Abstract
Lineage separation and divergence form a temporally extended process whereby populations may diverge genetically, morphologically, or ecologically, and these contingent properties of species provide the operational criteria necessary for species delimitation. We inferred the historical process of lineage formation in the coast horned lizard (Phrynosoma coronatum) species complex by evaluating a diversity of operational species criteria, including divergence in mtDNA (98 specimens; 2,781 bp) and nuclear loci (RAG−1, 1,054 bp; BDNF 529 bp), ecological niches (11 bioclimatic variables; 285 unique localities), and cranial horn shapes (493 specimens; 16 landmarks). A phylogenetic analysis of mtDNA recovers 5 phylogeographic groups arranged latitudinally along the Baja California Peninsula and in California. The 2 southern phylogeographic groups exhibit concordance between genetic, morphological, and ecological divergence; however, differentiation is weak or absent at more recent levels defined by phylogeographic breaks in California. Interpreting these operational species criteria together suggests that there are 3 ecologically divergent and morphologically diagnosable species within the P. coronatum complex. Our 3-species taxonomic hypothesis invokes a deep coalescence event when fitting the mtDNA genealogy into the species tree, which is not unexpected for populations that have diverged recently. Although the hypothesis that the 3 phylogeographic groups distributed across California each represent distinctive species is not supported by all of the operational species criteria evaluated in this study, the conservation status of the imperiled populations represented by these genealogical units remains critical.
Keywords: evolution, geometric morphometrics, niche modeling, phylogeography, speciation
Lineage separation and divergence form a temporally extended process that may render populations reciprocally monophyletic for haplotype variation, reproductively isolated, ecologically divergent, or morphologically distinctive (1, 2). These contingent properties serve as the operational criteria that systematists use as evidence to delimit species, and they can arise at different times and in different orders during the process of lineage formation (3). This view of speciation is consistent with the general metapopulation lineage concept (2), which defines species as segments of population-level evolutionary lineages. Applying this lineage-based framework to delimit species shifts focus away from adopting a single operational criterion, which often yields conflicting views of species numbers, and increases the relevance for studying lineage separation by using multiple lines of evidence (3). Species delimitation is notoriously difficult when operational criteria support discordant species boundaries, but this is to be expected in recent or adaptive radiations (4). Evaluating multiple operational criteria not only increases our ability to detect recently separated lineages (5), but it can also provide stronger evidence for lineage separation when in agreement (3). In this study, we quantify multiple operational species criteria, including divergence in genetic, ecological, and morphological characters, as well as presence or absence of gene flow, to investigate lineage diversification in the coast horned lizard (Phrynosoma coronatum) species complex.
The P. coronatum species complex is distributed across a diversity of ecological zones spanning >2,200 km from the Cape Region of Baja California to Northern California (6). Most of the proposed species boundaries within the P. coronatum complex occur at junctions separating different ecological regions in Baja California (7), which implies that ecological diversification has played a critical role during lineage formation in this system. Ecological divergence represents an important stage during the process of lineage separation that has become increasingly feasible to quantify by using geographical information systems (GIS), and it is furthermore now possible to integrate these data within a phylogeographic framework (8–10). Predicting the geographic distributions of lineages can help to identify barriers to dispersal that are important because they can restrict gene flow and facilitate population divergence on an evolutionary timescale (11). Alternatively, an area of predicted overlap between parapatric populations may signify a contact zone where gene flow is ongoing (12).
The presence of prominent cranial horns is one of the most conspicuous anatomical features of Phrynosoma lizards (6), and these horns are presumed to function as a defense against predators (13). The P. coronatum complex exhibits a considerable degree of geographic variation in occipital and temporal horn shape (14), and prior species boundary hypotheses have relied solely on this and other morphological sources of variation to delimit species. More than 20 attempts to partition this geographic variation into a discrete taxonomy underlie a turbulent taxonomic history, with 1–6 species recognized at various times (15, 16). Despite this long history of taxonomic inquiry, cranial horn shape variation remains to be quantified and compared among populations by using rigorous statistical methods. The most recent morphological study recognized 4 species within the P. coronatum complex (17), each of which was presumed to be genetically isolated and to qualify as distinct evolutionary (18) or phylogenetic species (19). Linear measurements of cranial horns were included in this study; however, standard linear measurements are limited to detecting changes in relative length and do not capture the extent to which complex shapes, such as curved cranial horns, vary among populations. Geometric morphometric techniques provide a viable alternative for analyzing complex shapes in multivariate space by retaining information about the relative spatial arrangements of the data throughout the analysis, which allows for the visualization of shape differences among groups (20). In this study, we conduct a geometric morphometric analysis of cranial horn shapes to determine whether the morphological evolution of these characters accompanies the formation of lineages in the P. coronatum species complex.
Results
Mitochondrial DNA Genealogy.
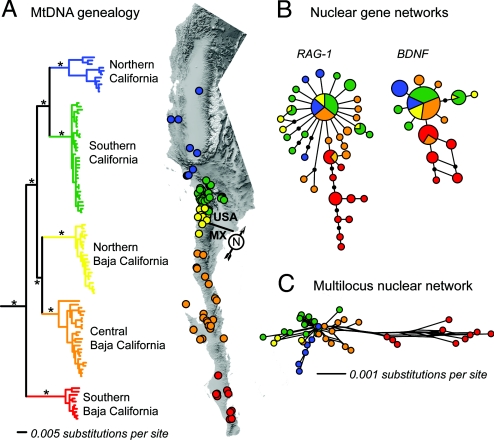
A partitioned Bayesian phylogenetic analysis of mtDNA data provides strong support (posterior probabilities ≥0.99) for 5 major clades corresponding to 5 geographically parapatric groupings of populations arranged latitudinally along the Baja California Peninsula and in California [Fig. 1A; supporting information (SI) Fig. S1]. Because our primary focus is on the population-level entities being diagnosed by these haplotype clades, we refer to them as phylogeographic groups. The distributions of the 5 phylogeographic groups of the P. coronatum complex, from south to north, are as follows:
Southern Baja California: distributed across the Isthmus of La Paz from the Cape Region of Baja California and across the Magdalena Plain to the southern edge of the Vizcaino Desert.
Central Baja California: distributed across the Vizcaino Desert and the Central Gulf Coast and into the southern portion of the California Floristic Province to Colonia Guerrero.
Northern Baja California: distributed from Ensenada, Mexico, to San Diego County, California, where it overlaps marginally with the Southern California phylogeographic group.
Southern California: overlaps marginally with the Northern Baja California phylogeographic group in San Diego County and extends north to the Los Angeles Basin and San Gabriel Mountains and east to the edge of the Mojave Desert.
Northern California: distributed from the Los Angeles Basin to the northern portion of the California Central Valley and Coast Ranges.
The average sequence divergence (uncorrected) among the 5 major clades within the P. coronatum complex is 0.0464. The highest level of average divergence is between Southern Baja California and Northern California (0.0568), whereas the lowest level is between Central and Northern Baja California (0.03753). Levels of sequence divergence within the 5 phylogeographic groups range from a low of 0.00657 in Southern California to a high of 0.01689 in Northern California (average = 0.01027).
We conducted a Shimodaira–Hasegawa (SH) test (21) to determine whether the mtDNA data support the currently recognized species boundaries in the P. coronatum complex by using topological constraints that enforce the monophyly of the currently recognized species. The SH test results indicate that the topological constraints imposed to reflect the current taxonomy are significantly worse (P < 0.0001) than the unconstrained maximum likelihood topology (MLbest = −9047.787297; MLconstraints = −9166.728912; D(LH) = −118.941615).
Fig. 1.
Mitochondrial DNA genealogy (A), nuclear allele networks (B), and multilocus nuclear network showing the genetic distance among specimens (C) for the P. coronatum species complex. The mtDNA genealogy is based on a partitioned Bayesian analysis of 2,781 base pairs, and asterisks denote major clades with posterior probability values ≥0.99. Allele networks for the RAG−1 and BDNF nuclear loci are based on statistical parsimony with a 95% connection significance. Specific locality data are provided in Table S5, and a detailed mtDNA genealogy and map illustrating the collecting locality of each sample is provided in Fig. S1.
Nuclear Gene Networks.
Two nuclear loci (RAG–1 and BDNF) show a substantial amount of allele sharing across the range of the P. coronatum complex, with each nuclear gene containing 1 allele that is widespread and shared among 4 of the 5 major phylogeographic groups (Fig. 1B). The nuclear alleles belonging to the Southern Baja California phylogeographic group cluster together, although several alleles from Central Baja California are shared with, or are only 1 mutational step removed from, this group (Fig. 1B). A multilocus nuclear gene network showing the genetic distance among specimens supports a clear separation between the Southern Baja California phylogeographic group and the remaining populations in the network (Fig. 1C). The resolution of the remaining portions of the multilocus network is too low to yield concordance between the multilocus network and the mtDNA genealogy (Fig. 1C).
Migration Estimation.
Migration is not evident across the 2 phylogeographic boundaries in Baja California, but signatures of bidirectional migration are recovered for those in California (Fig. S2). Likelihood ratio tests comparing nested demographic models (22) reveal that a model assuming no migration is rejected only for the phylogeographic boundary between Southern California and Northern Baja California (P = 0.040; Table 1 and Table S1), which are also the only phylogeographic groups that show evidence of mtDNA haplotype overlap (Fig. 1A; Fig. S1).
Table 1.
Phylogeographic comparisons of migration, morphological divergence, and niche differentiation in the P. coronatum complex
| Phylogeographic comparison | Migration profile (LRT significance) | Morphometrics (male) | Morphometrics (female) | Bioclimate data | Niche identity (simulated mean, CI) |
|---|---|---|---|---|---|
| Northern California vs.Southern California | Nonzero peakP ≥ 0.322 | F(1,91) = 0.0507P < 0.8223 | F(1,92) = 0.0386P < 0.8447 | F(1,303) = 1.9029P < 0.1688 | 0.546 (0.753, 0.0508) |
| Southern California vs.Northern Baja California | Nonzero peakP = 0.040 | F(1,47) = 7.3432P < 0.0094 | F(1,73) = 9.9786P < 0.0023 | F(1,159) = 76.7841P < 0.0001 | 0.567 (0.834, 0.0562) |
| Northern Baja California vs.Central Baja California | Peak at zeroP ≥ 0.308 | F(1,59) = 46.6623P < 0.0001 | F(1,72) = 34.1975P < 0.0001 | F(1,90) = 305.1521P < 0.0001 | 0.38(0.844, 0.0569) |
| Central Baja California vs.Southern Baja California | Peak at zeroP ≥ 0.200 | F(1,84) = 22.1170P < 0.001 | F(1,82) = 73.9413P < 0.0001 | F(1,82) = 12.3785P < 0.0007 | 0.382(0.832, 0.0561) |
The posterior probability distributions for migration rates have nonzero peaks for the 2 northern phylogeographic breaks (indicating presence of gene flow), but a nested demographic model assuming no migration cannot be rejected for the Southern vs. Northern California comparison using a likelihood ratio test (LRT). MANOVA results for morphological and environmental differences are significant (P < 0.05; shown in bold) for all but the Southern California vs. Northern California comparisons. Niche identity tests comparing the observed similarity values (Warren's I) to simulated distributions (100 pseudoreplicates) reject the hypothesis that any parapatric phylogeographic groups are distributed in identical climate space.
Niche Divergence.
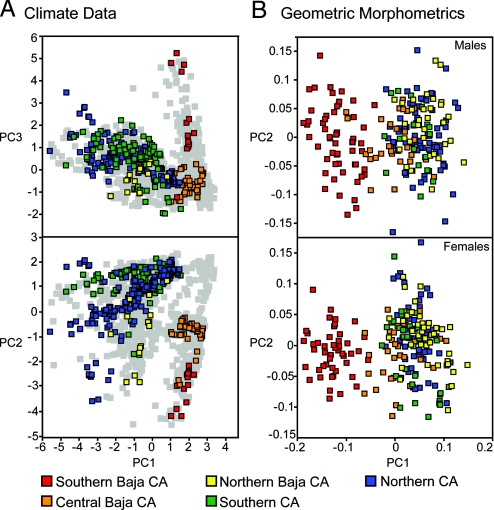
Principal components analysis (PCA) of 11 bioclimatic variables (Table S2) reveals a high degree of niche overlap between the phylogeographic groups in California and in Northern Baja California (Fig. 2A). Niche separation between Southern Baja California and Central Baja California with respect to the remaining phylogeographic groups is evident, whereas the separation between Southern Baja California and Central Baja California is strong, but incomplete (Fig. 2A). The PCA analysis revealed 4 components collectively explaining >87% of the variation, with the first vector responsible for >40% of the variation (Fig. 2A; Table S3).
Fig. 2.
Multivariate plots of the climate variables (A) and geometric morphometrics (B) for the P. coronatum complex. The climate data plots include 1,000 background points (shown in gray), drawn at random from the distribution of the P. coronatum complex (A). Variable loadings for the first, second, and third components for the climate data, respectively, are: mean temperature of coldest quarter, mean temperature of warmest quarter; temperature annual range, mean diurnal range; precipitation of driest month, precipitation of warmest quarter.
The climatic niche models predicted for the 5 phylogeographic groups exhibit varying degrees of overlap, with minimal overlap occurring at the extreme limits of the Southern Baja California and Central Baja California phylogeographic groups (Fig. S3). The 3 phylogeographic groups in California exhibit a substantial degree of overlap and overprediction beyond the known range of the species complex (Fig. S3). The relative influences of certain environmental parameters are often group specific. For example, the variable “mean temperature of coldest quarter” (bio11) makes a large contribution only to the model for Southern Baja California (88%), whereas the models for the groups that overlap spatially and show evidence of nuclear gene flow in Southern California (i.e., Northern Baja California and Southern California) share reliance on the “precipitation of driest month” variable (bio14; Table S4).
Multivariate analysis of variance (MANOVA) tests on adjacent phylogeographic groups suggest that among-group environmental differences are significant (P < 0.05) for all comparisons, with the exception of the differences between Southern California and Northern California (P < 0.168; Table 1). Niche identity tests comparing observed similarity values [Warren's I; (23)] between adjacent phylogeographic groups suggest that niche similarity is higher in California and lower between groups in Baja California (Table 1). However, comparing the observed similarity values to simulated null distributions (100 pseudoreplicates) rejects the hypothesis that any adjacent phylogeographic groups are distributed in identical climate space (Table 1).
Geometric Morphometrics of Cranial Horn Morphology.
The PCA plots generated by using geometric morphometric landmark data on cranial horns (Fig. 3A) show a strong, but incomplete, separation between Southern Baja California and Central Baja California, whereas the remaining phylogeographic groups overlap substantially (Fig. 2B). Variation in horn morphology is captured largely on the first vector (39%), whereas separation among groups on PC2 is limited, accounting for only 16% of the variation (Fig. 2B). Similar to the pattern observed with the environmental data, MANOVA tests comparing adjacent phylogeographic groups suggest that the differences in cranial horn shapes are significant (P < 0.05) for all comparisons, with the exception of the differences between Southern California and Northern California (P < 0.8; Table 1).
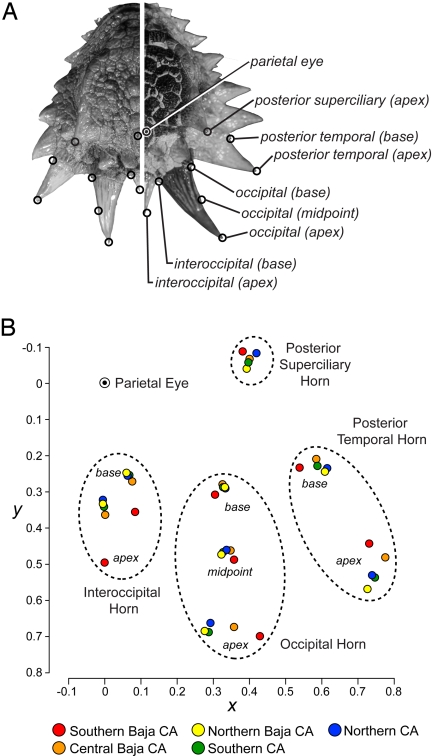
Fig. 3.
Geographic variation in cranial horn shapes. (A) Cranial horn shape divergence and geometric morphometric landmark positions between P. coronatum complex specimens from Contra Costa County, California (Left; MVZ 33623) and Southern Baja California (Right; MVZ 100473). (B) The Southern Baja California phylogeographic group differs notably from all others in cranial horn shape. Mean landmark positions (average Bookstein coordinates; CS = 1) for each phylogeographic group are shown in relation to the parietal eye (x = 0, y = 0). Males and females produced similar results, and only results for females are shown here.
There are clear differences in cranial horn shape between the phylogeographic groups (Fig. 3). The curvature of the occipital horn, the largest and most conspicuous cranial horn in the armament of the P. coronatum complex, curves outward in the Southern Baja California phylogeographic group and is either parallel or curves inward toward the midline of the body in the more northern groups (Fig. 3B). Southern Baja California is distinctive in other horn characteristics as well, including a noticeably longer interoccipital horn and a shorter, laterally pointing posterior temporal horn (Fig. 3B). The 3 phylogeographic groups in California overlap substantially in cranial horn shape, whereas Central Baja California is often intermediate between these and Southern Baja California (Fig. 3B).
Discussion
Species Delimitation.
Analyses of mtDNA, 2 nuclear loci, climatic niche models, and geometric morphometrics of cranial horn shapes in the P. coronatum species complex reveal that, at the deepest phylogenetic level, the geographical diagnoses of species boundaries are largely equivalent among the molecular genetic, ecological, and morphological criteria. The mtDNA genealogy (Fig. 1A) and multilocus nuclear network (Fig. 1C) support a congruent phylogeographic break in Southern Baja California, and there appears to be no nuclear gene flow across this boundary (Table 1). This phylogeographic break is accompanied by divergence in ecological niche (as measured by bioclimatic variables) and in cranial horn shapes (Fig. 2 A and B; Table 1). The morphometric study of Montanucci (17) supported the recognition of a distinctive Southern Baja California species, and the genetic, ecological, and geometric morphometric data presented here support that conclusion. These data add further evidence for recognizing multiple independent evolutionary lineages (i.e., separate species) within the P. coronatum species complex. We recognize the Southern Baja California phylogeographic groups as a distinct species in the P. coronatum complex and follow Montanucci (17) in recognizing this lineage as P. coronatum (Blainville), with a type locality of Cape San Lucas, Baja California (24). The timing of this initial divergence event in the P. coronatum complex may be as recent as 2.3 myr (25). This species exhibits a number of properties suggesting that it is a separately evolving evolutionary lineage, including ecological divergence (26), allelic monophyly and exclusivity (19, 27, 28), reproductive isolation [e.g., lack of gene flow; (29)], and phenotypic differences (30).
Unlike the sharp character concordance observed across the phylogeographic break in Southern Baja California, patterns of character divergence are more obscured among the 4 remaining phylogeographic groups. The nuclear loci do not support the exclusivity of these phylogeographic groups (Fig. 1C), although this may indicate that the slowly evolving nuclear exons used in this study are inadequate for resolving relationships at this shallow temporal level. Despite the lack of genealogical resolution provided by the nuclear loci, these data provide pertinent information regarding migration, which is rejected for all but the Northern Baja and Southern California phylogeographic groups (Table 1). Although the signature of nuclear gene flow detected at this phylogeographic break could reflect current or historical processes, the spatial overlap of mtDNA haplotypes observed across this break argues for a lack of reproductive isolation among populations, and these independent lines of evidence suggest that the Northern Baja California and Southern California phylogeographic groups are not distinct evolutionary lineages. Evidence for recognizing the Northern California and Southern California phylogeographic groups as distinct evolutionary lineages is also weak, because these populations are not ecologically or morphologically distinct from one another (Table 1). However, the Central Baja California phylogeographic group satisfies all of the operational species criteria evaluated in this study and merits recognition as a distinct evolutionary lineage. This interpretation also follows from the species delimitation framework proposed by Bond and Stockman (31), which relies on the cohesion species criterion (32). Under this framework, parapatric populations are considered conspecific if they are ecologically interchangeable, which does not appear to be the case for Central Baja California with respect to Southern California to the north, nor with P. coronatum to the south (Table 1). According to the principles of priority, the name Phrynosoma cerroense (Stejneger) is applied to the Central Baja California phylogeographic group, with a type locality of Cedros Island, on the Pacific Coast of Baja California. Phrynosoma blainvillii (Gray) is applied to the remaining populations belonging to the Northern Baja California, Southern California, and Northern California phylogeographic groups, with a type locality of San Diego, San Diego County, California (14).
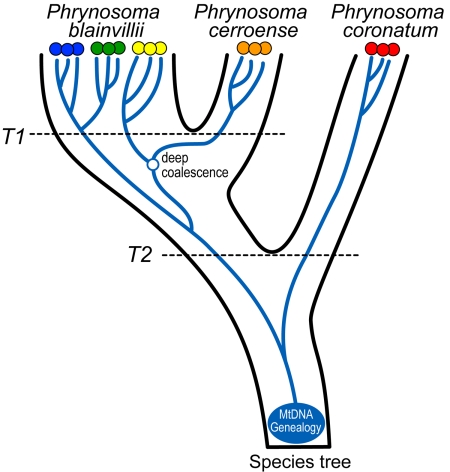
A cursory inspection of our 3-species hypothesis for the P. coronatum complex in relation to the mtDNA genealogy (Fig. 1A) reveals that haplotypes of P. blainvillii form a paraphyletic group, because the Northern Baja California haplotype clade of P. blainvillii is more closely related to P. cerroense than to the remaining clades contained within P. blainvillii. Although the mtDNA genealogy does provide strong support (≥0.99) for the inferred relationships, it is becoming increasingly appreciated that the genealogy inferred from any single locus can provide an inaccurate portrait of the species tree (33, 34). Reinterpreting the mtDNA genealogy within the context of the 3-species phylogeny illustrates that the case of P. blainvillii mtDNA paraphyly is rectified by invoking 1 instance of deep coalescence in the mtDNA locus (Fig. 4). Testing this “deep coalescence” hypothesis for the mtDNA genealogy requires the collection of additional multilocus nuclear data that are variable at this temporal level in conjunction with species tree inference methods that account for the process of lineage sorting (35, 36).
Fig. 4.
Proposed species tree for the P. coronatum species complex and the contained mtDNA genealogy composed of 5 phylogeographic groups. The failure of all mtDNA haplotype lineages within P. blainvillii to coalesce between T1 and the present results in an opportunity for deep coalescence between P. blainvillii and P. cerroense.
Conservation Implications.
The P. coronatum species complex is undergoing population declines and local extirpations that are most pronounced in agricultural and urban areas. A primary factor contributing to these declines is the destruction of appropriate native chaparral habitats with sand substrates (37). The introduction of the invasive Argentine ant (Linepithema humile) also contributes to population declines by displacing the natural prey source, which consists of large harvester ants [e.g., Pogonomyrmex; (38)]. Experimental studies confirm that a diet consisting of only the introduced species causes starvation of captive lizards (39). Historically, populations in Southern California were heavily commercialized (40), and concerns regarding the impacts of continued international trade led to CITES Appendix II listing in 1975 (www.cites.org). In California, the 3 phylogeographic groups discovered within P. blainvillii represent important components of diversity that should be treated as distinct management units (distinct population segments) in future conservation and management decisions. In Baja California, P. cerroense and P. coronatum require special conservation considerations. Although many populations of P. cerroense are protected by extensive habitat reserves, heavy agriculture across the Magdalena Plain has the potential to eliminate P. coronatum from a significant portion of their range and cause a repeat of the massive population declines documented in California. This habitat loss is unfortunate, given that Baja California harbors a unique component of Phrynosoma diversity.
Materials and Methods
Genetic Data.
We generated DNA sequence data for 98 specimens collected from throughout their range in California (n = 63) and Baja California (n = 35) (Table S5). We sequenced the entire mitochondrial ND1 (969 bp) and ND2 (1,033 bp) protein-coding genes and an 800-bp fragment of the 12S rRNA gene. We also sequenced 2 protein-coding nuclear genes, including 1,100 bp of recombination activating gene-1 (RAG−1) and 700 bp of brain-derived neurotrophic factor (BDNF). We collected complete data for most specimens (Table S5). New primer sequences developed for this study include an internal ND2 primer (phrynoND2int: 5′-CAACAGCCCTCCTAACAATAGCCATA-3′), an internal ND1 primer (phrynoND1int: 5′-CTAACAGACCTAAACCTAGGC-3′), and a modified version of the external ND1 primer tMet (phrynotMet: 5′-GGCTCATTAGTAGAGGAGGGGTTTAAACCAAC-3′). The remaining primer sequences, lab protocols, and DNA alignment methods for the 12S rRNA data are provided in Leaché and McGuire (41). We included 7 species to represent outgroup taxa, including Phrynosoma asio, Phrynosoma cornutum, Phrynosoma hernandesi, Phrynosoma mcallii, Phrynosoma modestum, Phrynosoma orbiculare, and Phrynosoma solare.
Phylogenetic Analysis.
We conducted phylogenetic analyses in a Bayesian framework by using mrBayes v3.1.2 (42). We separated the data into 7 partitions corresponding to the 12S rRNA gene (partition 1), first, second, and third codon positions for ND1 (partitions 2, 3, and 4, respectively), and first, second, and third codon positions for ND2 (partitions 5, 6, and 7, respectively). We used the Akaike information criterion (AIC) in MrModeltest v2.2 (43) to determine the best-fit nucleotide substitution model for each data partition (Table S6). Analyses were run with 4 chains (using default heating values) for 40 million generations, sampling every 1,000 generations. We implemented convergence diagnostics using the program AWTY (44).
We tested the morphometric-based taxonomy proposed by Montanucci (17) using the SH test (21). The maximum-likelihood tree constraining the monophyly of the 4 species recognized by Montanucci (17) was compared with an ML tree containing no topological constraints. We implemented the SH test using PAUP v.4.0b10 with 10,000 RELL bootstrap replicates and the GTR+I+Γ model of nucleotide substitution. We also implemented a partitioned SH test in RAxML v 7.0.4 (45) using the same data partitioning strategy used in our Bayesian phylogenetic analysis, with the exception that the GTR+Γ model was applied to each partition.
Allele and Multilocus Networks.
We constructed allele networks for the nuclear loci (BDNF and RAG−1) using statistical parsimony with a 95% connection significance in the program TCS v1.21 (46). To infer haplotypes, we determined the phase of nuclear genotypes computationally using the program PHASE v2.1 (47, 48). We constructed a multilocus genetic network representing the relationships among specimens by converting the distance matrices for alleles from the separate nuclear loci into an organism matrix using the program POFAD v1.03 (49). The reconstructed organism network was visualized by using the NeighborNet algorithm (50) in SplitsTree v4.6 (51). We present the reconstructed organism network based on uncorrected patristic distances, but we also explored networks that incorporated corrected distance matrices using models selected by using the AIC.
Migration Estimation.
We used the RAG−1 and BDNF nuclear loci to determine whether any phylogeographic groups are exchanging migrants by testing nested models of population divergence in the program IMa (22). Assumptions of the IM model are that the loci under study are neutral and have not undergone intragenic recombination. We tested for intragenic recombination using the difference of sums of squares (DSS) test in TOPALi v2.5 (52), and deviations from neutrality were tested with the HKA test (53) by using the program DnaSP v4.10.4 (54). HKA tests for neutrality were not significant, and there is no evidence for recombination in either nuclear locus according to the DSS test. We ran 10 replicate IMa analyses (each using different starting seeds and 12 concurrent chains) for 10 million steps after an initial burn-in phase of 100,000 generations. Nested models of population divergence were compared by using likelihood ratio tests (22).
Environmental Data.
Occurrence data were supplied by the samples from the genetic analyses augmented with museum records acquired from the database portal HerpNET (www.herpnet.org; accessed on November 29, 2007). Localities with a maximum georeferencing uncertainty of >10 km were excluded, leaving 285 unique sample localities used in the analysis. Minimum convex polygons circumscribing the geographic distribution of each mtDNA clade were used to assign HerpNET samples to phylogeographic groups and to define the area of analysis for climatic data sampling. All GIS tasks used ArcGIS 9.2 (ESRI) with bioclimatic variables from the Worldclim v1.4 dataset (55). Eight of the 19 bioclimatic variables were highly correlated [Pearson correlation coefficient (r) > 0.75; methods follow Rissler et al. (56)], and we used 11 bioclimatic variables for all analyses (Table S2).
Niche Modeling.
We used Maxent v3.1 (57, 58) to generate predictive distribution models based on known occurrence samples and their corresponding environmental variables. Maxent is an appropriate choice for our data compared with other distributional modeling methods, because it provides robust performance with small sample sizes of presence-only data (59). We used the genetic samples as our presence-only dataset (the training set) with the bioclimatic variables to create distribution models by using default parameters in Maxent. The modeling calculations used the environmental data from the minimum convex polygon study area and then projected onto a broader geographic area encompassing western North America. We conducted multivariate statistical analyses of the environmental variables using R v2.6.0 and JMP v7 (SAS). We performed PCA to test whether the phylogeographic groups are discernible based on the bioclimatic variables without a priori designation of groups, and we tested for significant differences between groups using MANOVA.
Niche Identity Tests.
We used niche identity tests (23) to test the null hypothesis that parapatric phylogeographic groups are distributed in identical environmental space. Given that environmental niche models make no biological assumptions of microhabitat use or species interactions, Warren et al. (23) proposed a mathematical approach that scales similarity from 0 (no overlap) to 1 (complete overlap), which we call Warren's I. We generated 100 simulated distribution models based on random replacement between samples of phylogeographic groups to test the null hypothesis that parapatric groups are distributed in identical environmental space.
Geometric Morphometrics of Cranial Horn Morphology.
We examined cranial horn morphology in 493 specimens (245 females; 248 males; Table S7). Digital images of each specimen were obtained by using a flat-bed scanner, and the x, y coordinates of 16 landmarks (Fig. 3A) from the cranial horns and head plus 2 ruler landmarks were recorded from each image by using TPSDIG (60). We used the IMP software package (www3.canisius.edu/sheets/morphsoft.html) for image visualization and statistical analysis. We calculated the average value for symmetric landmarks (e.g., landmark positions on opposite sides of the head) using the program BigFix, resulting in 9 landmarks for each specimen (2 baseline and 7 averaged landmarks). We then generated Procrustes superimpositions for each specimen using CoordGen (20). We used PCA to determine whether any morphological groupings were detectable without the a priori designation of groups, and we tested for significant differences in cranial horn shapes between parapatric phylogeographic groups using a 1-way MANOVA in JMP 7.0 (SAS).
Supplementary Material
Acknowledgments.
For tissue loans, we thank B. Hollingsworth and D. Wood (San Diego Society of Natural History, San Diego), R. Murphy and R. MacCulloch (Royal Ontario Museum, Toronto, ON, Canada), J. Vindum (California Academy of Sciences, San Francisco), G. Busteed and P. Johnson (U.S. National Park Service), W. Hodges, T. Reeder, E. Ervin, and R. Young. We are indebted to the collection managers at the California Academy of Sciences, Los Angeles County Museum, Museum of Vertebrate Zoology (MVZ) (Berkeley, CA), and the Smithsonian Institute (Washington, DC) for participating in the HerpNET data portal and for providing access to specimens. We thank H. Constable (HerpNET) for providing georeferenced data, N. Winters for scanning lizards at the MVZ, and D. L. Warren for modifying ENMtools to accommodate our data. We thank the Conservation Program of ESRI for donating GIS software. The use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government. This work was funded in part by National Science Foundation Grant DEB 0330750 (to J.A.M.) and grants from The Nature Conservancy, California Department of Fish and Game, California Department of Parks and Recreation, Metropolitan Water District, U.S. Fish and Wildlife Service, and U.S. Department of Defense (to R.N.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Nucleotide sequences have been deposited in the GenBank database (accession nos. GQ279396–GQ929730).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906380106/DCSupplemental.
References
- 1.Harrison . In: Endless Forms: Species and Speciation. Howard DJ, Berlocher SH, editors. Oxford: Oxford Univ Press; 1998. pp. 19–31. [Google Scholar]
- 2.de Queiroz K. In: Endless Forms: Species and Speciation. Howard DJ, Berlocher SH, editors. Oxford: Oxford Univ Press; 1998. pp. 57–75. [Google Scholar]
- 3.de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 4.Wake DB. Problems with species: Patterns and processes of species formation in salamanders. Ann Mo Bot Gard. 2006;93:8–23. [Google Scholar]
- 5.Sites JW, Marshall JC. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol Evol. 2003;18:462–470. [Google Scholar]
- 6.Sherbrooke WC. Introduction to Horned Lizards of North America. Berkeley: Univ of California Press; 2003. [Google Scholar]
- 7.Grismer LL. Amphibians and Reptiles of Baja California Including Its Pacific Islands and the Islands in the Sea of Cortez. Berkeley: Univ of California Press; 2002. [Google Scholar]
- 8.Kidd DM, Ritchie MG. Phylogeographic information systems: Putting the geography into phylogeography. J Biogeogr. 2006;33:1851–1865. [Google Scholar]
- 9.Richards CL, Carstens BC, Knowles LL. Distribution modeling and statistical phylogeography: An integrative framework for generating and testing alternative biogeographical hypotheses. J Biogeogr. 2007;34:1833–1845. [Google Scholar]
- 10.Rissler LJ, Apodaca JJ. Adding more ecology into species delimitation: Ecological niche models and phylogeography help define cryptic species in the black salamander (Aneides flavipunctatus) Syst Biol. 2007;56:924–942. doi: 10.1080/10635150701703063. [DOI] [PubMed] [Google Scholar]
- 11.Wiens JJ. Speciation and ecology revisited: Phylogenetic niche conservatism and the origin of species. Evolution (Lawrence, Kans) 2004;58:193–197. doi: 10.1111/j.0014-3820.2004.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 12.Cicero C. Barriers to sympatry between avian sibling species (Paridae: Baeolophus) in local secondary contact. Evolution (Lawrence, Kans) 2004;58:1573–1587. doi: 10.1111/j.0014-3820.2004.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 13.Young KV, Brodie ED, Jr, Brodei ED., III How the horned lizard got its horns. Science. 2004;304:65. doi: 10.1126/science.1094790. [DOI] [PubMed] [Google Scholar]
- 14.Klauber LM. The horned toads of the coronatum group. Copeia. 1936;1936:103–110. [Google Scholar]
- 15.Jennings MR. Phrynosoma coronatum. Cat Am Amphib Reptiles. 1988;428:1–5. [Google Scholar]
- 16.Brattstrom BH. Status of the subspecies of the coast horned lizard, Phrynosoma coronatum. J Herpetol. 1997;31:434–436. [Google Scholar]
- 17.Montanucci RR. Geographic variation in Phrynosoma coronatum (lacertilia, Phrynosomatidae): Further evidence for a peninsular archipelago. Herpetologica. 2004;60:117–139. [Google Scholar]
- 18.Wiley EO. The evolutionary species concept reconsidered. Syst Zool. 1978;27:17–26. [Google Scholar]
- 19.Cracraft J. Species concepts and speciation analysis. Curr Ornithol. 1983;1:159–187. [Google Scholar]
- 20.Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric Morphometrics for Biologists: A Primer. London: Elsevier Academic; 2004. [Google Scholar]
- 21.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 22.Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci USA. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution (Lawrence, Kans) 2008;62:2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- 24.Smith HM, Taylor EH. Type-localities of Mexican reptiles and amphibians. Univ Kansas Sci Bull. 1950;33:313–380. [Google Scholar]
- 25.Leaché AD, Crews SC, Hickerson MJ. Two waves of diversification in mammals and reptiles of Baja California revealed by hierarchical Bayesian analysis. Biol Lett. 2007;3:646–650. doi: 10.1098/rsbl.2007.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Valen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239. [Google Scholar]
- 27.Mishler BD. The morphological, developmental, and phylogenetic basis of species concepts in bryophytes. Bryologist. 1985;88:207–214. [Google Scholar]
- 28.Baum DA, Shaw KL. In: Experimental and Molecular Approaches to Plant Biosystematics. Hoch PH, Stevenson AG, editors. St. Louis: Missouri Botanical Garden; 1995. pp. 289–303. [Google Scholar]
- 29.Mayr E. Systematics and the Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 30.Sokal RR, Crovello TJ. The biological species concept: A critical evaluation. Am Nat. 1970;104:127–153. [Google Scholar]
- 31.Bond JE, Stockman AK. An integrative method for delimiting cohesion species: Finding the population-species interface in a group of Californian trapdoor spiders with extreme genetic divergence and geographic structuring. Syst Biol. 2008;57:628–646. doi: 10.1080/10635150802302443. [DOI] [PubMed] [Google Scholar]
- 32.Templeton AR. In: Endless Forms: Species and Speciation. Howard DJ, Berlocher SH, editors. Oxford: Oxford Univ Press; 1998. pp. 32–41. [Google Scholar]
- 33.Pamilo P, Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 34.Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:762–768. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddison WP. Gene trees in species trees. Syst Biol. 1997;46:523–536. [Google Scholar]
- 36.Edwards SV, Liu L, Pearl DK. High-resolution species trees without concatenation. Proc Natl Acad Sci USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher RN, Suarez AV, Case TJ. Spatial patterns in the abundance of the coastal horned lizard. Conserv Biol. 2002;16:205–215. doi: 10.1046/j.1523-1739.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- 38.Suarez AV, Bolger DT, Case TJ. The effects of fragmentation and invasion on the native ant community in coastal Southern California. Ecology. 1998;16:2041–2056. [Google Scholar]
- 39.Suarez AV, Case TJ. Bottom-up effects on persistence of a specialist predator: Ant invasions and horned lizards. Ecol Appl. 2002;12:291–298. [Google Scholar]
- 40.Jennings MR. Impact of the curio trade for San Diego horned lizards (Phrynosoma coronatum blainvillii) in the Los Angeles Basin, California: 1885–1930. J Herpetol. 1987;21:356–358. [Google Scholar]
- 41.Leaché AD, McGuire JA. Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading mitochondrial gene tree. Mol Phylogenet Evol. 2006;39:628–644. doi: 10.1016/j.ympev.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 43.Nylander JAA. MrModeltest v2. Uppsala Univ, Uppsala: Evolutionary Biology Centre; 2004. [Google Scholar]
- 44.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 45.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 46.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 47.Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joly S, Bruneau A. Incorporating allelic variation for reconstructing the evolutionary history of organisms from multiple genes: An example from Rosa in North America. Syst Biol. 2006;55:623–636. doi: 10.1080/10635150600863109. [DOI] [PubMed] [Google Scholar]
- 50.Bryant D, Moulton V. Neighbor-Net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 51.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 52.McGuire G, Wright F. TOPAL 2.0: Improved detection of mosaic sequences within multiple alignments. Bioinformatics. 2000;16:130–134. doi: 10.1093/bioinformatics/16.2.130. [DOI] [PubMed] [Google Scholar]
- 53.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 55.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 56.Rissler LJ, Hijmans RJ, Graham CH, Moritz C, Wake DB. Phylogeographic lineages and species comparisons in conservation analyses: A case study of California herpetofauna. Am Nat. 2006;167:655–666. doi: 10.1086/503332. [DOI] [PubMed] [Google Scholar]
- 57.Phillips SJ, Dudik M, Schapire RE. A maximum entropy approach to species distribution modeling. Proc Twenty-First Int Conf Machine Learn. 2004;69:83–99. [Google Scholar]
- 58.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190:231–259. [Google Scholar]
- 59.Elith J, et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- 60.Rohlf FJ. Comparative methods for the analysis of continuous variables: Geometric interpretations. Evolution (Lawrence, Kans) 50:2143–2160. doi: 10.1111/j.0014-3820.2001.tb00731.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.