Abstract
The nuclear pore complex (NPC) resides in circular openings within the nuclear envelope and serves as the sole conduit to facilitate nucleo-cytoplasmic transport in eukaryotes. The asymmetric distribution of the small G protein Ran across the nuclear envelope regulates directionality of protein transport. Ran interacts with the NPC of metazoa via two asymmetrically localized components, Nup153 at the nuclear face and Nup358 at the cytoplasmic face. Both nucleoporins contain a stretch of distinct, Ran-binding zinc finger domains. Here we present six crystal structures of Nup153-zinc fingers in complex with Ran and a 1.48 Å crystal structure of RanGDP. Crystal engineering allowed us to obtain well-diffracting crystals so that all ZnF-Ran complex structures are refined to high resolution. Each of the four zinc finger modules of Nup153 binds one Ran molecule in apparently non-allosteric fashion. The affinity is measurably higher for RanGDP than for RanGTP and varies modestly between the individual zinc fingers. By microcalorimetric and mutational analysis, we determine that one specific hydrogen bond accounts for most of the differences in the binding affinity of individual zinc fingers. Genomic analysis reveals that only in animals do NPCs contain Ran-binding zinc fingers. We speculate that these organisms evolved a mechanism to maintain a high local concentration of Ran at the vicinity of the NPC, using this zinc finger domain as a sink.
Keywords: nuclear pore complex, Nup153, Ran, zinc fingers, crystallography
Introduction
Nucleocytoplasmic transport is controlled and facilitated by macromolecular protein assemblies termed nuclear pore complexes (NPCs). NPCs reside in circular openings of the nuclear envelope where inner and outer nuclear membranes are fused. The NPC is a large macromolecular assembly with a calculated mass of ∼50 MDa 1. Based on electron-microscopy (EM) studies from X. laevis oocytes and S. cerevisiae, the general shape and structure of the pore is conserved across eukaryotes 2; 3. These EM studies define the pore as a ring embedded in the nuclear envelope, exhibiting 8-fold rotational symmetry around a central axis and imperfect 2-fold symmetry between the cytoplasmic and nucleoplasmic faces. Despite its size, the NPC is only made up of ∼30 proteins, or nucleoporins (Nups), arranged in a few biochemically defined subcomplexes that assemble the entire structure in a modular fashion 4. As the single constitutive barrier to regulate permeability, the NPC transports a wide range of substrates across the double membrane of the nuclear envelope 5; 6; 7. Active transport through the NPC is mediated by nuclear transport receptors (NTRs), also called karyopherins or importins/exportins 8; 9.
The small G protein Ran is the master regulator of NTR-mediated, nuclear protein transport 10. Ran selectively promotes binding or release of import or export cargos to NTRs by means of a chemical gradient. Ran binds mostly GDP in the cytoplasm, and mostly GTP in the nucleus. The GTPase activating protein (GAP), localized to the cytoplasmic face of the NPC, and the GTP exchange factor (GEF), RCC1, bound to chromatin inside the nucleus, together promote this asymmetry by modulating nucleotide hydrolysis and exchange. The established gradient provides directionality to protein transport. In the nucleus, RanGTP releases import-cargo from NTRs by competitive binding. RanGTP is recycled back to the cytoplasm via a trimeric complex formed with NTRs and export-cargo. At the cytoplasmic face of the NPC, RanGTP interacts with RanGAP to hydrolyze GTP and disrupt the trimeric NTR-mediated export complex. NTF2 (nuclear transport factor 2) recycles RanGDP back into the nucleus 11.
Nup153 and Nup358 (RanBP2) are large, metazoa-specific nucleoporins with multiple roles 12; 13; 14; 15; 16. Both interact with Ran through a zinc finger cassette composed of several individual zinc finger (ZnF) motifs 17; 18. Nup153 is predominantly localized to the nuclear face of the NPC, although recent studies suggest the three major domains of Nup153 are localized to different regions of the NPC 19; 20; 21; 22. The N-terminal portion contains a pore-targeting region and an RNA binding domain 23; 24; 25; 26; 27. The ZnF cassette is comprised of multiple zinc fingers and defines the center of Nup153. The C-terminal region of Nup153 harbors ∼30 phenylalanine-glycine (FG-) repeats, unstructured motifs found in several Nups lining the inner channel of the NPC that are responsible for NTR interaction 28; 29; 30; 31. Nup358 has several characterized domains including a cyclophilin homology domain, a SUMO ligase domain, a structural leucine-rich region, previously characterized Ran binding domains (RanBDs), and a cassette containing multiple zinc fingers 12; 15; 32; 33; 34. The ZnFs of Nup153 and Nup358 are representative of the “RanBP2-type” ZnF family, recognized by the conserved sequence pattern W-X-C-X(2,4)-C-X(3)-N-X(6)-C-X(2)-C (Fig. 1)35. RanBP2-type zinc fingers fold into a structure composed of two β-hairpin strands that sandwich a Zn2+ ion coordinated with four cysteine residues 36. The RanBP2-type zinc finger structure is distinct from other zinc fingers, however it only defines a common scaffold, not a common function. Ran binding has been reported for Nup153 and Nup358 zinc fingers and not for the other structural homologs 37; 38. It is unclear what exact role these Ran-binding zinc fingers play at the NPC, nor is it conclusively analyzed whether they bind Ran in a nucleotide dependent or independent manner 17; 18; 37; 39.
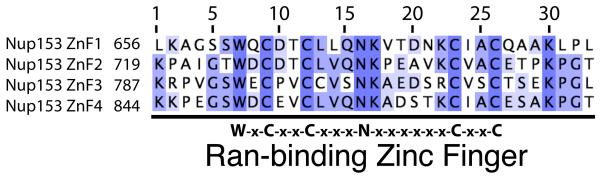
Figure 1.
Alignment of the four Ran-binding zinc fingers of Nup153 from R. norvegicus. The numbering used for individual zinc fingers is listed above. Identical residues are highlighted in dark blue, with decreasing levels of conservation highlighted in lighter colors of blue. Below is the consensus sequence for the“RanBP2” family of zinc finger proteins.
We have determined the crystal structures of all four ZnFs of Nup153 in complex with RanGDP. Our structural data suggests that all ZnF modules preferentially bind to RanGDP rather than RanGTP, supported by mutational and microcalorimetric data. While the primary sequence of the ZnF in the cassette is conserved, the number of ZnFs varies among species. Our data supports largely uncooperative binding of Ran to the individual ZnF modules within Nup153 or Nup358, explaining why the exact number of consecutive ZnFs is not conserved. Although we detect differences between ZnF binding to RanGDP vs. RanGTP, they are small and may not be of functional consequence. We propose that the ZnFs within Nup153 and Nup358 are primarily used to create a ‘Ran sink’ and thereby increase the local concentration of Ran at both the nucleoplasmic and cytoplasmic face of the NPC.
Results and Discussion
Crystallographic analysis of Nup153-ZnF•RanGDP complexes
All protein constructs used in this study were from Rattus norvegicus, except Nup358 from Homo sapiens. The central region of Nup153 (residues 658 – 885) contains four zinc fingers; ZnF1 (residues 658 – 686), ZnF2 (residues 723 – 750), ZnF3 (residues 790-817), and ZnF4 (residues 848 - 885). The individual ZnF domains were cloned and recombinantly expressed as GST fusion proteins in E. coli. In addition to individual ZnF domains, the tandem pairs of ZnF1 and ZnF2 (ZnF12, residues 658-750) as well as ZnF3 and ZnF4 (ZnF34, residues 790-885) were examined (see Table S1 for all protein constructs used in this study). Full-length Ran was cloned and expressed as a His-tag fusion protein in E. coli. RanGDP was separated from RanGTP using ion-exchange chromatography and the nucleotide-loaded state was validated with HPLC. Our RanGDP structure is solved at 1.48 Å resolution with two molecules in the asymmetric unit. Molecule A is well ordered with all residues in both switch regions defined. However, in molecule B a portion of the switch II region (residues 69 – 74) is disordered and cannot be resolved. This discrepancy arises from vdW interactions unique to molecule A between Phe77molA with Phe11molB and Leu168molB. Switch II in the absence of γ-phosphate from GTP or contact with a neighboring molecule, remains unresolved and serves as indication for it's significant disorder. Diffraction quality crystals of a Nup153-ZnF in complex with RanGDP were initially obtained for ZnF2, ZnF4, ZnF12, and ZnF34, but only the ZnF2 complex crystals diffracted satisfactorily. Well-diffracting crystals of all other complexes were obtained after introducing a structure-based, surface point mutation in Ran, F35S, to stabilize a crystal contact as described below. Despite significant effort, no crystals were obtained for any ZnF construct in complex with RanGTP (in the form of the GTPase deficient mutant, RanQ69L).
Overall structure of the ZnF•RanGDP complex
Each Nup153-ZnF and RanGDP bind with a 1:1 stoichiometry. ZnF-bound RanGDP is nearly identical to unbound RanGDP with an rmsd of 0.90 Å and 0.40 Å when compared to two RanGDP structures (Protein Data Bank (PDB) codes 1BYU 40 and 3GJ0 (this study) respectively). The bound ZnF also maintains a structure similar to the unbound ZnF (rmsd 0.77 Å compared to PDB code 2GQE 37). Four short β-strands form two orthogonal hairpins flanking the hydrophobic core containing the strictly conserved Trp7ZnF residue (for ZnF residue numbering scheme, see Fig. 1). A single Zn2+ ion is sandwiched between the two β-hairpins, and coordinated by four cysteine residues (Cys9, Cys12, Cys23, and Cys26). The side chain amide of Asn16 is highly conserved in all RanBP2-type zinc fingers, suggesting the hydrogen bond formed between Asn16 and the backbone amide group of residue 24 helps maintain the RanBP2-type ZnF fold. Nup153-ZnF binds to RanGDP at a region neighboring and partially encompassing switch I, residues 28 to 48 (residues 65 to 84 define the switch II region, Fig. 2) 41. The binding interface measures 469 Å2, with major hydrophobic interactions made with Ran structural elements β1, β3, and α4, as well as electrostatic interactions with the N-terminus and switch I region of RanGDP. Hydrophobic interactions mediated by the “LVA” motif of Nup153 ZnFs, have been highlighted in previous biochemical and structural studies 37; 39. Leu13Nup153, Val14Nup153, and Ala25Nup153 from Nup153-ZnF2 interact with RanGDP between switch I and strand β1 of RanGDP, with some interaction at the switch II region. Leu13Nup153 is situated in a hydrophobic pocket formed by the aliphatic carbon chain of Lys38Ran, Val47Ran, and Pro49Ran. Val14Nup153 interacts primarily with Trp64Ran, Lys12Ran, and with the carbon chain of Gln82Ran. Ala25Nup153 forms hydrophobic interactions with Ile81Ran, and Trp64Ran, and Leu43Ran. Critical hydrogen bonds include the backbone carbonyl group of residue 8Nup153 binding with the side chain Lys38Ran, and the backbone carbonyl of Cys26Nup153 binding with the side chain of Thr42Ran. Both Ran residues reside in the switch I region, and are known to undergo significant rearrangement in the RanGTP conformation. To summarize, the Nup153-ZnF2•RanGDP structure is principally stabilized by hydrophobic interactions with a constant region of Ran, and two hydrogen bonds with the switch I region of Ran.
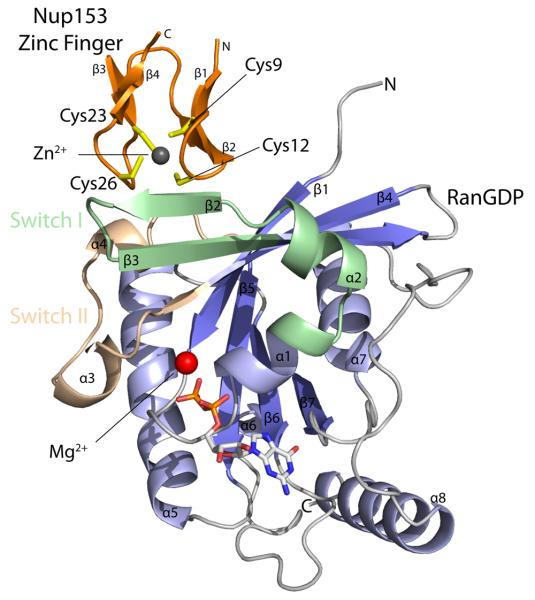
Figure 2.
Ribbon diagram representation of the Nup153-ZnF2•RanGDP complex. RanGDP β-sheets are colored darkblue with α-helices colored light blue. The Nup153-ZnF molecule is colored orange. Both the switch I and switch II regions of Ran are highlighted. Representative of the “RanBP2” family of protein-binding zinc fingers, the Nup153 ZnF contains two orthogonal β-hairpins with four cysteine residues, colored yellow, to coordinate a single Zn2+ ion colored grey. The zinc finger binds near the switch I region of RanGDP, indicated here by the secondary structural element β2. The GDP nucleotide is shown in the center of Ran with a bound Mg2+ ion.
Our 1.8 Å Nup153-ZnF2•RanGDP structure superimposes well with the 2.1 Å crystal structure from Schrader et al., solved in an unrelated space group (rmsd 0.55 Å compared to PDB code 3CH5 39). However, the alternative crystal-packing observed in this study results in a significantly different interpretation of parts of the ZnF•RanGDP interface. The contact between Phe714Nup153–ZnF2 and a hydrophobic pocket of Ran near β1 and β4, has been suggested to be a significant contact between Nup153-ZnF2 and RanGDP 39. In our Nup153-ZnF2•RanGDP structure, Phe72Ran from a symmetry-related Ran molecule occupies the previously described hydrophobic pocket. The ZnF2 construct used in our study does not include residues 703-722, N-terminal to ZnF2 as used in Schrader et al., but is representative of the ZnF2 construct used to determine the unbound Nup153-ZnF2 NMR structure 37. Although our ZnF2 structure does not include the N-terminal extension, the constructs and solved structures of ZnF12 and ZnF34 do include this region and are described in detail below. Our structural data, in accordance with our binding data, suggests the linker region between the ZnF domains has a rather small contribution in ZnF binding with RanGDP.
Engineering improved crystal contacts
The structures of RanGDP in complex with ZnF1, ZnF3, ZnF4, ZnF12 and ZnF34, respectively, were solved at high resolution only after re-engineering a crystal contact involving Ran, to improve crystal packing. In the initial orthorhombic Nup153-ZnF2•RanGDP crystals the packing interaction was very weak along one crystallographic axis, where a single contact point was observed. We reasoned that this sub-optimal packing might be the cause of weak diffraction, high temperature factors, and high mosaicity initially observed (data not shown). The weak contact is formed by a Ran•Ran interaction, involving a single hydrogen-bond and a small, strained vdW interface between Phe35Ran and Pro58Ran' (Ran' denotes the symmetry mate) (Fig. 3). Several studies have shown that site-directed surface mutagenesis can be used to improve crystal quality 42; 43; 44. In our case, we reasoned that changing Phe35Ran to serine would reduce the entropic cost of a clash between Phe35Ran and Pro58Ran', and be compatible with the hydrophilic character of the neighboring His53Ran'. In similar fashion Phe35Ran was mutated to aspartic acid in hopes of forming an additional H-bond with His53Ran'. Although the RanF35D mutation proved unable to crystallize, the RanF35S point mutation crystallized and improved the diffraction limit of ZnF4, ZnF12, and ZnF34 crystals by 1 Å and enabled crystallization of ZnF1 and ZnF3.
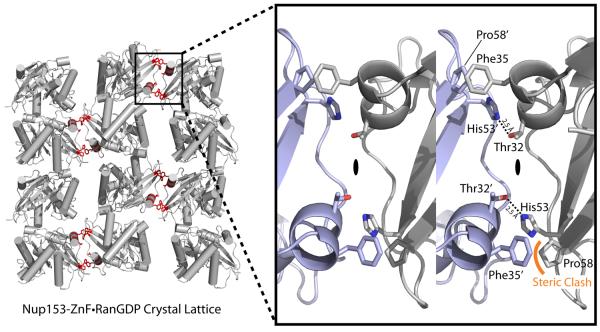
Figure 3.
Crystallographic lattice formed in the crystallization of a Nup153-ZnF•RanGDP complex. Residues highlighted in red indicate contacts made between neighboring Ran molecules down a single plane of the P21 lattice. This plane represents the smallest crystallographic interface and was chosen for crystal engineering to stabilize packing, in hopes of improving diffraction. The enlarged 3D image on the right depicts residues binding at the interface. Thr32' and His53 make a single hydrogen bond, mirrored across a 2-fold symmetrical face as indicated. A clash between Phe35' and Pro58, though tolerated, was theorized to partially counteract the strong interaction between Thr32' and His53. A point-mutation of Phe35' to Serine stabilizes the crystal contact and increases the resolution of our crystallographic experiments by 1 Å.
The modified interface between neighboring RanF35S molecules generates a stronger network of hydrogen bonds to stabilize the Nup153-ZnF•RanGDP crystal lattice, and although Ser35Ran is just out of range for making an H-bond with His53Ran', the predominant contact remains between Thr32Ran and His53Ran'. By removing Phe35Ran we have alleviated the clash with neighboring Pro58Ran' and stabilized the critical hydrogen bond between Thr32Ran and His53Ran'. The overall structure of Nup153-ZnF•RanGDP is unchanged by the F35S mutation, but the packing of complexes with RanF35S results in slightly different cell dimensions and orientations of symmetry axes (reducing the orthorhombic to a monoclinic space group with a non-crystallographic symmetry axis).
Comparison of the four Nup153 zinc fingers bound to RanGDP
The RanF35S mutant enabled us to individually crystallize all four ZnF modules of Nup153 with Ran, allowing for a comprehensive analysis of the interaction. Data collection and refinement statistics for all constructs are listed in Table 1. Crystals of RanGDP alone were obtained and the structure refined to 1.48 Å (Rwork/Rfree = 18.0%/20.3%). Each of the ZnF structures in complex with RanGDP contains one Nup153-ZnF molecule bound to one molecule of RanGDP (Fig. 4a). The orientations of each Nup153-ZnF with respect to RanGDP are quite similar in the various structures (Table S2). ZnF1 demonstrated the highest deviation (mean rmsd value of 1.07 Å) from the other ZnFs. Of note, the ZnF moiety is not involved in crystal contacts, thus similar orientation of the different ZnFs is very likely representative of the in vivo situation.
Table 1.
X-ray crystallographic data collection and refinement statistics
| Data Set | RanGDP | Nup153ZnF12 RanF35S-GDP |
Nup153ZnF34 RanF35S-GDP |
Nup153ZnF1 RanF35S-GDP |
Nup153ZnF2 RanGDP |
Nup153ZnF3 RanF35S-GDP |
Nup153ZnF4 RanF35S-GDP |
|---|---|---|---|---|---|---|---|
|
PDB Code Data Collection* |
3GJ0 | 3GJ7 C |
3GJ8 B |
3GJ6 A |
3GJ3 A |
3GJ4 B |
3GJ5 B |
| Space group | P41212 | P21 | P21 | P21212 | P21212 | P21 | P21 |
| Cell dimensions: | |||||||
| a,b,c (Å) | 81.5, 81.5, 130.5 | 58.5, 61.1, 80.2 | 74.3, 61.7, 70.6 | 60.1, 80.3, 54.6 | 59.7, 80.2, 58.0 | 68.4, 61.6, 72.1 | 70.4, 61.3, 74.0 |
| β (°) | 90 | 93.7 | 112.3 | 90 | 90 | 110.0 | 112.6 |
| Unique reflections | 71,145 | 39,366 | 52,824 | 7,528 | 26,894 | 29,511 | 53,894 |
| Resolution (Å) | 50-1.48 (1.53-1.48) |
50-1.95 (2.02-1.95) |
50-1.82 (1.89-1.82) |
50-2.70 (2.80-2.70) |
50-1.80 (1.86-1.80) |
50-2.15 (2.23-2.15) |
50-1.79 (1.85-1.79) |
| Rsyma | 7.5 (99.9) | 9.6 (82.7) | 6.9 (51.6) | 12.3 (47.1) | 7.2 (73.3) | 9.4 (45.8) | 6.3 (33.3) |
| Completeness | 95.6 (93.0) | 93.3 (90.1) | 99.3 (99.3) | 98.3 (92.3) | 99.6 (99.4) | 95.3 (92.4) | 98.1 (97.2) |
| Redundancy | 9.7 (7.7) | 2.3 (1.7) | 3.2 (3.0) | 4.6 (4.4) | 7.8 (7.7) | 2.7 (2.8) | 2.7 (2.7) |
| I/σ | 32.3 (1.8) | 12.9 (1.2) | 19.2 (2.0) | 16.8 (2.6) | 30.2 (2.9) | 15.2 (2.5) | 18.6 (3.0) |
| Wilson B factor (Å2) | 19.6 | 37.0 | 24.3 | 61.4 | 26.4 | 34.9 | 23.2 |
| Refinement | |||||||
| Resolution (Å) | 30 - 1.48 | 30 - 1.93 | 30 - 1.82 | 30 - 2.70 | 30 - 1.78 | 30 - 2.15 | 30 - 1.79 |
| Nonhydrogen Atoms | 3264 | 3689 | 3714 | 1851 | 1854 | 3652 | 3654 |
| Water molecules | 478 | 294 | 548 | 34 | 250 | 206 | 458 |
| Rworkb / Rfreec (%) | 18.0 / 20.3 | 18.6 / 22.9 | 17.0 / 20.3 | 20.5 / 27.9 | 16.1 / 19.4 | 20.5 / 25.7 | 19.9 / 23.3 |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.005 | 0.008 | 0.007 | 0.007 | 0.007 |
| Bond angles (°) | 1.24 | 1.06 | 0.943 | 1.14 | 1.11 | 1.02 | 1.07 |
| B factors (Å2) | |||||||
| ZnF | - | 86.0 | 58.2 | 203.3 | 50.7 | 71.5 | 67.0 |
| Ran | 24.8 | 51.1 | 32.9 | 71.8 | 33.4 | 48.5 | 40.5 |
| GDP | 18.9 | 40.1 | 24.1 | 60.2 | 24.4 | 39.1 | 34.0 |
| Water | 37.2 | 54.8 | 45.7 | 62.0 | 43.8 | 46.8 | 53.1 |
| Ramachandran Plot (%)d | |||||||
| Most Favored | 93.3 | 89.0 | 90.1 | 80.1 | 88.9 | 88.7 | 90.3 |
| Allowed | 6.7 | 10.5 | 9.1 | 18.9 | 11.1 | 10.8 | 9.3 |
| Generously Allowed | 0 | 0.3 | 0.7 | 1.0 | 0 | 0.5 | 0.5 |
| Disallowed | 0 | 0.3 | 0 | 0 | 0 | 0 | 0 |
Rsym = Σ|Ii - <Ii>|/ΣIi, where Ii is the intensity of the ith observation and <Ii> is the mean intensity of the reflection.
Rwork = Σ(||Fobs| - |Fcalc|| / Σ|Fobs|)
Rfree = R value for a randomly selected subset (5%) of the data that were not used for minimization of the crystallographic residual.
Calculated with the program PROCHECK 1.
Highest resolution shell (10% of data) shown in parenthesis.
Related crystal groups
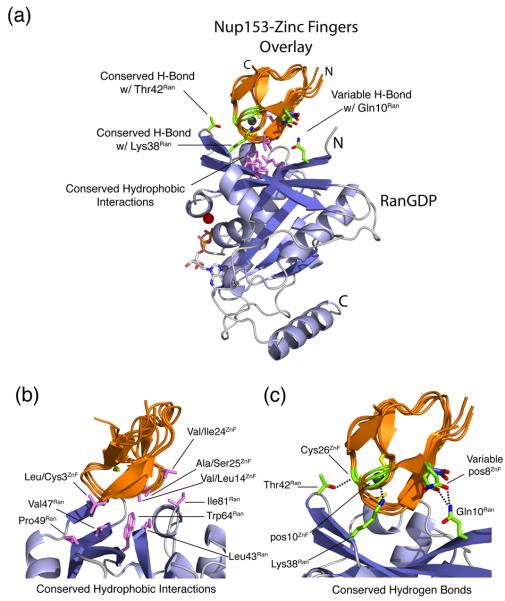
Figure 4.
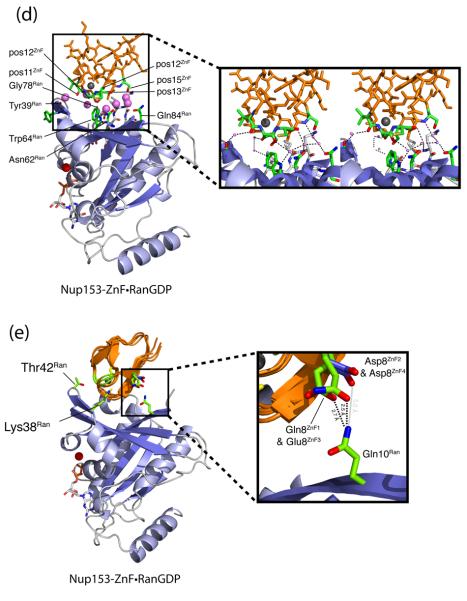
Comparison of the four individual Nup153 zinc fingers (ZnFs), colored orange, in complex with RanGDP, colored blue. The four individual ZnFs are overlaid to highlight differences and similarities in binding with Ran. Residues that make intermolecular hydrogen bonds are colored in green, while residues that facilitate hydrophobic interactions are colored violet. (a) A overview of the Nup153-ZnF•RanGDP complex showing that each ZnF binds independently and at the same location with RanGDP. Hydrogen bonds conserved amongst the four ZnFs are labeled. The variable hydrogen bond at residue 8ZnF with Gln10Ran is only formed between Ran and ZnF1 or ZnF3. (b) An enlarged view of all residues responsible for the hydrophobic interaction at the Nup153-ZnF•RanGDP interface. (c) An enlarged view of hydrogen bonds, in green, that facilitate the Nup153-ZnF•RanGDP interaction. Conserved H-bonds are made between the carbonyl of Cys26ZnF with the sidechain hydroxyl group of Thr42Ran, and Lys38Ran with the carbonyl at residue10ZnF. A variable H-bond is present at residue 8ZnF binding with Gln10Ran. A stable bond is formed between Gln8ZnF1 and Gln10Ran, as well as between Glu8ZnF3 and Gln10Ran. Residue 8 of ZnF2 and ZnF4, colored dark-blue, contain an Asp residue, too short to make the H-bond with Gln10Ran. (d) A conserved water network at the interface between Nup153-ZnF and RanGDP. Residues facilitating these contacts are labeled and highlighted in green, with water molecules colored violet. (e) A more detailed view of the non-conserved hydrogen bond made with Gln10Ran by Gln8ZnF1 and Glu8ZnF3, colored green. Asp8ZnF4 and Asp8ZnF4, colored blue, are unable to bridge the distance necessary to make the H-bond with Gln10Ran, a calculated distance of ∼5.0 Å.
In addition to conserved hydrophobic interactions, the backbone carbonyl of residue 10ZnF and the side chain of Lys38Ran, as well as the backbone carbonyl of Cys26ZnF and the hydroxyl group of Thr42Ran form two conserved H-bonds (Fig. 4a-c). A water network, identically observed in each of the four individual ZnF•RanGDP structures, further stabilizes the ZnF•Ran interaction by mediating hydrogen bonds (Fig. 4d).
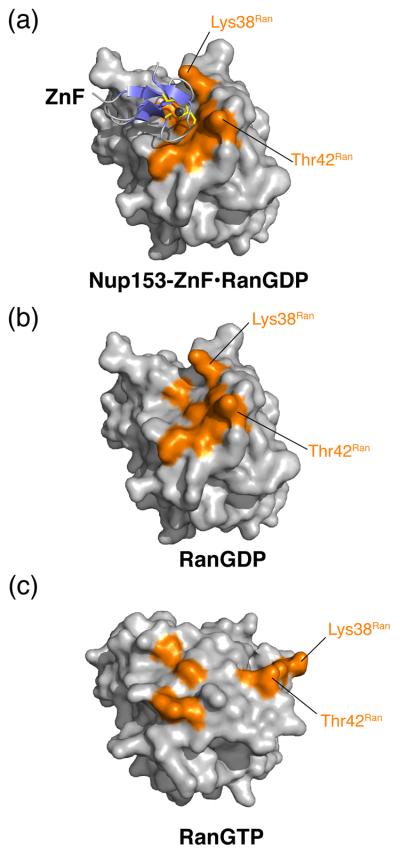
Nucleotide-dependent changes in the conformation of Ran occur at the switch I and switch II regions 41. When bound to GDP, Ran is in the open conformation with switch I swung out, away from the nucleotide and closer to β1 (Fig. 2). Upon binding of GTP, the two switch regions close to accommodate the γ-phosphate. As highlighted in Fig. 5, residues of Ran critical for hydrogen bonding with Nup153-ZnF shift away from the ZnF binding site when Ran is in the GTP-bound conformation. Superimposing RanGTP (PDB code 1WA5) and our RanGDP structure bound to ZnFs, shows that Lys38Ran is shifted 26 Å away from the ZnF binding site when Ran is bound to GTP. Thr42Ran is buried in the RanGTP structure to interact with the magnesium ion and thus the H-bond with ZnF cannot be maintained. To highlight the influence of the conformational shift between RanGDP/RanGTP and binding of ZnFs, we have modeled a putative Nup153-ZnF2•RanGTP complex, replacing RanGDP in our structure with RanGTP (PDB code 1WA5) (Fig. 5) 45. In this modeled complex, the calculated binding interface area is reduced by 27% to 345 Å2. H-bonds to switch I residues are not only lost, but in the case of Thr42Ran, are mutually exclusive between the RanGDP and RanGTP conformations, suggesting a mechanism for preferential binding of ZnF to RanGDP over RanGTP.
Figure 5.
Surface representation of RanGDP or RanGTP, grey, with or without Nup153-ZnF,blue ribbon diagram. Residues involved in intermolecular contact are colored orange. (a) ZnF•RanGDP complex. Lys38Ran and Thr42Ran, located in the switch I region, form conserved H-bonds with the ZnF, and are, known to undergo significant conformational changes upon binding of GTP. Both residues are positioned to interact with ZnF in this conformation. (b) RanGDP interaction surface in the absence of ZnF. (c) RanGTP in absence of modeled ZnF. Lys38Ran and Thr42Ran are displaced up to 30 Å, and are no longer able to interact with the Nup153-ZnF in this conformation.
Apart from these interactions in the switch I region, some interactions remain unique to the individual ZnFs. The principle distinction between the binding modes of the Nup153-ZnFs is the ability or inability to form a H-bond with Gln10Ran. Both Gln8ZnF1 and Glu8ZnF3 H-bond via the side chain carbonyl, while Asp8ZnF2 and Asp8ZnF4 do not, because the shorter aspartic acid side chain does not reach far enough (Fig. 4e). This single hydrogen bond accounts in large part for the differences in binding affinity measured by ITC as described below and in Table 2.
Table 2.
Thermodynamic parameters for ZnF-Ran binding in Isothermal Titration Calorimetry experiments.
| Titrand | [Titrand] (μM) |
Titrant | [Titrant] (mM) |
T (K) |
Kd 1 (μM) |
N1 | Kd 2 (μM) |
N2 | ΔH1 / ΔH2 (kJ/mol) |
TΔS1 / TΔS2 (kJ/mol) |
|---|---|---|---|---|---|---|---|---|---|---|
| Nup153ZnF1234 | 12 | RanGDP | 0.31 | 293 | 1 | 2 | 8 | 2 | −40.8 / 3.9 | −7.38 / 32.7 |
| ” | 12 | RanQ69L | 0.40 | 293 | 20 | 2 | - | - | −39.3/ - | −12.9 / - |
| Nup153-ZnF12 | 15 | RanGDP | 0.29 | 293 | 4 | 1 | 57 | 1 | −41.0 / 20.4 | −10.3 / 44.2 |
| Nup153-ZnF34 | 23 | RanGDP | 0.35 | 293 | 3 | 1 | 59 | 1 | −17.2 / −2.2 | 14.0 / 21.6 |
| Nup153-ZnF1 | 13 | RanGDP | 0.30 | 293 | 6 | 1 | - | - | −7.8 | 21.4 |
| Nup153-ZnF1Q8D | 20 | RanGDP | 0.36 | 293 | 8 | 1 | - | - | −11.2 | 17.6 |
| Nup153-ZnF2 | 32 | RanGDP | 1.25 | 278 | 49 | 1 | - | - | 16.8 | 39.8 |
| Nup153-ZnF2D8Q | 18 | RanGDP | 0.34 | 293 | 4 | 1 | - | - | −5.5 | 24.9 |
| Nup153-ZnF3 | 29 | RanGDP | 0.30 | 293 | 7 | 1 | - | - | −3.1 | 26.0 |
| Nup153-ZnF3E8D | 17 | RanGDP | 0.34 | 293 | - | - | - | - | - | - |
| Nup153-ZnF4 | 50 | RanGDP | 0.95 | 278 | 47 | - | - | - | 12.6 | 35.7 |
| Nup153-ZnF4D8Q | 19 | RanGDP | 0.36 | 293 | 3 | 1 | - | - | −6.6 | 24.3 |
| Nup358-ZnF12 | 25 | RanGDP | 1.25 | 278 | 15 | 2 | - | - | −3.4 | 22.5 |
Protein constructs containing tandem ZnF pairs (ZnF12, ZnF34) have been crystallized in the same condition as the individual zinc finger domains. The electron density map for each crystal shows only one ZnF bound to RanGDP in agreement with the structural interactions described above for single ZnF constructs. In these structures, predominantly the second ZnF (ZnF2 and ZnF4) is seen to bind with RanF35SGDP, even though, considering the crystal packing, ZnF1 and ZnF3 could be accommodated as well. To distinguish which ZnF was bound, both possibilities were modeled and refined. R factors were considerably lower for the correct ZnF and the difference density showed reduced noise. Interestingly, and in contrast to the study by Schrader et al., we do not observe any interaction of the linker residues between ZnF1 and ZnF2 with Ran.
Isothermal titration calorimetry
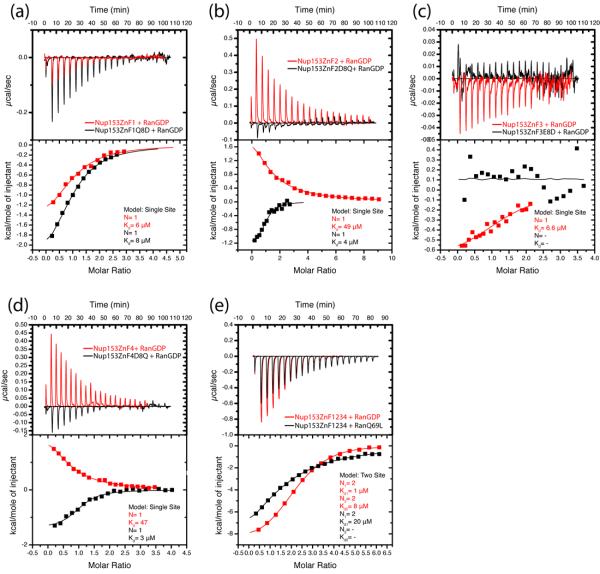
To expand upon our crystallographic analysis of the Nup153-ZnF interaction with RanGDP, we performed binding assays using isothermal titration calorimetry (ITC). These experiments demonstrate that individual Nup153-ZnF domains bind to RanGDP with modestly varying affinity (Table 2). ZnF1 / ZnF3 have comparable binding characteristics that differ from those measured for ZnF2 / ZnF4. ZnF1 is shown to bind RanGDP with a measured KD of 6.5 μM and ZnF3 binds with a KD of 6.6 μM, in an exothermic reaction. ZnF2 / ZnF4 have a lower binding affinity for RanGDP, with 49 μM and 47 μM respectively, in an endothermic reaction. The enthalpic differences in RanGDP binding of these ZnFs is likely due to conformational contributions, thus not readily explainable based on our structures. The variation in enthalpy between sites is also measured with the ZnF pairs and the full Nup153 ZnF domain as described below and in Table 2.
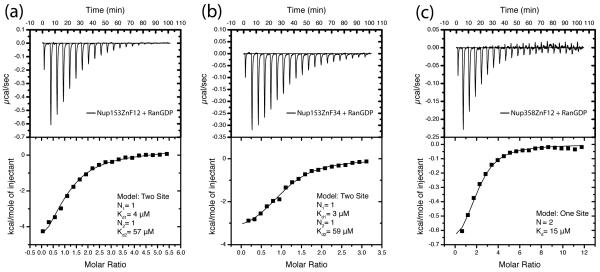
Structural data suggests that the missing hydrogen bond between Gln10RanGDP and residue 8ZnF may be responsible for this differences. In ZnF2 / ZnF4 Asp8 is too short to H-bond with Gln10RanGDP, in contrast to Glu/Gln8 in ZnF1 / ZnF3 which forms this bond (Fig. 4d). When residue 8ZnF was mutated to a glutamine in ZnF2 and ZnF4, the binding affinity for RanGDP increased to values very close to those measured for ZnF1 and ZnF3 (Fig. 6a, 6b, 6c, 6d). Nup153 was also tested to compare the differences in affinity for RanGDP vs. RanGTP (in the form of the GTPase deficient mutant, RanQ69L) (Fig. 6e). We next asked whether individual ZnF domains exhibit an allosteric effect on their neighbors. Pairs of ZnFs from Nup153 and Nup358 (Nup153ZnF12, Nup153ZnF34, and Nup358ZnF12) were assayed for RanGDP binding (Fig. 7). A two-site model best fit the data in each experiment with Nup153. The two binding sites in the tandem constructs exhibit affinities that are similar to the values measured for the individual ZnFs, indicating no significant allosteric effect. The data also suggests that the flexible linker between ZnF domains does not significantly contribute to binding affinity for Nup153ZnF2, as our data measured with and without the linker agree with previously measured affinity 39. Additionally, the binding of Nup358ZnF12 to RanGDP was measured to characterize ZnFs of Nup358 in comparison with those of Nup153. Based on the high level of sequence conservation between Nup153 and Nup358 (Fig. S1), it is not surprising to find that ZnFs from Nup358 behave similarly. The ITC data for Nup358ZnF12 is best fit to a single site model, with two molecules of RanGDP binding per molecule of Nup358ZnF12. This corresponds to each ZnF binding independently, similar to Nup153, however differs from Nup153 because both ZnFs of Nup358 lack the Glu/Gln8 residue needed to H-bond with Gln10RanGDP. To summarize the data from our calorimetric experiments, we compared binding of the entire ZnF regions of Nup153 RanGDP or RanQ69L (Fig 6). Nup153 binds RanGDP in a 2-site model with affinities of 1 μM and 8 μM, with two molecules of RanGDP binding per site. This accounts for each ZnF binding with RanGDP independently for a total of four molecules of RanGDP binding with the ZnF domain of Nup153, in agreement with published data 37. RanQ69LGTP binds with lower affinity to Nup153, with one site showing negligible binding and the other exhibiting a KD of 20 μM in an independent 2-site model. The Nup358 ZnF casette binds both GDP- and GTP-bound Ran, and again, the interaction with RanGDP is measurably stronger (Fig. 6).
Figure 6.
Isothermal calorimetry (ITC) for each individual ZnF binding RanGDP. Point-mutations are used to probe the variable H-bond interaction with Gln10Ran, determined to be the interaction responsible for discrepancies in affinities measured between ZnFs and RanGDP. Data for independent ZnFs is fit to a single site model with N=1. (a) WT-ZnF1 shown in red compared with mutated ZnF1 in black. Mutating Gln6 to Asp is shown to have a minor effect on binding with Gln10Ran. (b) WT-ZnF2 shown in red with the mutant protein shown in black. WT-ZnF2 shows no detectable signal, however by mutating Asp6 to Gln signal is restored. (c) WT-ZnF3 is shown in red and mutant ZnF3 is shown in black. Mutating Glu6 to Asp is enough to abolish signal by disrupting a single H-bond with Gln10Ran. (d) WT-ZnF4 is shown in red and has no detectable signal. However mutating Asp6 to Gln introduces a new H-bond with Gln10Ran and restores signal, similar to results seen with ZnF2. (e) Binding of the WT Nup153-ZnF domain to GDP (in red) is compared to binding with RanGTP (in black). In agreement with our structural observations, RanGDP binds ZnFs with strong and weak sites, while RanGTP binds with the same stoichiometry, but only with weak affinity. Experimental values for N, Kd, enthalpy, ΔH, and entropy, TΔS, are listed in Table 2. Constructs are described in Supplementary Table 1.
Figure 7.
ITC illustrating low μM affinity between RanGDP and two tandem ZnF pairs from the Nup153-and Nup358-ZnF domain. (a) Interaction between RanGDP and ZnF12. The data is best fit to a two-site model suggesting each ZnF binds independently, but with different affinities. (b) Interaction between ZnF34 and RanGDP, again suggesting two independent sites with different affinities. (c) Interaction between Nup358ZnF12 and RanGDP. As with Nup153, the ZnFs of Nup358 bind with low μM affinity to RanGDP. The data is best fit to a single site model in accordance with both ZnFs from Nup358 lacking the residue nececcary at pos8 to H-bond with Gln10Ran. Experimental values for N, Kd, enthalpy, ΔH, and entropy, TΔS, are listed in Table 2. Constructs are described in Supplementary Table 1.
Conservation of RanBP2-type ZnF cassettes in nucleoporins
We have performed a phylogenetic analysis to put our structural and biochemical data in an evolutionary context. The RanBP2-type ZnF domain can be readily recognized due to its characteristic signature sequence motif (Fig. 1). The ZnF domain is found in all Nup153 and Nup358 homologs, which are exclusively present in animals, but absent in plants and fungi. RanBP2-type ZnF cassettes usually contain 4 repeats in Nup153, and 8 repeats in Nup358. Exceptions are however not infrequent. The number in sequenced Nup153 homologs varies between 2 and 6, while in Nup358 we found between 2 and 8 ZnFs (data not shown). These variations match our observation that the ZnF modules function largely independently and not in a cooperative fashion. Similarly, we fail to find a specific signature that would define an order in which the ZnF modules are arranged within the ZnF region. We also asked whether there is a distinction between ZnFs present in Nup153 compared to those in Nup358. Given the asymmetric distribution of Nup153 and Nup358 at the nucleoplasmic and cytoplasmic side of the NPC, respectively, one could hypothesize that the ZnFs exhibit locus-specific tasks. However, we do not find such differences. The sequence conservation of the Ran-binding ZnF matches well with those residues that are structurally and functionally important. The structurally important residues Trp7, Cys9, Cys12, Asn16, Cys23 and Cys26 are strictly conserved (Fig. S1). The residues involved in hydrophobic interactions with Ran Leu13, Val14, and Ala25, are also quite well conserved across all animals and the functional significance has been recognized even before structural data was available 37. Comparing just the linker between individual ZnFs shows no sequence conservation, consistent with it not having a significant role in the protein interaction.
Comparison of binding interactions amongst RanBP2-type zinc fingers
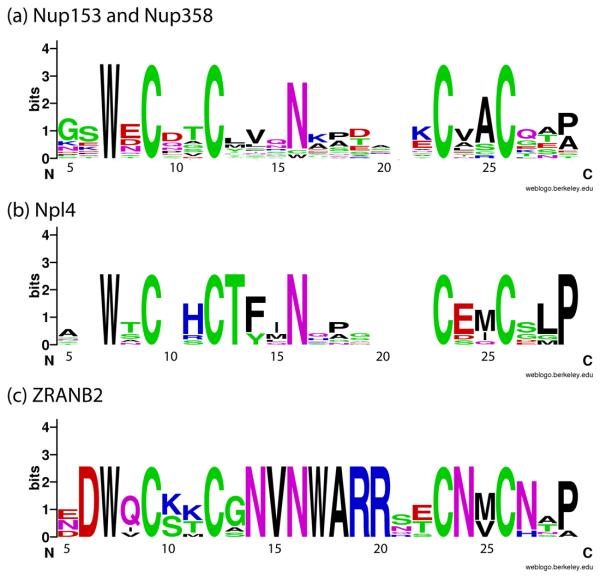
RanBP2-type zinc fingers are recognized by the conserved sequence W-X(2)-CX(3)-N-X(6)-C-X(2)-C as shown in Fig. 1. This sequence signature only defines the structural scaffold, but excludes the residues important for binding interactions. Currently three different binding partners are known. The Nup153/Nup358 class binds Ran, the Npl4 class binds ubiquitin and the ZRANB2 class binds ssRNA 39; 46; 47. Phylogenetic analysis of these RanBP2-ZnF classes readily reveals that each type of ZnF has additional conserved residues (Fig. 8). For Nup153/Nup358, these are the discussed residues Leu13, Val14 and Ala25 37. In the Npl4-type, the same ZnF surface binds ubiquitin involving the conserved residues Thr13, Phe14 and Met25. By switching these residues the Nup153 ZnF can be converted into a ubiquitin-binding moiety 37. The ssRNA-binding splicing factor ZRANB2 is the best conserved of the three known RanBP2-ZnF classes. The publication of the ssRNA-bound ZRANB2 structure (PDB code 3G9Y) details ssRNA bound at a different surface of the scaffold via the highly conserved residues between residues 14 and 20 47. Considering the sequence signature of the Nup153/Nup358-ZnF class it is unlikely that it can bind nucleic acids, as was reported when Nup153 was first described 13. In summary, the RanBP2-type zinc finger emerges as a scaffold for various binding partners and it will be interesting to learn about the interacting partners of the additional, less characterized classes, including sharpin, Mdm2, and Mdm4 48; 49.
Figure 8.
Weblogos from members of the RanBP-2 class of zinc fingers known to have unique binding partners. The logos highlight similarities and differences amongst the three groups of zinc fingers: the Ran binding Nup153/Nup358 group, the ubiquitin binding Npl4 group, and the single-stranded RNA binding zn256 group. The logos demonstrate the overall conservation of the canonical RanBP2 type zinc finger sequence W-X-C-X(2,4)-C-X(3)-N-X(6)-C-X(2)-C, while highlighting regions important for facilitating binding with a unique binding partner. (a) ZnFs from Nup153 and Nup358 that bind with Ran. Only the first finger from each protein has been used in the alignment. The previously described “LVA,” in positions 13, 14, and 25, have low information content, although position 25 has been shown influence Ran binding 37. (b) The logo for Npl4 again demonstrates the elevated conservation of positions 13, 14, and 15, previously shown to modulate interaction with ubiquitin. (c) The ZRANB2 logo highlights the strong conservation this sequence in eukaryotes. A structure of ZRANB2 (PDB codes 3G9Y and 2K1P), demonstrates the strict conservation and elevated level of positive charges at positions 14-20 that regulate binding of single-stranded RNA 47. Each logo is based on alignments, shown in Fig S2, of sequences from a diverse group of eukaryotes.
Conclusion
Here we present a comprehensive analysis of the interaction of the Nup153/Nup358 ZnF class with Ran. We identify individual residues within the four ZnFs of mammalian Nup153 that modulate the binding affinity to Ran. The G protein is preferentially bound in the GDP-bound form, because switch I residues are involved in critical hydrogen bond interactions with ZnF and because switch I contributes about 1/3 of the total interaction surface. Nonetheless, binding to RanGTP is also observed and the local RanGTP to RanGDP ratio likely determines the nucleotide-binding state of ZnF-bound Ran. We cannot detect a significant difference between the binding behavior of Nup358 and that of Nup153 fingers, arguing for a common function for both proteins. Neither Nup358 nor Nup153 are universally conserved nucleoporins, but are specific to animals. We suggest that the ZnF moieties are used to increase the local concentration of Ran at both the nucleoplasmic and cytoplasmic face of the NPC. This may accelerate nucleocytoplasmic protein transport by keeping Ran close to the NPC, a sophistication that may be dispensable in unicellular eukaryotes, or potentially replaced by a separate mechanism, as proposed for plants 50. Our phylogenetic analysis suggests that other RanBP2-type ZnFs do not bind Ran, since they share only the structurally important residues and use distinct binding interfaces for their respective protein and nucleic acid interactions.
Materials and Methods
Protein purification
Bacterial expression constructs for Nup153 domains from Rattus norvegicus and Nup358 from Homo sapiens were cloned as glutathione-S-transferase (GST) fusion proteins in the pGEX-6P1 vector (GE Healthcare) (Table 1). Ran was expressed as a 6xHis-tag fusion protein from a pET-28a vector, engineered to contain a protease 3C site after the N-terminal affinity tag (EMD Biosciences). All proteins were expressed in Escherichia coli strain BL21(DE3) RIL (Stratagene).
Bacterial cell pellets harboring GST fusion proteins were suspended in 20 mM potassium phosphate pH 7.0, 150 mM NaCl, 2 mM DTT and lysed using a french press. The crude lysate was supplemented with 200 μM phenylmethanesulfonyl fluoride (PMSF) and centrifuged at 15 000 g for 15 minutes. Soluble protein was mixed with 0.5ml of glutathione Sepharose beads (4 Fast Flow, GE Healthcare) per 1000 ODs for 2 hrs at 277 K. After three batch washes in resuspension buffer, resin was washed in ZnCl2 buffer (10 mM Tris/HCl pH 8.0, 100 mM NaCl, 10 μM ZnCl2, 2 mM DTT), and Nup153 proteins were eluted directly from the resin by incubating with protease HRV-3C overnight at 277 K. The eluted protein was purified by anion exchange chromatography on a HiTrapQ column (GE Healthcare) via a linear NaCl gradient and size exclusion chromatography using a Superdex S75 26/60 column (GE Healthcare) run in 15 mM Tris/HCl pH 8.0, 150 mM NaCl 1 mM MgCl2, 5 μM ZnCl2, 1.5 mM DTT. All truncations of the full-length Nup153-ZnF domain, as well as point mutations, were generated with PCR mutagenesis and purified as described above.
Bacterial cell pellets harboring His-tagged Ran were suspended in 20 mM Tris/HCl pH 8.0, 200 mM NaCl, 5 mM imidazole, 3 mM β-mercaptoethanol (β-ME), and lysed using a french press. Crude lysate was supplemented with 200 μM phenylmethanesulfonyl fluoride and clarified by centrifugation at 15 000 g for 15 minutes. The soluble fraction was then incubated with 1ml Ni-NTA per 1000 ODs for 1 hour at 277 K and loaded onto a disposable column (Pierce). The column was washed with 4 bed volumes of 10 mM Tris/HCl pH 8.0, 400 mM NaCl, 10 mM imidazole, 3 mM β-ME, and eluted with 6 bed volumes of 10 mM Tris/HCl pH 8.0, 50 mM NaCl, 150 mM imidazole, 3 mM β-ME. Eluted protein was dialyzed against 10 mM Tris/HCl pH 8.0, 100 mM NaCl, 1 mM DTT, for 1 hour before the 6xHis-tag was cleaved. RanGDP was separated from RanGTP using anion exchange chromatography on a HiTrapQ column (GE Healthcare) via a linear NaCl gradient, followed by purification with size exclusion chromatography using a Superdex S200 26/60 column (GE Healthcare) in 15 mM Tris/HCl pH 8.0, 150 mM NaCl 1 mM MgCl2, 5 μM ZnCl2, 1.5 mM DTT. RanQ69L and RanF35S were generated with PCR mutagenesis and purified as described above. The nucleotide-bound state of Ran was confirmed by HPLC, on a analytical C18 column. The nucleotide was released from Ran by heat denaturation at 369 K and centrifugation to separate coagulated protein from soluble nucleotide. The soluble fraction was loaded on a HPLC and GTP was separated from GDP with a linear acetonitrile gradient in 50 mM TEA/HCl pH 7.5, 10 mM tetrabutylammonium-sulfamate (TBA).
Crystallization
Ran was concentrated to 15 mg/ml using Vivaspin 20 concentrators (Sartorius) and mixed with Nup153-ZnF protein at equimolar concentrations. All Nup153-ZnF fragments in complex with RanGDP were crystallized in the same condition (0.1 M BisTris/HCl pH 6.5, 18-20% PEG3350) using the hanging-drop method and mixing 1 μl protein with 1 μl of crystallization condition at 291 K. Crystals of wtRanGDP in complex with ZnF2 grew in 5-7 days forming birefringent plates of 400 × 200 × 30 μm. RanGDP crystals formed on the edges of Nup153-RanGDP complex crystals after two weeks, as small 150 × 150 × 150 μm bipyramidal crystals. Crystals of Nup153 ZnF constructs in complex with RanF35S-GDP grew in 1-2 days. All crystals were cryoprotected by adding 12% (v/v) glycerol to the reservoir solution before flash freezing in liquid nitrogen. All datasets were collected at the NE-CAT beamlines 24ID-C and 24ID-E at Argonne National Laboratory. Crystal parameters and data collection statistics are listed in Table 1.
Structure determination
Data reduction was carried out using HKL2000 51. The initial Nup153-ZnF2•RanGDP structure was solved by molecular replacement, using RanGDP as the search model (PDB code 1BYU). All other structures were solved by molecular replacement using our 1.48 Å RanGDP structure as the search model. Model building was carried out using COOT 52. PHENIX 53 was used for all refinement steps. The ZnF present in each tandem ZnF construct was determined by analysis of the difference maps and refinement statistics with the respective ZnF sequences. Proper assignment of the correct ZnF correlates with better refinement statistics, including a drop in the R/Rfree factors. Data collection and refinement statistics are summarized in Table 1. All figures were made using PyMOL 54.
Isothermal titration calorimetry
RanGDP and all ZnF constructs were purified and dialyzed into the same buffer containing 15 mM Tris/HCl pH 8.0, 150 mM NaCl 1 mM MgCl2, 5 μM ZnCl2, 1.5 mM DTT, and either 2.5 μM GDP or 2.5 μM GTP as indicated, prior to ITC. Protein concentrations were determined spectrophotometrically at 280 nm immediately before the experiment. ITC was performed using a VP-ITC microcalorimeter (MicroCal, Northhampton, MA). Titrations were performed at 293 K, or 278 K, by injecting 14 μl aliquots of RanGDP, or RanQ69LGTP (Table 2), into the ITC cell containing 1.43 ml of Nup153 or Nup358 ZnF proteins (Table 2). Binding stoichiometry, enthalpy and entropy, as well as the equilibrium binding dissociation constant was determined using a “one site model” for individual zinc fingers and Nup358 ZnF12, or a “two sets of independent sites” model for all other experiments, to define molecular association in the software suite, MicroCal Origin 2.9 (MicroCal).
Sequence analysis
Sequences were aligned using MUSCLE 55 and edited in JalVIEW 56. Sequence logos were made using WebLogo 57; 58.
Protein Data Bank Accession Numbers
Coordinates and structure factors for all crystal structures have been deposited in the PDB (IDs 3GJ0, 3GJ3, 3GJ4, 3GJ5, 3GJ6, 3GJ7, 3GJ8).
Supplementary Material
Sequence alignment of the human Nup358/RanBP2 and Nup153 ZnFs, including their respective C-terminal linker regions. (a) Conservation of the “RanBP2-like” ZnF motif between Nup358 and Nup153. Strict conservation is shown in dark-blue, while lighter shades of blue depict decreasing levels of conservation. The linker regions between these ZnFs seem to have to have no conserved sequence or length. (b) Weblogo of the same alignment showing conservation in the ZnF motif, and lack thereof in the linker region.
Alignment of sequences from divergent eukaryotes, of the “RanBP2-like” family of zinc finger proteins that have known binding partners, Nup153, Nup358, Npl4, and ZRANB2. The sequences are colored from red to white to represent a decrease in conservation. Below the alignments is a sequence logo calculated using the alignment above and WebLogo 57; 58.
Experimental constructs, abbreviations, and descriptions.
Mean rmsd values from overlays of the individual zinc fingers. Coordinates are taken from crystal structures of ZnFs in complex with RanGDP from this study to highlight consistency amongst the ZnF structures.
Acknowledgements
We thank the staff at beamlines 24-ID-C/-E at Argonne National Laboratory and X6A at the National Synchrotron Light Source for assistance and data collection, past and present members of the Schwartz lab for discussions and editing. We would like to thank Katie Ullman for the kind gift of Nup358. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH-GM68762) provided instrumentation. Supported by a Pew Scholarship (T.U.S.) and the Herman Eisen Fellowship in Biology (J.R.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 2.Kiseleva E, Allen TD, Rutherford S, Bucci M, Wente SR, Goldberg MW. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J Struct Biol. 2004;145:272–88. doi: 10.1016/j.jsb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Kiseleva E, Rutherford S, Cotter LM, Allen TD, Goldberg MW. Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci. 2001;114:3607–18. doi: 10.1242/jcs.114.20.3607. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–6. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–53. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–51. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 7.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–66. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–71. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 9.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–15. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 10.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 11.Stewart M, Kent HM, McCoy AJ. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–46. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–13. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 13.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 14.Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 2005;114:319–30. doi: 10.1007/s00412-005-0019-3. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–8. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 16.Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr Biol. 1998;8:1376–86. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- 17.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–95. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaseen NR, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5516–21. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell. 2004;15:4261–77. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–67. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 21.Pante N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–17. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–14. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimaano C, Ball JR, Prunuske AJ, Ullman KS. RNA association defines a functionally conserved domain in the nuclear pore protein Nup153. J Biol Chem. 2001;276:45349–57. doi: 10.1074/jbc.M102592200. [DOI] [PubMed] [Google Scholar]
- 24.Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 25.Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A. 2003;100:981–5. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullman KS, Shah S, Powers MA, Forbes DJ. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–64. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–56. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–5. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribbeck K, Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–71. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–30. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–20. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 33.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–7. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 35.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–30. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Higa MM, Alam SL, Sundquist WI, Ullman KS. Molecular characterization of the Ran-binding zinc finger domain of Nup153. J Biol Chem. 2007;282:17090–100. doi: 10.1074/jbc.M702715200. [DOI] [PubMed] [Google Scholar]
- 38.Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–21. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrader N, Koerner C, Koessmeier K, Bangert JA, Wittinghofer A, Stoll R, Vetter IR. The crystal structure of the Ran-Nup153ZnF2 complex: a general Ran docking site at the nuclear pore complex. Structure. 2008;16:1116–25. doi: 10.1016/j.str.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Stewart M, Kent HM, McCoy AJ. The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2) J Mol Biol. 1998;284:1517–27. doi: 10.1006/jmbi.1998.2204. [DOI] [PubMed] [Google Scholar]
- 41.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 42.Banatao DR, Cascio D, Crowley CS, Fleissner MR, Tienson HL, Yeates TO. An approach to crystallizing proteins by synthetic symmetrization. Proc Natl Acad Sci U S A. 2006;103:16230–5. doi: 10.1073/pnas.0607674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper DR, Boczek T, Grelewska K, Pinkowska M, Sikorska M, Zawadzki M, Derewenda Z. Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr D Biol Crystallogr. 2007;63:636–45. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 44.Mizutani H, Saraboji K, Malathy Sony SM, Ponnuswamy MN, Kumarevel T, Krishna Swamy BS, Simanshu DK, Murthy MR, Kunishima N. Systematic study on crystal-contact engineering of diphthine synthase: influence of mutations at crystal-packing regions on X-ray diffraction quality. Acta Crystallogr D Biol Crystallogr. 2008;64:1020–33. doi: 10.1107/S0907444908023019. [DOI] [PubMed] [Google Scholar]
- 45.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–7. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem. 2003;278:20225–34. doi: 10.1074/jbc.M300459200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM, Mackay JP. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proc Natl Acad Sci U S A. 2009;106:5581–6. doi: 10.1073/pnas.0802466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim S, Sala C, Yoon J, Park S, Kuroda S, Sheng M, Kim E. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci. 2001;17:385–97. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 49.Yu GW, Allen MD, Andreeva A, Fersht AR, Bycroft M. Solution structure of the C4 zinc finger domain of HDM2. Protein Sci. 2006;15:384–9. doi: 10.1110/ps.051927306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meier I, Xu XM, Brkljacic J, Zhao Q, Wang HJ. Going green: plants' alternative way to position the Ran gradient. J Microsc. 2008;231:225–33. doi: 10.1111/j.1365-2818.2008.02038.x. [DOI] [PubMed] [Google Scholar]
- 51.Minor Z. O. a. W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, editor. Methods in Enzymology. Vol. 276. Academic Press; New York: 1997. J. a. R. M. S. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 54.DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the human Nup358/RanBP2 and Nup153 ZnFs, including their respective C-terminal linker regions. (a) Conservation of the “RanBP2-like” ZnF motif between Nup358 and Nup153. Strict conservation is shown in dark-blue, while lighter shades of blue depict decreasing levels of conservation. The linker regions between these ZnFs seem to have to have no conserved sequence or length. (b) Weblogo of the same alignment showing conservation in the ZnF motif, and lack thereof in the linker region.
Alignment of sequences from divergent eukaryotes, of the “RanBP2-like” family of zinc finger proteins that have known binding partners, Nup153, Nup358, Npl4, and ZRANB2. The sequences are colored from red to white to represent a decrease in conservation. Below the alignments is a sequence logo calculated using the alignment above and WebLogo 57; 58.
Experimental constructs, abbreviations, and descriptions.
Mean rmsd values from overlays of the individual zinc fingers. Coordinates are taken from crystal structures of ZnFs in complex with RanGDP from this study to highlight consistency amongst the ZnF structures.