.85 | Sex, weight | 0.40 |
| Combined hippocampi (cc) | 0.72 (0.07) | 0.76 (0.06) | 0.71 (0.07) | 0.49–0.93 | Sex, weight | 0.13 |
| Corpus callosum (mm2) | 88.9 (12.0) | 90.3 (12.6) | 88.5 (11.7) | 60.2–136.5 | Sex, age, weight | 0.21 |
Figure 3.
Plots showing the relationship of age and weight and total brain volume of the sample. Total brain volume is plotted against age in the top. There is a significant difference between the total brain volumes of males (circles, dashed line) compared with females (triangles, solid line), but the flat lines indicate the lack of age effect on total brain volume. The lower plot also demonstrates an effect of sex, but not weight on total brain volume.
Figure 4.
Plot showing the relationship between the area of the corpus callosum and age. The area of the corpus callosum has been corrected for the effects of sex and weight. The dashed line shows the slope of the regression coefficient estimated using standard linear regression analysis and the solid-line shows the results of using first-order splines to model the data in younger animals (<14 years) and older animals (>14 years).
Table 3.
Partial correlation of each structure with mother's dominance rank during year of birth
| Partial correlation (p value) |
|||||
|---|---|---|---|---|---|
| Total brain volume | Cerebrum | Cerebellum | Hippocampus | Corpus callosum | |
| Mother's rank | 0.004 (0.959) | 0.004 (0.956) | 0.006 (0.931) | −0.039 (0.583) | 0.073 (0.296) |
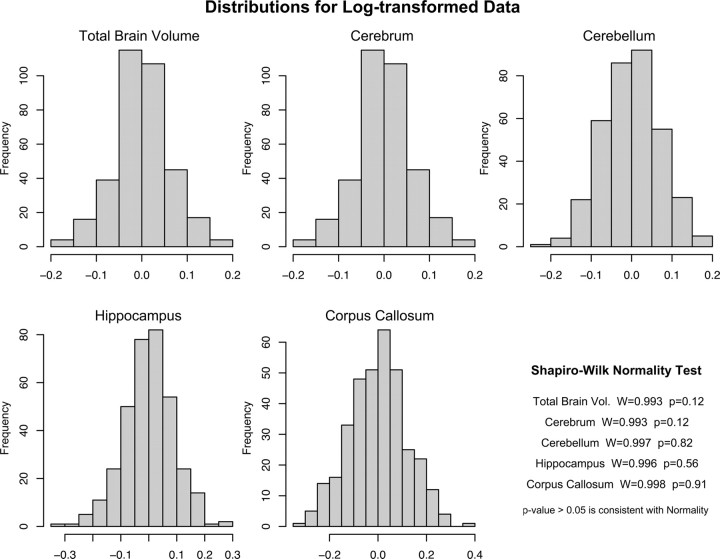
The distributions of the log-transformed trait values after accounting for significant covariates are shown in Figure 5. The Shapiro–Wilk test for normality showed that none of the log-transformed trait distributions were significantly different from the normal distribution (Fig. 5, bottom right), important for the heritability estimate, and subsequent heritability estimations were made on these log-transformed data.
Figure 5.
Distributions of log-transformed variables after correcting for effect of significant covariates.
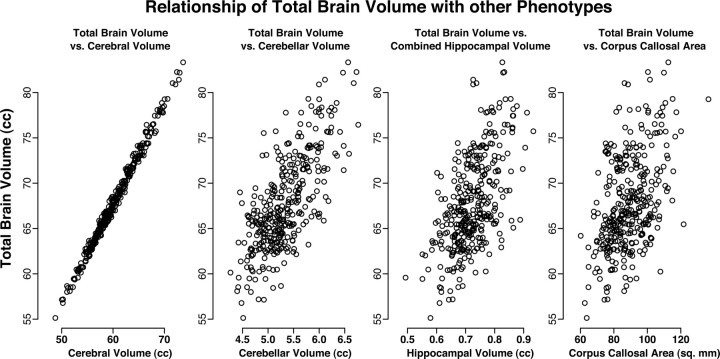
Total brain volume correlated strongly with all other phenotypes (Fig. 6, Table 4). Likewise, cerebral volume had high correlations with the other phenotypes, especially the other two volume measurements (hippocampus and cerebellum). Corpus callosal area had the lowest correlations with the other phenotypes, especially with cerebellar volume.
Figure 6.
Scatter plots showing relationship of total brain volume to the other phenotypes.
Table 4.
Interphenotypic correlations
| Total brain volume | Cerebrum | Cerebellum | Hippocampus | Corpus callosum | |
|---|---|---|---|---|---|
| Total brain volume | 1.0 | 0.996 | 0.72 | 0.56 | 0.47 |
| Cerebrum | 1.0 | 0.66 | 0.58 | 0.47 | |
| Cerebellum | 1.0 | 0.43 | 0.23 | ||
| Hippocampus | 1.0 | 0.40 | |||
| Corpus callosum | 1.0 |
Heritability
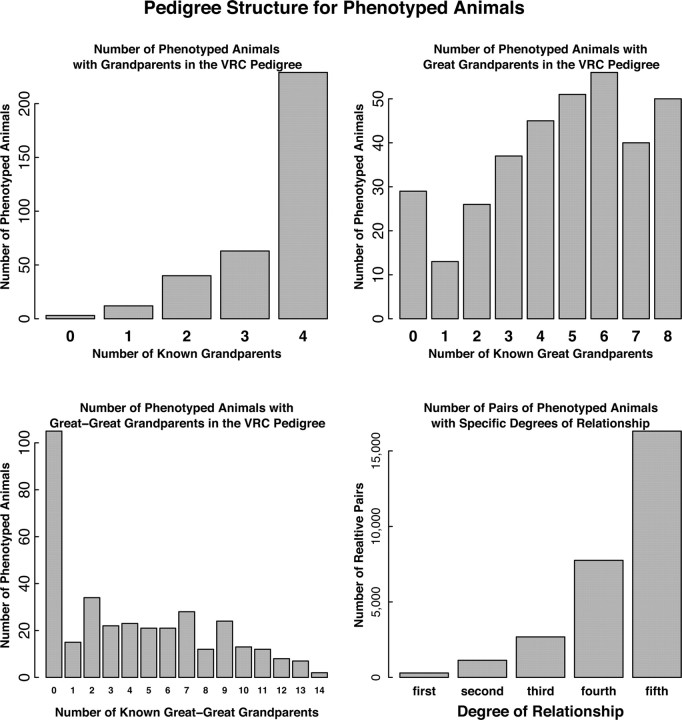
For estimations of heritability, we used considerable prior information regarding the positions in the VRC pedigree of the 347 phenotyped animals. The mothers were known for all but one animal, and fathers were known for 328 of the animals. At least one grandparent was known for 344 animals and all four grandparents were known for 229 animals (Fig. 7, top left). All four sets of great grandparents were known for 50 of the animals, and at least one great grandparent was known for 318 of the animals (Fig. 7, top right). There were 242 animals with at least one known great-great grandparent (Fig. 7, bottom left). Among the >60,000 possible pairings of individuals within the phenotyped subset of the pedigree, there were 294 first-degree relative pairs, 1137 second-degree relative pairs, 2682 third-degree relative pairs, 7754 fourth-degree relative pairs, and 16,313 fifth-degree relative pairs (Fig. 7, bottom right).
Figure 7.
Pedigree structure in the VRC. Distribution of number of phenotyped animals (total = 347) that had grandparents (top left), great grandparents (top right), and great-great grandparents (bottom left) in the full pedigree. Bottom right, Number of pairs of first-, second-, third-, fourth-, and fifth-degree relatives among the phenotyped animals.
All phenotypes demonstrated heritabilities >0.86 (Table 5). Heritability estimates for total brain volume and cerebral volume were >0.98, indicating that almost all of the phenotypic variance was attributable to additive genetic sources. Because the maximum value of h2 is 1.0 and given the potential difficulty estimating parameters near a boundary, the heritability for cerebral volume was also estimated as a ratio, using total brain volume to scale the value. As expected, this reduced the trait heritability estimate, but the estimate remained high (h2 = 0.83, SE = 0.08). Likewise, heritability estimates for the other phenotypes after scaling by total brain volume remained high and were very similar to estimates made using total brain volume as a covariate (described below and shown in the last column of Table 5).
Table 5.
Summary of heritability estimates for brain phenotypes
| h2* (SE) | 95% confidenceinterval | Included covariates | h2 (SE) after including a factor for log total brain volume | |
|---|---|---|---|---|
| Total brain volume | 0.99 (0.06) | 0.87–1.0 | Sex | NA |
| Cerebral volume | 0.98 (0.06) | 0.86–1.0 | Sex | 0.77 (0.08) |
| Cerebellar volume | 0.86 (0.09) | 0.68–1.0 | Sex, weight | 0.85 (0.07) |
| Combined hippocampal volume | 0.95 (0.07) | 0.81–1.0 | Sex, weight | 0.86 (0.08) |
| Corpus callosal area | 0.89 (0.07) | 0.75–1.0 | Sex, age, weight | 0.58 (0.09) |
*p < 10−16. NA, Not applicable.
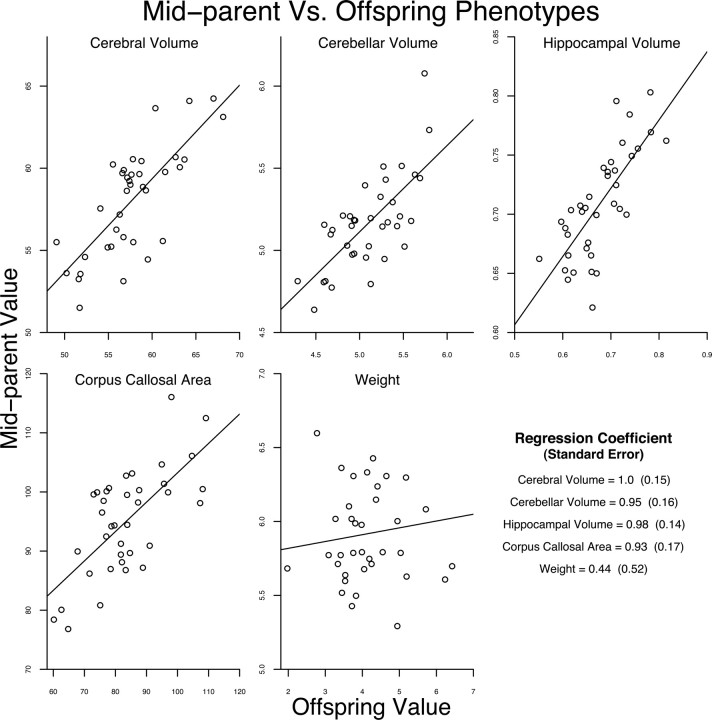
These high heritability estimates can be illustrated graphically in a subset of the sample by comparing the offspring phenotypic values with the average of the parents' phenotypic value (mid-parent phenotype). For 37 nuclear families, we had complete phenotype data for offspring and both parents. These families included 32 unique mothers, 14 unique fathers and seven full sibling pairs. After adjusting for the effects of sex, the phenotypic values for the parents were averaged and plotted against the values for the offspring. There is a strong linear relationship between mid-parent and offspring measurements for all brain phenotypes (Fig. 8). For comparison, the parent-offspring relationship in this sample for the moderately heritable trait weight (h2 = 0.43) is also shown. The line plotted on each panel shows the slope of the regression coefficient that is listed in the last panel and mirrors the heritability estimates. Due to the smaller sample size and limited familial relationships in this subsample, the SEs for the regression slopes are substantially larger than the SEs for heritability estimates listed in Table 5, which incorporate all animals in the pedigree.
Figure 8.
Mid-parent versus offspring plots. After regressing out the effect of sex, phenotypic values were calculated for the 37 complete parent-offspring trios. The averaged phenotypic value for the two parents (mid-parent value) was plotted against the offspring's phenotypic value. The middle panel of the second row shows a plot for weight, a trait with lower heritability (h2 = 0.43).
Given the significant correlations between the phenotypes (Fig. 6, Table 3), the high heritability estimates for each phenotype could simply reflect the heritability of the more general trait of total brain volume. To dissect the overlap between each phenotype and total brain volume, we also estimated the heritabilities for each phenotype after including total brain volume as a covariate. These estimates (last column of Table 5), indicate that each phenotype is highly heritable independent of the genetic component of total brain volume.
Discussion
The VRC was originally established in 1975 to study brain-behavior relationships. Vervets from this colony have been used to study serotonin-behavior relationships (McGuire and Raleigh, 1985; Raleigh et al., 1985), maternal behavior and development (Fairbanks, 1996), substance use and abuse (Melega et al., 2008), and behavioral and cognitive impulsivity (Fairbanks, 2001; James et al., 2007). Heritability, using the VRC pedigree, has been reported for impulsivity and aggressiveness (Fairbanks et al., 2004), novelty seeking (Bailey et al., 2007) and risk factors for diabetes and metabolic syndrome (Kavanagh et al., 2007), and a QTL for variation in dopamine metabolism was recently mapped at a genomewide significance level using the VRC pedigree (Freimer et al., 2007). In this study, we extended the neurobehavioral characterization of the VRC through high-resolution three-dimensional brain MRIs and estimated high heritabilities in this sample for total brain volume, cerebral volume, cerebellar volume, hippocampal volume, and corpus callosal cross-sectional area.
A wide range of values was observed for each phenotype. Within each sex, there was an almost 15cc difference between the minimum and maximum values for total brain volume, in a sample with a mean of 72.8 cc for males and 65.9 cc for females (Fig. 3, Table 2). Thus, despite the inbred structure and common environment of the pedigree, substantial trait variability exists and provides a good substrate for the identification of the genetic sources of this variance. There was no detectable effect of age on total brain volume in our sample (Fig. 3). This finding is similar to that obtained in other Old World monkeys including baboons, macaques and mangabeys, whose brains reach mature size by age three to six years (Leigh, 2004). Given that vervets have been observed to accumulate Alzheimer's-like amyloid-β plaques (Lemere et al., 2004), it is notable that we found little or no loss of brain volume with increasing age. This finding may reflect the longitudinal impact of the low fat diet of the VRC.
The cross-sectional area of the corpus callosum showed a gradual increase until age 13–15 years and then leveled off to show little change in older years (Fig. 4). These findings are similar to imaging studies in humans (Giedd et al., 1999) and capuchins (Phillips and Sherwood, 2008), a new world monkey. In both of these studies, cross-sectional area showed a gradual increase into the second decade of life. These studies hypothesized that the increase in cross- sectional area reflects increases in the amount of myelination.
The heritability estimates for brain structures in the VRC sample are higher and more precise than estimates for similar measures from previous studies of human and NHP samples. The estimates are specific to the VRC and cannot be generalized to other populations. Several factors may contribute to the high heritability estimates in the VRC. Most importantly, the VRC is maintained in a controlled and relatively homogenous environment that reduces the contribution of environment to overall trait variance, leading to a larger proportional contribution of individual genotypes; the animals live in similarly composed groups, housed in uniform enclosures with standard diets. They are exposed to identical climate, day-night cycles, air quality, and noise levels. An examination of animals living in different environments would likely result in lower heritability estimates. There are no comparable studies of brain morphometry in wild vervet populations, however, Cardini and Elton (2008) examined detailed measurements from >300 vervet skulls collected from various locations on Sub-Saharan Africa and found strong geographical and environmental correlations with morphological variation.
Another factor contributing to high heritability estimates is the reliability of the phenotype measurements. The phenotypes were measured using high quality brain images that were carefully processed to minimize measurement error. The variance component analysis method used to estimate heritabilities, constrains the total variance to 1.0 and assigns all components of variance to either a genetic component or an environmental component. Because measurement error is a component of variance that will not follow any systematic pattern within the pedigree, it is modeled as environmental variance, thereby decreasing the portion of variance attributable to a genetic component (i.e., lowering the heritability estimate). Reduction of measurement error likely explains at least a portion of our relatively high heritability estimates for the hippocampus compared with similar studies in humans and other nonhuman primates. In previous studies, the hippocampus was segmented by automated methods such as voxel-based morphometry, which is limited in its ability to accurately capture individual-specific variation, or by manual segmentation, which is prone to inter-rater variability. The hybrid-discriminative learning algorithm implemented in this study combined the anatomic accuracy and ability to identify individual-specific variability of manual tracing with the high reliability of automated methods.
The precision of the heritability estimates, i.e., the narrow confidence intervals (Table 5), reflect the large number of individuals included in this study, as well as the extent and structure of the vervet pedigree (Fig. 7), which is up to eight generations deep, and which adds considerable power to genetic analysis (Freimer and Sabatti, 2004). The phenotyped sample spanned seven generations and included a variety of close and distant familial relationships. The availability of close relationships (full-siblings, half-siblings, and parent-offspring) improves precision because the SE of the heritability estimate is scaled by the coefficient of relationship (Falconer and Mackay, 1996). Heritability analyses limited to close relationships such as full-sibs are susceptible to bias because of confounds introduced by effects of genetic dominance (Falconer and Mackay, 1996). The addition of more distant relationships reduces this bias.
In this study, we modeled the additive genetic component of phenotypic variance, which is the sole contributor to narrow sense heritability. Dominance and epistatic components are not explicitly modeled in these analyses and, if present, would contribute to the nongenetic variance. The high estimates of heritability in this study do not rule out the possibility of higher order contributions from dominance and epistatic sources, but indicate that the majority of the genetic variance for the examined phenotypes is due to additive genetic sources. The existence of mutations in multiple genes leading to autosomal recessive microcephaly in humans (Woods et al., 2005) indicates that recessive alleles of large effect are a potential source of variation for brain structural phenotypes. Despite the possibility that the extensive inbreeding in the VRC could enrich for the effects of recessive alleles, the observed high additive heritability suggests that such mutations do not account for substantial variability in the VRC.
There are a few potential factors which, if present, would lead to an overestimation of heritability. Gene-environment covariances occur if an individual's genotype influences the environmental contributions to a trait (Falconer and Mackay, 1996). For example, a correlation between genotype and environment would arise in the VRC if genetically determined differences in brain volume also resulted in differential exposure to environmental sources of variation such as nutrition, which then additionally affect brain volume. Because these sources of variation are systematically associated with individual genotypes, gene-environment covariances are confounded with genetic contributions and lead to an overestimation of trait heritability. The well controlled environment of the VRC likely reduces the influence of gene-environment covariances, which, in general, make small contributions to the total phenotypic variance and at most would result in a small error in the estimation of heritability. One variable that could inflate heritability estimates is female dominance rank, which is socially inherited in vervet matrilines. The rearing environment provided by subordinate mothers is different from that provided by dominate mothers (Fairbanks, 1996) and could be hypothesized to impact brain development. However, our data from a subset of the phenotyped animals shows no significant correlation between mother's dominance rank during gestation and early infancy and any of the brain phenotypes (Table 3).
Another possible source of heritability overestimation is the presence of cryptic relationships in the pedigree. Heritability is estimated by scaling the observed trait covariance between two individuals by their coefficient of relationship (Falconer and Mackay, 1996). Relationships, unaccounted for in the pedigree structure would lead to lower coefficients of relationship and result in an overestimation of heritability. Careful colony management and detailed records show extensive connections and depth in the pedigree (Fig. 7), which minimize the possibility of cryptic relationships.
We observed high heritability for most of the specific brain structural measures even after including as a covariate the effects of the strong correlations between these measures (Table 5, last column). This finding suggests that two categories of genetic variant contribute to the genetic architecture of brain structural variability; variants contributing to general brain morphometry, and variants acting in a region-specific manner. The wide range of highly heritable structural phenotypes in the VRC suggest that this pedigree may be a powerful resource for QTL mapping of genes contributing to general and region specific brain morphometry.
Heritability summarizes the additive genetic contributions from all loci contributing to variability in a specific trait. Thus high trait heritability does not necessarily translate into high specific locus heritability, which is important for mapping individual QTL. However, the experimental parameters that have added power to the current analysis of brain phenotypes including genetic homogeneity, extended pedigree structure and well controlled environmental sources of variation also make the VRC an excellent model system to pursue linkage analysis. Together with the anticipated results of QTL mapping studies in the VRC of several neurophysiologic and neurobehavioral phenotypes, these investigations offer the prospect of correlating structural and functional variation in a single model organism that is closely related to humans.
Footnotes
This work was supported by the National Institutes of Health (NIH) Roadmap for Biomedical Research, Grant U54 RR021813; the University of California, Los Angeles (UCLA) Semel Institute; and Grants NCRR 2R01RR016300-03, P40RR019963, and 5P20RR020750-03. Support was also provided by Grants RR12169, RR13642, and RR00865 from the National Center for Research Resources (NCRR), a component of the NIH. Scott Fears was supported by the Training Grant for Neurobehavioral Genetics, 5T32NS048004. We gratefully acknowledge the many individuals whose contributions made this project possible, including David Jentsch, John Mazziotta, and Peter Whybrow. Members of the VRC management and MRI scanning team included Kelli Barnet, Stephanie Groman, Jennifer Kay, and Pes Morton. The manual segmentation of cerebellum, hippocampi, and corpus callosum area was done by Zvart Abaryan, Alenoush Aramians, Eric Chen, Andrew Cheng, Richard Chhuon, Pedram Daraei, James Anstey, Mario Feghali, Eliza Fayz, Jenny Hardy, Dominic Hovsepian, Nil Kaymaz, Anastasia Kosenko, Brian Koski, Kimberly Lajcik, Michelle Lee, Thuy Nguyen, Tuyet Nguyen, Rhusheet Patel, Tina Phan, Vandad Saadat, Elaine Tring, Yeseul Yoo, and Charles Yu.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Atwood LD, Wolf PA, Heard-Costa NL, Massaro JM, Beiser A, D'Agostino RB, DeCarli C. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35:1609–1613. doi: 10.1161/01.STR.0000129643.77045.10. [DOI] [PubMed] [Google Scholar]
- Bailey JN, Breidenthal SE, Jorgensen MJ, McCracken JT, Fairbanks LA. The association of DRD4 and novelty seeking is found in a nonhuman primate model. Psychiatr Genet. 2007;17:23–27. doi: 10.1097/YPG.0b013e32801140f2. [DOI] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. Genetic variability of human brain size and cortical gyral patterns. Brain. 1997;120:257–269. doi: 10.1093/brain/120.2.257. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Altshuler LL, Greider T, Curran J, Keen B, Dixon WJ. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Res. 1998;82:11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Cardini A, Elton S. Variation in guenon skulls (I): species divergence, ecological and genetic differences. J Hum Evol. 2008;54:615–637. doi: 10.1016/j.jhevol.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Falk D, Vannier M, Konigsberg L, Helmkamp RC, Hildebolt C. Heritability of brain size and surface features in rhesus macaques (Macaca mulatta) J Hered. 1990;81:51–57. doi: 10.1093/oxfordjournals.jhered.a110924. [DOI] [PubMed] [Google Scholar]
- Eckert M. Neuroanatomical markers for dyslexia: a review of dyslexia structural imaging studies. Neuroscientist. 2004;10:362–371. doi: 10.1177/1073858404263596. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA. Individual differences in maternal style of old world monkeys. Adv Study Behav. 1996;25:579–611. [Google Scholar]
- Fairbanks LA. Individual differences in response to a stranger: social impulsivity as a dimension of temperament in vervet monkeys (Cercopithecus aethiops sabaeus) J Comp Psychol. 2001;115:22–28. doi: 10.1037/0735-7036.115.1.22. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, McGuire MT. Determinants of fecundity and reproductive success in captive vervets. Amer J Primatol. 1984;7:27–38. doi: 10.1002/ajp.1350070106. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, Newman TK, Bailey JN, Jorgensen MJ, Breidenthal SE, Ophoff RA, Comuzzie AG, Martin LJ, Rogers J. Genetic contributions to social impulsivity and aggressiveness in vervet monkeys. Biol Psychiatry. 2004;55:642–647. doi: 10.1016/j.biopsych.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Ed 4. Harlow, Essex, UK: Longmans Green; 1996. Introduction to quantitative genetics. [Google Scholar]
- Freimer N, Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat Genet. 2004;36:1045–1051. doi: 10.1038/ng1433. [DOI] [PubMed] [Google Scholar]
- Freimer NB, Service SK, Ophoff RA, Jasinska AJ, McKee K, Villeneuve A, Belisle A, Bailey JN, Breidenthal SE, Jorgensen MJ, Mann JJ, Cantor RM, Dewar K, Fairbanks LA. A quantitative trait locus for variation in dopamine metabolism mapped in a primate model using reference sequences from related species. Proc Natl Acad Sci U S A. 2007;104:15811–15816. doi: 10.1073/pnas.0707640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Rajapakse JC, Vaituzis AC, Liu H, Berry YC, Tobin M, Nelson J, Castellanos FX. Development of the human corpus callosum during childhood and adolescence: a longitudinal MRI study: new trends in developmental neuroimaging in psychiatry. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:571–588. doi: 10.1016/s0278-5846(99)00017-2. [DOI] [PubMed] [Google Scholar]
- Grady L. Random walks for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2006;28:1768–1783. doi: 10.1109/TPAMI.2006.233. [DOI] [PubMed] [Google Scholar]
- Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baaré WF, van Oel C, van Haren NE, Collins DL, Evans AC, Amunts K, Bürgel U, Zilles K, de Geus E, Boomsma DI, Kahn RS. Genetic contributions to human brain morphology and intelligence. J Neurosci. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AS, Groman SM, Seu E, Jorgensen M, Fairbanks LA, Jentsch JD. Dimensions of impulsivity are associated with poor spatial working memory performance in monkeys. J Neurosci. 2007;27:14358–14364. doi: 10.1523/JNEUROSCI.4508-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Fairbanks LA, Bailey JN, Jorgensen MJ, Wilson M, Zhang L, Rudel LL, Wagner JD. Characterization and heritability of obesity and associated risk factors in vervet monkeys. Obesity. 2007;15:1666–1674. doi: 10.1038/oby.2007.199. [DOI] [PubMed] [Google Scholar]
- Kelly AM, Margulies DS, Castellanos FX. Recent advances in structural and functional brain imaging studies of attention-deficit/hyperactivity disorder. Curr Psychiatry Rep. 2007;9:401–407. doi: 10.1007/s11920-007-0052-4. [DOI] [PubMed] [Google Scholar]
- Leigh SR. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol. 2004;62:139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- Lemere CA, Beierschmitt A, Iglesias M, Spooner ET, Bloom JK, Leverone JF, Zheng JB, Seabrook TJ, Louard D, Li D, Selkoe DJ, Palmour RM, Ervin FR. Alzheimer's disease abeta vaccine reduces central nervous system abeta levels in a non-human primate, the Caribbean vervet. Am J Pathol. 2004;165:283–297. doi: 10.1016/s0002-9440(10)63296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DM, Yang C, Sawyer-Glover AM, Moseley ME, Schatzberg AF. Early life stress and inherited variation in monkey hippocampal volumes. Arch Gen Psychiatry. 2001;58:1145–1151. doi: 10.1001/archpsyc.58.12.1145. [DOI] [PubMed] [Google Scholar]
- Mahaney MC, Williams-Blangero S, Blangero J, Leland MM. Quantitative genetics of relative organ weight variation in captive baboons. Hum Biol. 1993;65:991–1003. [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- McGuire MT. Basel, New York: S Karger; 1974. Contributions to primatology, vol 1: the St. Kitts vervet. [Google Scholar]
- McGuire MT, Raleigh MJ. Serotonin-behavior interactions in vervet monkeys. Psychopharmacol Bull. 1985;21:458–463. [PubMed] [Google Scholar]
- Melega WP, Jorgensen MJ, Laćan G, Way BM, Pham J, Morton G, Cho AK, Fairbanks LA. Long-term methamphetamine administration in the vervet monkey models aspects of a human exposure: brain neurotoxicity and behavioral profiles. Neuropsychopharmacology. 2008;33:1441–1452. doi: 10.1038/sj.npp.1301502. [DOI] [PubMed] [Google Scholar]
- Narr KL, Woods RP, Thompson PM, Szeszko P, Robinson D, Dimtcheva T, Gurbani M, Toga AW, Bilder RM. Relationships between IQ and regional cortical gray matter thickness in healthy adults. Cereb Cortex. 2007;17:2163–2171. doi: 10.1093/cercor/bhl125. [DOI] [PubMed] [Google Scholar]
- Newman TK, Fairbanks LA, Pollack D, Rogers J. Effectiveness of human microsatellite loci for assessing paternity in a captive colony of vervets (Chlorocebus aethiops sabaeus) Am J Primatol. 2002;56:237–243. doi: 10.1002/ajp.1078. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. A twin MRI study of size variations in human brain. J Cogn Neurosci. 2000;12:223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum Brain Mapp. 2007;28:464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE, Carmelli D. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiol Aging. 2000;21:63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Sherwood CC. Cortical development in brown capuchin monkeys: a structural MRI study. Neuroimage. 2008;43:657–664. doi: 10.1016/j.neuroimage.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJ, Neale MC, Hulshoff Pol HE, Baaré WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30:311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, Brammer GL, McGuire MT, Yuwiler A. Dominant social status facilitates the behavioral effects of serotonergic agonists. Brain Res. 1985;348:274–282. doi: 10.1016/0006-8993(85)90445-7. [DOI] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Lancaster J, Shelledy W, Glahn D, Blangero J, Fox P. Heritability of brain volume, surface area and shape: an MRI study in an extended pedigree of baboons. Hum Brain Mapp. 2007;28:576–583. doi: 10.1002/hbm.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. Neuroimage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- The R Development Team. Vienna: R Foundation for Statistical Computing; 2008. R: a language and environment for statistical computing. [Google Scholar]
- Tu Z, Narr KL, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008;27:495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP. Characterizing volume and surface deformations in an atlas framework: theory, applications, and implementation. Neuroimage. 2003a;18:769–788. doi: 10.1016/s1053-8119(03)00019-3. [DOI] [PubMed] [Google Scholar]
- Woods RP. Multitracer: a Java-based tool for anatomic delineation of grayscale volumetric images. Neuroimage. 2003b;19:1829–1834. doi: 10.1016/s1053-8119(03)00243-x. [DOI] [PubMed] [Google Scholar]
- Woods RP. Spatial transformation models. In: Bankman I, editor. Handbook of medical imaging: processing and analysis. Ed 2. Vol 1. Burlington, MA: Elsevier; 2009. pp. 541–568. [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]