Abstract
The proper glycosylation of proteins trafficking through the Golgi apparatus depends upon the conserved oligomeric Golgi (COG) complex. Defects in COG can cause fatal congenital disorders of glycosylation (CDGs) in humans. The recent discovery of a form of CDG, caused in part by a COG4 missense mutation changing Arg 729 to Trp, prompted us to determine the 1.9 Å crystal structure of a Cog4 C-terminal fragment. Arg 729 is found to occupy a key position at the center of a salt bridge network, thereby stabilizing Cog4's small C-terminal domain. Studies in HeLa cells reveal that this C-terminal domain, while not needed for the incorporation of Cog4 into COG complexes, is essential for the proper glycosylation of cell surface proteins. We also find that Cog4 bears a strong structural resemblance to exocyst and Dsl1p complex subunits. These complexes and others have been proposed to function by mediating the initial tethering between transport vesicles and their membrane targets; the emerging structural similarities provide strong evidence of a common evolutionary origin and may reflect shared mechanisms of action.
Keywords: congenital disorder of glycosylation, Golgi apparatus, multi-subunit tethering complex, vesicle trafficking, X-ray crystallography
Multiple lines of evidence implicate the conserved oligomeric Golgi (COG) complex in retrograde transport within the Golgi apparatus (1, 2). For example, partial knockdown of mammalian COG3 permitted exocytosis, while simultaneously blocking the retrograde transport of Shiga toxin from endosomes to the ER (3). COG was initially isolated from bovine brain on the basis of its ability to stimulate an in vitro Golgi transport assay (4). Two of its 8 subunits, Cog1/ldlB and Cog2/ldlC, had previously been identified in a genetic screen for compromised cell surface receptor stability (5, 6). In the yeast Saccharomyces cerevisiae, the COG complex was discovered through studies of one of its subunits, Cog8p/Dor1p. Implicated in vesicle targeting to the Golgi apparatus, Cog8p was found to co-immunoprecipitate with 7 other polypeptides (7). The 7 associated polypeptides included two (Cog2p/Sec35p and Cog3p/Sec34p) that had earlier been identified in genetic screens for temperature-sensitive mutations causing secretion defects (8). In further support of a role in trafficking to and within the Golgi, COG interacts physically with a large number of Golgi-localized trafficking factors, including (in yeast) the Rab family G protein Ypt1p, the SNAREs Sed5p, Ykt6p, Gos1p, and Sec22p, and COPI vesicle coat proteins (9).
COG is one of several large protein complexes proposed to function as multisubunit tethering factors, mediating the initial interaction between transport vesicles and their target membranes (2, 10–12). For many of these complexes, a dearth of structural information has slowed the development of mechanistic models and has made it difficult to know how—or whether—to apply lessons learned from studies of one complex to another. Subunits of the COG, exocyst, Dsl1p, and Golgi-associated retrograde protein (GARP) complexes share limited sequence homology (7, 13) that has been suggested to reflect convergent evolution (13). Structures, most of them partial, have been reported for one COG complex subunit, two Dsl1p complex subunits, and four exocyst subunits (14–20); these structures contain one (Cog2p), two (Dsl1p, Sec15, Exo84p), three (Sec6p), four (Exo70p), or five (Tip20p) helix bundle domains. In each of the exocyst and Dsl1p complex structures, the tandem helical domains form an extended, rod-like array (20, 21). Structural studies of a fourth multisubunit tethering complex, the transport protein particle I (TRAPP I) complex (22), demonstrate that it is structurally unrelated to the COG/Dsl1p/exocyst family.
Among the known multisubunit tethering complexes, only COG (23–28) and TRAPP (29) have been directly implicated in human genetic disease. Mutations in the Cog1 (24), Cog7 (26–28), and Cog8 (23, 25) subunits cause type II congenital disorders of glycosylation (CDGs). Type II CDGs lead to a wide variety of neurological and developmental abnormalities brought on by defects in the processing of N-linked glycan chains [for review, see (30)]. Most recently, a Cog4 missense mutation that converts Arg 729 to Trp (R729W) has been reported to cause, in conjunction with a submicroscopic deletion of the COG4 gene in the other allele, a type II CDG (31). To investigate the structural basis for the CDG-II caused by the R729W mutation, we have now determined the crystal structure of the C-terminal 250 residues of human Cog4, representing ≈30% of the polypeptide chain. Arg 729 is observed to participate in a network of salt bridges that stabilize a small C-terminal domain, a conclusion reinforced by functional studies in HeLa cells. More generally, the Cog4 X-ray structure establishes an unambiguous link between the COG, Dsl1p, and exocyst complexes, providing strong evidence that they are derived from a common evolutionary precursor.
Results
Structure of Cog4-(525–785).
In our initial attempts to determine the crystal structure of human Cog4, we focused on the full-length polypeptide (residues 1–785). Full-length Cog4, overproduced in insect cells, was monomeric as judged by ultracentrifugation but failed to crystallize despite extensive efforts. To identify Cog4 fragments that retained the critical Arg 729 residue (31) and might be better suited for structural studies, we used limited proteolytic digestion. Fragments with an apparent molecular weight of ≈27 kDa were observed in digests using several different proteases. One such fragment, generated by thermolysin cleavage, was identified as Cog4 residues 524–785 using mass spectrometry and N-terminal sequencing. We overproduced a slightly shorter fragment (residues 525–785) in Escherichia coli. This fragment, unlike the full-length protein, is dimeric as judged by analytical ultracentrifugation. We determined the X-ray structure of the Cog4-(525–785) dimer by MAD phasing and refined it to 1.9 Å resolution (Fig. 1A and Table S1). The final model includes residues 536–785 of one monomer and 537–785 of the other monomer, with the remaining N-terminal residues, for which no interpretable electron density was observed, likely disordered in the crystals. The two monomers adopt very similar conformations (Fig. S1), each consisting of 11 α-helices numbered H1–H11.
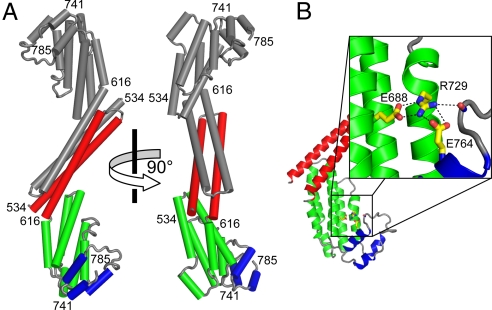
Fig. 1.
X-ray crystal structure of Cog4-(525–785). (A) H. sapiens Cog4, including residues 536–785. (B) Ionic interaction network centered around Arg 729.
Helices H1 and H2 from one monomer (red in Fig. 1A) pair with H1 and H2 of the other monomer (gray in Fig. 1A) to form a 4-helix bundle. This 4-helix bundle is evidently responsible for the dimerization of Cog4-(525–785) in solution. In full-length Cog4, which is monomeric, these intermolecular contacts may be replaced by intramolecular contacts, presumably involving regions of Cog4 not present in the fragment we crystallized. Helices H3–H8 (green) and H9–H11 (blue) each form compact helix bundles. These bundles are termed domains D and E, respectively, based on structural comparisons with other tethering complex subunits (see below).
Based on multiple sequence alignment, the most highly conserved region of Cog4 lies within the H7 and H8 helices of domain D (Fig. S2). Helix H8, which contains Arg 729, fits into a groove on the surface of domain E. Polar and hydrophobic contacts with domain E are mediated by conserved residues in H8, including Arg 729 as well as Gln 732 and Ile 736. The lengthy side chain of Arg 729 traverses the width of domain D, with its guanidinium group forming the central residue in a network of salt bridges involving Glu 688 (in domain D) and Glu 764 (in domain E) (Fig. 1B). Clearly, this salt bridge network is disrupted upon replacing Arg 729 with Trp. Moreover, the bulky and relatively inflexible Trp side chain might itself cause additional perturbations in the interface between domains D and E. Thus, one hypothesis that emerges from the X-ray structure is that domain E is important for COG complex function, and that the R729W mutation affects the stability and/or positioning of this domain.
Functional Analysis in HeLa Cells.
The importance of domain E is strongly supported by functional studies. As a first step, we knocked down Cog4 in HeLa cells using a COG4-specific shRNA plasmid. Glycosylation of cell surface proteins was aberrant, as judged using the fluorescently labeled lectin Galanthus nivalus lectin (GNL)-Alexa-647, which specifically recognizes terminal mannose (see SI Materials and Methods). We next tested the ability of mutant Cog4 proteins to rescue this defect. C-terminal truncations removing domain E (residues 741–785), domains D and E together (618–785), or the entire crystallized region (525–785) were evaluated using siRNA-resistant constructs containing a C-terminal Myc tag. Whereas full-length Cog4, as expected, was able to rescue the glycosylation defect, thereby abolishing GNL staining, the truncated Cog4 subunits displayed glycosylation defects equivalent to those observed upon transfection with an empty control vector (Fig. 2 A and B). The truncated subunits are properly incorporated into COG complexes based on their quantitative co-immunoprecipitation with Cog6, a subunit that, within the hetero-octameric complex, is positioned far from Cog4 (32) (Fig. 2C). We conclude that truncating as few as 45 residues, corresponding to domain E, from the C terminus of the Cog4 subunit severely compromises the function of the mammalian COG complex.
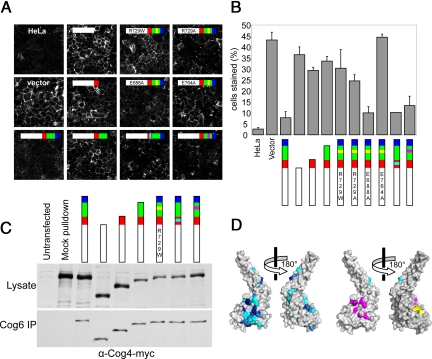
Fig. 2.
Mutational analysis of Cog4 function in Cog4-silenced HeLa cells. (A) Cells transfected with the indicated siRNA-resistant allele were incubated with GNL-Alexa-647, which labels mis-glycosylated cell surface proteins. All constructs (except “vector”) include Cog4 residues 1–524, indicated by the white bars. Red, green, and blue bars represent the protein regions color-coded as in Fig. 1A. Cyan and magenta boxes indicate sets of mutations as indicated in Fig. S2. A yellow box indicates mutations of R729 or interacting residues as noted on the bar itself (e.g., R729W). (B) Quantification of the percentage of cells [± SD, n = 3 (>100 cells each)] as shown in Fig. 2A with detectable GNL staining of plasma membrane glycoproteins. (C) To assess protein expression level, cells transfected with silencing-resistant Cog4 alleles carrying C-terminal 3× Myc tags were analyzed by western blotting. To assess the integrity of the COG complex, the quantity of tagged Cog4 present in Cog6 immunoprecipitation reactions was evaluated by western blotting. (D) Surface representations of the Cog4-(525–785) monomer. In the left-hand pair of images, the surface is color-coded by sequence conservation (darker blue is more conserved). In the right-hand pair, the residues that were studied by site-directed mutagenesis are color-coded to match the triangles in Fig. S2.
Next, we examined the impact of the R729W mutation in HeLa cells. Full-length Cog4 containing the R729W mutation failed to rescue the glycosylation defect observed in Cog4 knockdown cells (Fig. 2 A and B). As discussed above, mutation of Arg 729 will abolish the salt bridge network linking the arginine guanidinium group with the carboxylates of Glu 688 and Glu 764. To selectively disrupt the salt bridge network, without introducing a bulky side chain such as Trp that might cause steric disruption, we replaced Arg 729 with Ala. This substitution compromised COG complex function to a degree comparable to that observed for the R729W mutation. Changing Glu 764 to Ala had a similar effect on function. Thus, disrupting the interdomain salt bridge in three different ways (R729W, R729A, or E764A) had deleterious effects similar to that observed upon deleting domain E altogether, strongly supporting its functional importance. On the other hand, although it might have been anticipated that the intradomain salt bridge between Glu 688 and Arg 729 would help to position Arg 729 for optimal interaction with Glu 764, the mutation E688A had at most a mild impact on GNL staining. Overall, we conclude that the functionally important Arg 729-Glu 764 salt bridge most likely serves to position and/or stabilize domain E.
Domain D contains, in addition to Arg 729, a patch of conserved surface-exposed residues (magenta in Fig. 2D). Truncating these residues to alanine or replacing them with charged residues did not significantly affect the function of human Cog4 as judged by GNL staining (Fig. 2 A and B). We hypothesized that the incomplete removal of endogenous Cog4 in the knockdown strain might be masking weaker phenotypes. We therefore took advantage of the superior genetics of S. cerevisiae to further analyze mutations in domains D and E. These yeast studies also allowed us to evaluate whether the critical function of domain E is conserved, especially since Arg 729's salt-bridging partner, Glu 764, is not (Fig. S2).
Functional Analysis in S. cerevisiae.
Mutations were engineered in a CEN plasmid containing yeast COG4 expressed under the control of its endogenous promoter (Tables S2 and S3). These plasmids were tested, using a plasmid shuffling assay, for their ability to complement a genomic deletion of the essential COG4 gene. Deleting domains D and E abolished function (Fig. S3A). Surprisingly, removal of domain E only, which caused severe glycosylation defects in human Cog4, did not cause an observable growth defect in yeast. Therefore, domain E is dispensable to the essential function of Cog4p in S. cerevisiae. Moreover, substituting Trp for Lys 810, the yeast equivalent of human Arg 729, did not result in a significant growth defect (Fig. S3B). These findings indicate that the function of domain E, which is compromised in humans by the R729W mutation, may be unique to higher eukaryotes. Consistent with this conclusion, the sorting of the vacuolar protein carboxypeptidase Y (CPY), a sensitive indicator of vacuolar protein sorting (vps) trafficking defects caused by COG mutations (9), was normal in both the K810W mutant and the domain E deletion mutant.
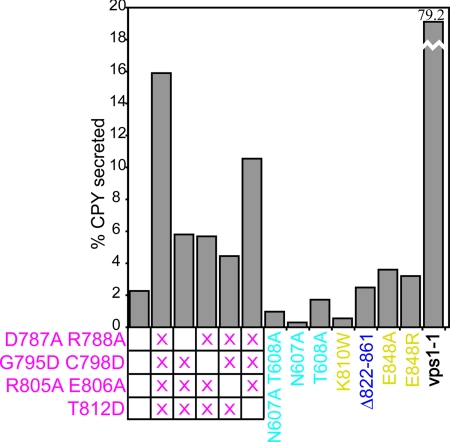
Next, we tested single and double amino acid substitutions designed to alter the conserved surface patch in domain D. Changing four residues to Ala, and replacing three small residues with Asp, led to a strong temperature-sensitive (Ts) growth defect (Fig. S3B). Vps defects were also evident; yeast expressing the mutant protein secreted about 7-fold more CPY than those expressing wild-type Cog4p at temperatures at which growth was unaffected (Fig. 3 and Fig. S3C). Thus, although we have not rigorously excluded the possibility that this cluster of conserved residues is important for protein stability, we hypothesize that it instead serves as a binding site for as-yet-unidentified cellular factors important for COG function.
Fig. 3.
Mutational analysis of Cog4p function in S. cerevisiae. The percentage CPY secreted was calculated by measuring intracellular and extracellular CPY by western blotting (see Fig. S3C). The vacuolar sorting mutant vps1–1 was included as a control.
Structural Homology Between Cog4 and Other Tethering Complex Subunits.
The Cog4-(525–785) X-ray structure bears an unexpectedly strong resemblance to the structures of subunits belonging to 2 other multisubunit tethering complexes, the exocyst complex and the Dsl1p complex (Fig. 4A). The striking resemblance between Cog4-(525–785) and the known exocyst and Dsl1p structures, particularly those of Sec6p (residues 411–805) (18) and Tip20p (20), provides strong confirmation that at least some of the subunits composing these complexes are descended from a common progenitor. We therefore adopt the domain nomenclature initially proposed by Dong et al. (15) and later extended by Tripathi et al. (20), according to which the domains of the exocyst and Dsl1p complexes are denoted A–E.
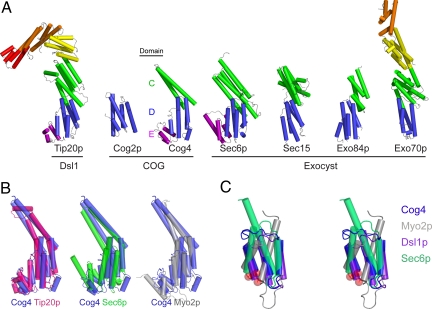
Fig. 4.
Structural alignment of Cog4-(525–785) to known COG, exocyst and Dsl1p subunits. (A) Shown are S. cerevisiae Tip20p (PDB ID 3FHN, residues 4–701 out of 701) (20), Cog2p (2JQQ, residues 109–262 out of 262) (14), Sec6p (2FJI, residues 411–805 out of 805) (18), Drosophila melanogaster Sec15 (2A2F, residues 382–699 out of 766) (19), S. cerevisiae Exo84p (2D2S, residues 525–753 out of 753) (15), and S. cerevisiae Exo70p (2PFV, residues 67–623 out of 623) (15–17). Pairwise alignment was performed with the program DaliLite (47) to match each of the other structures to Cog4-(525–785). The DaliLite Z scores for the alignments shown were 12.3 (Cog4-Tip20p), 3.8 (Cog4-Cog2p), 13.1 (Cog4-Sec6p), 10.1 (Cog4-Sec15), 6.2 (Cog4-Exo84p), and 8.0 (Cog4-Exo70p). (B) Cog4- (525–785) superimposed on proteins containing similar domains C, D, and E. (C) Stereoview of E domains, aligned using DaliLite and with the N terminus of each domain indicated by a red sphere. Included, in addition to the structures cited above, are the E domains of the cargo-binding domain of S. cerevisiae Myo2p (2F6H) (33) and the Dsl1p complex subunit Dsl1p (Y. Ren, P.D.J., and F.M.H., personal communication). No significant alignment was discernable for domain E of Tip20p.
The fragment of Cog4 studied here was generated by cleavage with the protease thermolysin, and comparison with other known structures reveals that the protease cleaved Cog4 in the middle of domain C. In the Cog4-(525–785) X-ray structure, the final two helices of domain C mediate nonphysiological dimerization. These helices occupy essentially the same positions as the corresponding helices in Sec6p-(411–805) (Fig. S4). Full-length Cog4 might contain helices N-terminally adjacent to the thermolysin cleavage site that would complete domain C according to a similar topology. Consistent with this idea, a pair of conserved residues in domain C, Asn 607 and Thr 608 (cyan in the right-hand panels of Fig. 2D), caused a Ts growth defect when truncated to Ala (Fig. 3). While Thr 608 may stabilize the interface between domains C and D, Asn 607 appears likely to form interactions with other domain C helices absent in the present structure (Fig. S5).
Domain E, which plays a critical role in the function of human Cog4, is present in some of the structurally characterized COG/Dsl1p/exocyst family subunits (Tip20p and Sec6p) but not in others (Exo70p, Exo84p, and Cog2p). An E domain is also present in the yeast myosin V motor Myo2p (33), the only other structure currently in the Protein Data Bank that is homologous to the COG/Dsl1p/exocyst family subunits (17, 20). Nonetheless, although the known E domains occupy a relatively uniform location relative to their respective D domains, they are more heterogeneous in structure than the D domains (Fig. 4C).
Discussion
The crystal structure of the C-terminal domains of Homo sapiens Cog4 reveals that Arg 729, whose mutation to Trp contributes to a human CDG (31), forms a crucial salt bridge that anchors domain E to domain D (Fig. 1). Our results establish that the E domain is essential for proper cell surface glycosylation in HeLa cells (Fig. 2). Likewise, mutating either Arg 729 (in domain D) or its salt bridge partner Glu 764 (in domain E) causes severe glycosylation defects. Because neither the D nor the E domain is important for formation of the COG complex, we conclude that these domains likely harbor sites that mediate key functional interactions with other components of the Golgi trafficking machinery. These partners presumably collaborate with the COG complex in vesicle tethering; e.g., by anchoring COG to the vesicle and/or Golgi membranes. No proteins have yet been reported that interact with domains D or E of Cog4; identifying such partners thus remains as an important goal for future efforts to understand COG complex function.
The Cog4-(525–785) X-ray structure also establishes a definitive homology among subunits of three tethering complexes (COG, Dsl1p, and exocyst) that function in discrete intracellular trafficking pathways. Each of these complexes contains at least one subunit with a recognizable E domain (Fig. 4C), highlighting the possibility that E domains play key functional roles not only in human Cog4 but more broadly across this multisubunit tethering complex family. Nonetheless, we also uncovered significant differences between human and yeast Cog4. While domain E is functionally critical for human Cog4, no phenotypic consequences were observed as a result of deleting this domain from the yeast protein. Conversely, we found that a surface patch of conserved residues is not important for Golgi function in HeLa cells, as judged by our GNL staining assay, but is nonetheless required in yeast for normal growth and CPY sorting. Thus, while Cog4 domains D and E are apparently important for interactions between the COG complex and its functional partners in both yeast and humans, the details of these interactions, and/or their phenotypic consequences, appear to have diverged.
Despite progress in characterizing the structures of individual COG/Dsl1p/exocyst subunits, relatively little is known about how the subunits within each complex interact with one another (32, 34–36). One exception is the Dsl1p and Tip20p subunits of the Dsl1p complex, which were recently shown to interact via N-terminal α-helices (20). In general, to fulfill a tethering function, it seems likely that binding sites for different partner proteins (or lipids) must be widely separated within each complex. For Cog4, we have shown here that the C-terminal-most region is functionally important. It is appealing to speculate that the Cog4 C terminus resides at the tip of one of the extended arms visualized in quick freeze/deep etch cryoelectron micrographs of unfixed COG samples (37). According to this hypothesis, the more N-terminal regions of Cog4, which would then mediate its incorporation into COG complexes, would be more centrally located. It is even possible that this is a general architectural principle; that is, that functionally important regions of tethering complexes are located at the end of extended arms. Nonetheless, despite tantalizing clues, further investigation will be needed to determine whether or not the large-scale structures of the COG, exocyst, and Dsl1p complexes are similar, and if so, whether function follows form.
Materials and Methods
Protein Expression and Purification.
Recombinant H. sapiens Cog4, residues 525–785, was overexpressed as a GST fusion protein using pGEX-4T1 (GE Healthcare) in E. coli Rosetta (Novagen). Further details are provided in the SI Materials and Methods.
Crystallization Structure Determination.
The native and selenium-derivatized crystals used for data collection were grown by hanging drop vapor diffusion; further details are provided in the SI Materials and Methods. Diffraction data were collected at NSLS beamline X29 and processed using the HKL software package (38). The structure of Cog4-(525–785) was determined using multiwavelength anomalous dispersion (MAD) at 2.4 Å resolution, with phases calculated using SHELX (39) and refined with SHARP (40). The structure was iteratively built using WinCoot (41) and refined using REFMAC (42). Starting phases for the 1.9 Å native data were obtained from a rigid body refinement of the resulting model to the native amplitudes, followed by iterative cycles of building and refinement.
Cog4 Knockdown and Rescue in HeLa Cells.
To generate a stable knockdown of Cog4, HeLa T2-GFP cells (3) were transfected with the shRNA plasmid (OriGene) (target sequence TGACATCTTGGACCTGAAGTTCTGCATGG) using Lipofectamine 2000 according to a protocol from the manufacturer (Invitrogen). Medium containing 1 μg/mL puromycin was used to select for the plasmid. Colonies were chosen based on Cog4 down-regulation as judged by immunoblotting. The silencing-resistant vector shCog4 was constructed by Pfu-based PCR using hCOD1-myc (7) and the primers TATTCTAGATTTAAAATTTTGTATGGATGGAGTTC and AAATCTAGAATATCGTCAGCTCTCTGAATGGC. Stable COG4 KD HeLa cells (clone no. 39, expressing Cog4 at <15% of endogenous levels; Fig. S6) were transfected with silencing-resistant COG4 alleles constructed in shCog4. Quantitative co-immunoprecipitation and antibodies used are described in SI Materials and Methods.
Analysis of Glycosylation Defects Using GNL.
In preliminary experiments, we found that peanut agglutinin staining, used previously to detect cell-surface glycosylation defects in cells derived from CDG-II patients (28, 43), was not useful for analyzing HeLa cell knock-downs because control cells stained positive. We therefore tested other lectins and discovered that Galanthus nivalis lectin (GNL; Vector Labs) labeled with Alexa-647 (Invitrogen, protocol supplied by manufacturer) stained the plasma membrane of Cog3-, Cog4-, and Cog7-knockdown HeLa cells but not mock-transfected cells. Unlike other terminal-mannose-binding lectins (e.g., Con A), GNL is highly specific for binding the α-1,3 linkage of terminal α-D-mannose (44–46). Use of GNL staining to analyze glycosylation defects is described in SI Materials and Methods.
S. cerevisiae Strain Production and Growth Analysis.
The parent strain BCR13 used for genetic analysis was derived from Research Genetics strain 25521 (Invitrogen), a diploid COG4/cog4Δ::KanMX mutant in a BY4743 background. The coding sequence of COG4, together with 400 bp upstream and downstream, was amplified from wild-type yeast by PCR and inserted into the pRS416 low-copy URA3 vector (pBCR114) for transformation into RG25521. Transformants were sporulated and dissected to yield BCR13, a cog4Δ::KanMX strain complemented by pBCR114. The same genomic region was cloned into pRS415, a low copy LEU2 vector, and subjected to targeted mutagenesis using the Quikchange protocol (Stratagene). Finally, mutant strains were produced by transforming the resulting plasmids into BCR13, growing single colonies overnight in SC -LEU, and selecting for strains having lost pBCR114 by plating on 5-fluoroorotic acid (5FOA)-containing media and, in parallel, on YPD. Mutants failing to grow on 5FOA were identified as lethal. All growth after loss of pBCR114 was conducted at room temperature to avoid selection for repressors of potential Ts phenotypes. Mutants Ts for growth were identified by growing overnight cultures in YPD of the strains isolated from 5FOA and spotting half-log dilutions onto YPD pre-equilibrated at room temperature or 37 °C, starting from initial dilution with OD600 normalized to ≈0.5, and observed after 2 days of growth. The CPY secretion assay is described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank E. Reynders, W. Annaert, and G. Matthijs for communicating results before publication; B. Kokona and R. Fairman for sedimentation velocity analytical ultracentrifugation; the staff of the NSLS X29 beamline for assistance with X-ray data collection; T. Kudlyk and R. Willett for technical assistance; S. Shen and M. Rose for advice and technical assistance; and Y. Ren for previously unpublished results depicted in Fig. 4C. This work was supported by National Science Foundation Grant MCB-0645163 (to V.V.L.) and National Institutes of Health Grants GM083144 (to V.V.L.) and GM071574 (to F.M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.M. is a guest editor invited by the Editorial Board.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HR0).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901966106/DCSupplemental.
References
- 1.Lupashin V, Sztul E. Golgi tethering factors. Biochim Biophys Acta. 2005;1744:325–339. doi: 10.1016/j.bbamcr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter DM, Paul KS, Waters MG. Purification and characterization of a novel 13 S hetero-oligomeric protein complex that stimulates in vitro Golgi transport. J Biol Chem. 1998;273:29565–29576. doi: 10.1074/jbc.273.45.29565. [DOI] [PubMed] [Google Scholar]
- 5.Chatterton JE, et al. Expression cloning of LDLB, a gene essential for normal Golgi function and assembly of the ldlCp complex. Proc Natl Acad Sci USA. 1999;96:915–920. doi: 10.1073/pnas.96.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Podos SD, Reddy P, Ashkenas J, Krieger M. LDLC encodes a brefeldin A-sensitive, peripheral Golgi protein required for normal Golgi function. J Cell Biol. 1994;127:679–691. doi: 10.1083/jcb.127.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 8.Wuestehube LJ, et al. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Pfeffer SR. Transport-vesicle targeting: Tethers before SNAREs. Nat Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 12.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 13.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanaugh LF, et al. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 15.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 16.Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal structure of the S. cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 17.Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J Mol Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 22.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Foulquier F, et al. A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1-Cog8 interaction in COG complex formation. Hum Mol Genet. 2007;16:717–730. doi: 10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- 24.Foulquier F, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci USA. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kranz C, et al. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- 26.Morava E, et al. A common mutation in the COG7 gene with a consistent phenotype including microcephaly, adducted thumbs, growth retardation, VSD and episodes of hyperthermia. Eur J Hum Genet. 2007;15:638–645. doi: 10.1038/sj.ejhg.5201813. [DOI] [PubMed] [Google Scholar]
- 27.Ng BG, et al. Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol Genet Metab. 2007;91:201–204. doi: 10.1016/j.ymgme.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, et al. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med. 2004;10:518–523. doi: 10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- 29.Matsui Y, et al. Loss of the SEDL gene product (Sedlin) causes X-linked spondyloepiphyseal dysplasia tarda: Identification of a molecular defect in a Japanese family. Am J Med Genet. 2001;99:328–330. doi: 10.1002/ajmg.1179. [DOI] [PubMed] [Google Scholar]
- 30.Jaeken J, Matthijs G. Congenital disorders of glycosylation: A rapidly expanding disease family. Annu Rev Genomics Hum Genet. 2007;8:261–278. doi: 10.1146/annurev.genom.8.080706.092327. [DOI] [PubMed] [Google Scholar]
- 31.Reynders E, et al. Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp262. Jun 3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem. 2005;280:32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 33.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem. 2005;280:27613–27623. doi: 10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- 35.Loh E, Hong W. The binary interacting network of the conserved oligomeric Golgi tethering complex. J Biol Chem. 2004;279:24640–24648. doi: 10.1074/jbc.M400662200. [DOI] [PubMed] [Google Scholar]
- 36.Oka T, et al. Genetic analysis of the subunit organization and function of the conserved oligomeric golgi (COG) complex: Studies of COG5- and COG7-deficient mammalian cells. J Biol Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- 37.Ungar D, et al. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1998;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Steet R, Kornfeld S. COG-7-deficient human fibroblasts exhibit altered recycling of Golgi proteins. Mol Biol Cell. 2006;17:2312–2321. doi: 10.1091/mbc.E05-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaku H, Goldstein IJ, Oscarson S. Interactions of five D-mannose-specific lectins with a series of synthetic branched trisaccharides. Carbohydr Res. 1991;213:109–116. doi: 10.1016/s0008-6215(00)90602-5. [DOI] [PubMed] [Google Scholar]
- 45.Shibuya N, Goldstein IJ, Van Damme EJ, Peumans WJ. Binding properties of a mannose-specific lectin from the snowdrop (Galanthus nivalis) bulb. J Biol Chem. 1988;263:728–734. [PubMed] [Google Scholar]
- 46.Van Damme EJM, Allen AK, Peumans WJ. Isolation and characterization of a lectin with exclusive specificity towards mannose from snowdrop (Galanthus nivalis) bulbs. FEBS Lett. 1987;215:140–144. [Google Scholar]
- 47.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.