Abstract
In the crystal structure of the complex between the soluble extracellular domain of tissue factor (sTF) and active-site-inhibited VIIa, residues 91 and 92 in the Pro79-Pro92 loop of sTF interact with the catalytic domain of VIIa. It is not known, however, whether this loop has a role in allosteric activation of VIIa. Time-resolved fluorescence anisotropy measurements of probes covalently bound to sTF mutants E84C and T121C show that binding uninhibited Factor VIIa affects segmental motions in sTF. Glu84 resides in the Pro79-Pro92 loop, and Thr121 resides in the turn between the first and second antiparallel β-strands of the sTF subdomain that interacts with the Gla and EGF1 domains of VIIa; neither Glu84 nor Thr121 makes direct contact with VIIa. Probes bound to T121C report limited segmental flexibility in free sTF, which is lost after VIIa binding. Probes bound to E84C report substantial segmental flexibility in the Pro79-Pro92 loop in free sTF, which is greatly reduced after VIIa binding. Thus, VIIa binding reduces dynamic motions in sTF. In particular, the decrease in the Pro79-Pro92 loop motions indicates that loop entropy has a role in the thermodynamics of the protein-protein interactions involved in allosteric control of VIIa activation.
Introduction
Tissue factor (TF), an integral membrane protein, initiates blood coagulation when its 219-residue extracellular domain binds factor VIIa, a serine protease that circulates in blood both as zymogen (VII) and as active enzyme (VIIa) (1–5). When bound to TF, the protease activity of VIIa toward its natural substrate, factor X (X), increases by several orders of magnitude (6). Thus, TF is the essential cofactor for activation of catalysis by VIIa (7). This activation is mediated by allosteric interactions with the extracellular domain of TF that promote conformational changes, including reorganization of the active site of VIIa, which convert the enzyme from a catalytically inefficient, zymogen-like state to a catalytically efficient state (8).
Allosteric interactions could cause structural and dynamic changes in VIIa, TF, or both. We showed previously, by time-resolved fluorescence anisotropy, that segmental motion of free VIIa correlates with minimally constrained motion of the 30-kDa catalytic domain, and that formation of the complex with recombinant soluble TF (residues 1–218, sTF) eliminates this motion (9). We now show, by time-resolved anisotropy measurements of fluorescence-probe-labeled mutants of sTF, that formation of the sTF:VIIa complex is also accompanied by reduction of segmental motions in sTF. Specifically, we find that a disordered loop between residues Pro79 and Pro92 of the extracellular domain looses mobility on formation of the complex with VIIa.
Comparison of the crystal structures of free sTF and its complex with active-site-inhibited VIIa indicate that loop between Pro79 and Pro92 is a region of high mobility (Fig. 1 A). For example, the atoms of the loop residues are characterized by high thermal factors (10–12), and one structure does not report the data for residues 87–90 (11,12), and furthermore, to obtain the first crystal structure of the sTF/VIIa complex (13), recombinant sTF was subjected to limited subtilisin digestion, removing loop residues 81–90.
Figure 1.
(A) PyMOL ribbon representation of the sTF:VIIa complex by Banner et al. (13), showing sTF residues Pro79 and Pro92 space filled. The VIIa heavy chain is red, the VIIa light chain is green, sTF is yellow and the calcium atoms are white. (B) Superposition of the sTF structure by Muller et al. (11,12), in light blue, and sTF complexed with VIIa, showing residues Glu94 and Thr121 space filled. (C) Interface of sTF and the VIIa heavy chain showing sTF residues Glu91, Pro92, Tyr94, and VIIa residues Arg162 and Leu163 (65).
When the loop residues are not considered, free sTF and sTF bound to VIIa seem to have highly similar structures (Fig. 1 B). A structural comparison omitting amino and carboxyl termini residues 1–5 and 211–219 and residues 81–90 within the Pro79-Pro92 loop showed 0.53 Å root mean-square deviation for 170 of 219 α-carbons (13,14). Accordingly, TF might seem to be a rigid receptor that undergoes no major conformational changes on binding VIIa.
However, we reasoned that because allosteric linkage requires thermodynamic reciprocity (15), VIIa binding would likely alter functionally important structural dynamics of sTF at regulatory interfaces, including residues in the Pro79-Pro92 loop (Fig. 1 C). In particular, Eigenbrot et al. (16) in their analysis of the structure of zymogen VII, noted that if free VIIa were to have a zymogen-like (VII-like) conformation, complex formation with TF would not be favored due to steric clash of α-helix residues Arg162 and Leu163 in the catalytic domain of VII(a) (chymotrypsin numbering convention) with sTF residues Glu91, Pro92, and Tyr94. These five residues are among those whose structural rearrangement might have a role in allosteric regulation of VIIa activity. However, the dynamic changes in sTF that regulate binding to VIIa are not understood because critical residues are missing or unresolved in available crystal structures.
We use time-resolved fluorescence anisotropy to measure global and segmental dynamics reported by fluorescent probes covalently bound to mutants sTF-E84C and sTF-T121C. In sTF-E84C, Glu84 was replaced with Cys, which was then site-specifically modified with fluorescent probes. Because Glu84 resides in the disordered loop located between Pro79 and Pro92, the probe was expected to report change in flexibility of the disordered region of sTF after binding to VIIa. Also, it has been shown previously that mutation of Glu84 to Ala does not significantly alter either the affinity for VIIa or the catalytic properties of the complex (17,18). In the second mutant, sTF-T121C, Thr121 was modified with Cys, which was then site-specifically modified with a fluorescent probe. Because residue Thr121 is located at the opposite end of the extracellular domain in a region with atoms that have average thermal factors, a probe in this position would be expected to exhibit little or no segmental flexibility. Also, it has been shown previously that the mutant T121A has wild-type functional properties (18). The time-resolved fluorescence anisotropy results provide the first, to our knowledge, documentation that significant dynamic changes occur in sTF when it binds to VIIa. Thus, formation of the cofactor-enzyme complex not only causes changes in conformation and dynamics of VIIa, as shown previously (9), but also causes reduced segmental flexibility in the Pro79-Pro92 loop of sTF. The linkage between VIIa-sTF association and sTF loop dynamics indicates that decreased entropy in the sTF disordered loop contributes to the thermodynamics of the protein-protein interactions involved in the allosteric control of VIIa activation.
Materials and Methods
Reagents
The dyes 5-({[(2-iodoacetyl)amino]ethyl}amino)naphthalene-1-sulphonic acid (IAEDANS) and fluorescein 6-iodoacetamide (F6IA) were purchased from Invitrogen (Carlsbad, CA). Chromozym tPA was purchased from Roche Applied Science (Indianapolis, IN). All other chemicals were reagent grade.
Proteins
Human recombinant VIIa was a generous gift from Novo Nordisk (Denmark). Wild-type sTF was expressed and purified as described previously (19–21). The vector for bacterial expression of sTF, p6TF219, was a gift from W. Konigsberg (Yale University). Protein concentrations were determined by absorbance at 280 nm using molar extinction coefficients of 58,400 cm−1 M−1 for VIIa and 37,450 cm−1 M−1 for sTF and its mutant variants (20). The following oligonucleotides were used for mutation of Glu84 or Thr121 to Cys in p6TF219 to generate the single-site mutants E84C-p6TF219 (E84C) and T121C-p6TF219 (T121C), respectively; the triplet base changes coding are shown in lowercase:
-
(1)
E84C, 5′-CAGGGAATGTGtgcAGCACCGGTTC-3′; and
-
(2)
T121C, 5′-GAGTTTTGAACAGGTGGGAtgcAAAGTGAATGTGACCGTA-3′.
Mutant T121C was expressed and purified in the same manner as wild-type sTF. Purification of E84C, however, followed a modified protocol. For wild-type sTF, the first chromatography was cation-exchange on S-Sepharose SP (pH 5.2, 25 mM sodium acetate buffer) and the second was anion-exchange on Q-Sepharose XL (pH 7.5, 25 mM Tris-HCl, 0.2 mM EDTA buffer). For E84C, the order of chromatography was reversed, and the cation-exchange step was buffered at pH 5.4. E84C tended to precipitate at pH 5.2, probably as a result of an increase in isoelectric point due to the loss of the carboxyl side chain; the small increase in pH improved the solubility of this mutant significantly. The salt gradients were as described previously (20). Protein purification was confirmed by SDS-PAGE as well as by matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry.
Labeling of Cys mutants
The single-Cys sTF mutants tended to form intermolecular dimers, particularly E84C. Before labeling (0.1 M phosphate buffer, pH 7.0), dimers were converted to monomers by reduction for 1 h with 1 mM β-mercaptoethanol (ME). Then probe was added (either IAEDANS or F6IA at 4–5 mM final concentration), and the labeling reaction proceeded overnight at 4°C. Protein and unreacted dye were then separated by gel filtration on a Sephadex G25 column eluted with a pH 7.0 buffer (50 mM HEPES, 0.1 M NaCl, 1 mM NaN3). The eluted protein was concentrated by centrifugation using Amicon (Millipore, Billerica, MA) Ultra-4 filters (10K MWCO). The labeling efficiency was calculated from absorption spectra, as described below, by assuming that the extinction coefficient of either probe at its lowest energy maximum was unaffected by conjugation. To determine the probe contribution to the absorbance of the dye-conjugated protein at 280 nm, we prepared ME adducts of each dye in the buffer used for gel filtration; the adduct absorption spectra were similar to those of the unreacted dyes. Then the extinction coefficients at 280 nm were calculated from the ratio of the adduct absorbance at 280 nm to its absorbance at its lowest energy maximum, using the adduct extinction coefficient at that wavelength. Accordingly, based on the absorption maximum of the IAEDANS adduct at 337 nm, using an extinction coefficient of 5800 M−1 cm−1 (22), the calculated extinction coefficient of the adduct at 280 nm is 1400 M−1 cm−1. Based on the absorption maximum of the F6IA adduct at 491 nm, using an extinction coefficient of 82,000 M−1 cm−1 (Molecular Probes, Invitrogen), the calculated extinction coefficient at 280 nm is 10,700 M−1 cm−1. These values were used to account for the contribution of the dye to the absorbance of the dye-conjugated protein at 280 nm.
In using site-specific Cys mutants to introduce sulfhydryl-reactive fluorescence probes, we were concerned that sTF has two intramolecular disulfide bonds that might be reactive under the labeling conditions. However, examination of the fluorescent protein bands on nonreducing SDS PAGE before staining with Coomassie Blue showed that only mutant monomer bands were labeled; bands corresponding to both remaining mutant dimers were nonfluorescent. We subjected wild-type protein to the same labeling protocol, and the protein band was also nonfluorescent. The monomer labeling of the Cys mutants was always substoichiometric (50–80% labeling). The reaction-site specificity was confirmed by matrix-assisted laser desorption/ionization mass spectrometry of the peptides generated by trypsin digestion.
Functional assay of cofactor affinity
A particular mutation can change the binding of cofactor to VIIa. It can also change the enzymatic activity of the complex. The apparent equilibrium dissociation constant and the maximum velocity for conversion of substrate to product reflect these effects, respectively. The apparent dissociation constant and maximum velocity for each cofactor:VIIa complex were determined by measuring VIIa activity in an amidolytic assay as a function of increasing concentrations of wild-type or mutant cofactor. The VIIa concentration used was 15 nM, which is ∼10-fold greater than the dissociation constant for unmodified sTF and active-site inhibited VIIa, determined by pressure perturbation (20), or ∼ 3-fold greater than the dissociation constant for unmodified sTF and VIIa, determined by surface plasmon resonance (23). The concentration range of wild-type or mutant sTF was 0–120 nM. The assay was carried out in a pH 7.4 buffer (25 mM Hepes, 100 mM NaCl, 5 mM CaCl2, 0.1% w/v bovine serum albumin) with 1 mM Chromozym t-PA as substrate. Substrate hydrolysis at 30°C was monitored by the absorbance increase at 405 nm using a SPECTRA Max M2 plate-reader (Molecular Devices, Toronto, Canada). The velocity observed, Vobs, as a function of cofactor concentration, reflects the concentration of cofactor-VII complex:
| (1) |
where kcat is the catalytic rate constant. A maximum velocity, Vmax cofactor, is achieved when VIIa is saturated with cofactor. Then the expression that defines the equilibrium concentration of the cofactor:VIIa complex is:
| (2) |
where [VIIao] is the total VIIa concentration. Apparent dissociation constants, Kd, were obtained by least squares analysis of the titration data using the appropriate root of the quadratic equation:
| (3) |
where [VIIao] is the total VIIa concentration and [cofactoro] is the total cofactor concentration at each point in the titration. Comparison of Vmax mutant to Vmax sTF reflects the catalytic efficiency of each mutant complex relative to the wild-type sTF complex.
Time-resolved fluorescence anisotropy
Time-resolved fluorescence anisotropy measurements were carried out by time-correlated single-photon counting (TCSPC), using the FLASC 1000 sample chamber (Quantum Northwest, Liberty Lake, WA), which has a unique T format for simultaneous detection of horizontal (H), vertical (V) and variable polarization components of the emission. A beam-splitting Glan-Thompson polarizer (Karl Lambrecht, Chicago, IL) separates the H and V components. This optical arrangement allows simultaneous detection of up to three polarization components of the emission—typically H, V and magic angle—by separate photomultipliers, which assures collection of decay curves under identical excitation conditions. Pulsed excitation was either at 360 nm and a repetition rate of 4.7 MHz from a frequency-doubled ps Mira 900 Ti:Sapphire laser (Coherent, Santa Clara, CA) for IAEDANS conjugates or at 470 nm and a repetition rate of 5.0 MHz from a LDH-P-C 470 laser diode (PicoQuant, Berlin, Germany) for F6IA conjugates. The H and V emission components were isolated through matched bandpass filters, using 480/40 nm (Chroma, Rockingham VT) or 495/25 nm (Andover, Salem NH) for IAEDANS conjugates and either 525/50 nm (Chroma, Rockingham VT) or 530/25 nm (Andover, Salem, NH) for F6IA conjugates. The V and H 162decay curves were collected for equal lengths of time, using the TimeHarp 200 PCI board (PicoQuant, Berlin), until 4 × 104 counts were obtained at the maximum of the V curve. The timing resolution was either 35 ps/channel or 70 ps/channel.
The time-resolved anisotropy, r(t), is given by
| (4) |
where IVV(t) and IVH(t) represent the vertical and horizontal decays, respectively, obtained using vertical excitation. The denominator of Eq. 4 is the total intensity decay, I(t),
| (5) |
where τi is the lifetime and αi is the amplitude of the ith component in the intensity decay, I(t).
The anisotropy decay for an immobile fluorophore attached to a rigid, general ellipsoid molecule is described by the sum of five exponential terms (24–26),
| (6) |
where the preexponential factors, βj, are trigonometric functions of the angles between the excitation and emission transition dipole moments of the probe and the symmetry axes of the ellipsoid (27), and the sum of βj is the limiting anisotropy at zero time, r0. The rotational correlation times, ϕj, depend on the size and shape of the molecule to which the probe is attached as well as the temperature and viscosity of the surrounding medium. Calculations of the relative values of rotational correlation times for ellipsoids of varying ratios show that two pairs of correlation times are essentially degenerate and, as a result, three correlation times at most might be resolved (28). However, excluding local probe motions, only a single exponential correlation time is observed for most proteins. The rotational correlation time at early times in the decay is a harmonic mean (29),
| (7) |
that depends on the orientation of the probe with respect to the molecular axes as well as the size and shape of the protein. This approximation assumes that 〈ϕ〉, also referred to as the global rotational correlation time (ϕ global), is longer than the average lifetime, 〈τ〉, which is an intensity-weighted average (30,31),
| (8) |
If there is segmental motion in a region of the protein containing the probe, there will be additional depolarization of the fluorescence, which will produce a component in the anisotropy decay, ϕsegmental, that is shorter than ϕglobal. In this case, the appropriate equation for the anisotropy decay is
| (9) |
where γ is a weighting factor between 0 and 1. Because anisotropy decay data are typically fit as a sum of exponentials, when there is segmental motion, the decay law becomes
| (10) |
where pshort = γ and plong = 1 − γ. Accordingly,
| (11) |
The individual vertical and horizontal decay curves, IVV(t) and IVH(t), respectively, were fit simultaneously, using the analysis software package FluoFit Pro V4.2.1 (PicoQuant, Berlin) according to the following relationships:
| (12a) |
| (12b) |
where r∞ is the anisotropy at infinite time (30), and is a factor, obtained using horizontal excitation, that corrects for the difference in the efficiencies of the V and H detection channels; under ideal conditions G = 1 (29,32,33).
Results
Cofactor activities and VIIa-binding affinities of sTF-E84C and sTF-T121C
The cofactor activities of wild-type sTF and the single-Cys mutants sTF-E84C and sTF-T121C were compared using an amidolytic-based assay, as described in Materials and Methods. Under the assay conditions, the catalytic efficiency as a function of cofactor concentration reflects directly the degree of saturation of VIIa by cofactor. This yielded apparent Kd values at 30°C of 2, 7, and 6 nM (the estimated uncertainty is Kd ÷/× 2) for wild-type sTF, sTF-E84C, and sTF-T121C, respectively. At cofactor concentrations where VIIa was saturated, the mutants stimulated hydrolysis of the peptide substrate Chromozyme-tPA to the same extent as wild-type sTF (the estimated uncertainty is Vmax ± 20%). Accordingly, although the mutants had slightly decreased affinities for VIIa, the catalytic efficiencies of the wild-type and mutant complexes were indistinguishable. In addition, covalent modification of sTF-E84C and sTF-T121C with either IAEDANS or F6IA affected neither the equilibrium dissociation constants nor the catalytic efficiencies of the cofactor:VIIa complexes. Thus, covalent modification with the fluorescence probes had no impact on functional properties.
Fluorescence emission spectra and lifetimes of IAEDANS-labeled sTF-E84C and sTF-T121C and their complexes with VIIa
To investigate the dynamics of the Pro79-Pro92 loop in sTF alone and in complex with VIIa, we selected the Cys-reactive probe IAEDANS. Thiol adducts of IAEDANS have emission spectra and lifetimes that are sensitive to the local environment and the average fluorescence lifetime ranges from 15–20 ns at ambient temperature (22). This lifetime range is appropriate because it is sensitive to the correlation times expected for segmental dynamics of a loop (a few nanoseconds) and to the global motions of sTF unbound (tens of nanoseconds) and complexed with VIIa (50–150 ns, depending on temperature). For example, at 25°C, the global correlation time reported for sTF-E84C labeled with Alexa-488, whose 〈τ〉 is ∼4 ns, is ∼15 ns (9,34,35) and that for VIIa active-site labeled with DANSYL-d-Phe-l-Phe-l-Arg chloromethyl ketone, whose 〈τ〉 is ∼11 ns, is ∼100 ns (9,34,35). In addition, IAEDANS has been used previously by others (36–39) as a steady-state anisotropy probe to investigate the contact regions between other sTF Cys mutants and VIIa. The two sTF Cys mutants described here, however, are at locations that do not make direct contact with VIIa. Therefore, when VIIa binds, any changes in the local environments of the fluorescence probes bound to sTF-E84C or sTF-T121C are expected to be due to indirect perturbations.
The fluorescence emission maxima of the IAEDANS-labeled sTF mutants E84C-AEDANS and T121C-AEDANS are 497 and 492 nm, respectively, (pH 7.0, 0.1 M NaCl, 1 mM NaN3, 50 mM HEPES buffer); the precision of the peaks is ±1 nm. For comparison, the emission maximum for the IAEDANS ME adduct, ME-AEDANS, is at 497 nm in the pH 7.0 HEPES buffer and 458 nm in 95% ethanol, in close agreement with the wavelengths reported by Hudson and Weber (22). Thus, the E84C IAEDANS adduct seems to be fully solvent exposed whereas the T121C IAEDANS adduct seems to be in a somewhat less polar environment. Addition of a threefold excess of VIIa (the μM concentrations of cofactor and presence of 5 mM calcium assure saturation of the cofactor) had no significant effect on the emission of T121C-AEDANS. However, saturation with VIIa caused a blue shift in the emission peak of E84C-AEDANS to ∼490 nm, indicating a change in the local environment of the probe. This suggests that VIIa binding induces a conformational rearrangement of the Pro79-Pro92 loop, and this rearrangement alters the local environment of Glu84, which resides within the loop.
From 5°–35°C, the average fluorescence lifetimes, 〈τ〉, varied from 22.4 to 20.9 ns for sTFT121C-AEDANS and 15.7 to 13.7 ns for E84C-AEDANS. Consistent with its lack of effect on emission wavelength of T121C-AEDANS, addition of VIIa also had no effect on 〈τ〉, indicating that binding of VIIa does not significantly alter the local environment of T121C-AEDANS. By contrast, addition of VIIa to E84C-AEDANS was accompanied by about a 3 ns increase in 〈τ〉. Consistent with the blue shift in the emission spectrum of E84C-AEDANS observed on addition of VIIa, the increase in 〈τ〉 indicates a VIIa-induced conformational rearrangement in the Pro79-Pro92 loop that changes the local environment of the IAEDANS adduct of E84C.
Anisotropy decays of IAEDANS-labeled sTF-E84C and sTF-T121C: unconstrained analyses
Fitting the anisotropy decays of the IAEDANS-labeled mutants for a single correlation time, which is the simplest hydrodynamic model, yielded unsatisfactory statistical parameters—reduced χ2 value, residuals, and autocorrelation of the residuals—indicating that the probe on both mutants had significant local and/or segmental motion. This was most evident for E84C-AEDANS: the values for the single-fit correlation times, ϕ, and the limiting anisotropy values, r0, were much smaller than those recovered for T121C-AEDANS, and the χ2 values were 2.0, or larger, versus ∼1.4 for T121C-AEDANS. For example, at 15°C, ϕE84C ∼7 ns whereas ϕT121C ∼21 ns, and r0,E84C ∼0.16 whereas r0,T121C ∼0.22. For comparison, the anisotropy decay of ME-AEDANS in glycerol at 15°C, yielded ϕ = 136 ns and r0 = 0.318. This value of r0 is comparable to the frozen anisotropy r = 0.320, calculated from the steady-state polarization P = 0.414 obtained by Hudson and Weber (22) for AEDANS in 1:2 propanediol at −55°C with excitation at 360 nm (from their Fig. 4); r = 2P/(3 − P).
A double exponential decay law for the time-resolved anisotropy gave statistically better fits for both mutants, particularly E84C-AEDANS. This was reflected by 15–20% reductions in χ2 for the T121C data sets versus a twofold, or greater, reduction in χ2 for the E84C data sets. The long correlation times for T121C-AEDANS increased by <5%, whereas the long correlation times recovered from the E84C data sets increased by a factor of two, or more. Also, the fitting statistics for the E84C data were improved by inclusion of an r∞ term (Eq. 12), which was not the case for the T121C data; the r∞ term eliminated a systematic offset in the residuals for the V and H decay curves of the E84C data sets. We suspected the anisotropy offset—typically in the range 0.01–0.02—might be the result of sample aggregation. This was supported by the observation that after IAEDANS-labeled E84C samples were maintained at higher temperatures (i.e., 35°C) for extended periods of time (several hours), the systematic offset in the residuals increased, consistent with formation of higher-order aggregates driven by hydrophobic interaction. When the r∞ term was included in the analyses, the long correlation times obtained were self-consistent although shorter than those obtained for T121C-AEDANS.
The uncertainties for the long correlation times of E84C-AEDANS and T121C-AEDANS, determined at the 95% confidence limit using the support plane-method (40), were correlated with the relative fluorescence intensity contributions of the long correlation times of the mutants. For example, at 25°C, ϕ long, T121C = 19.3 (+2.0, −1.6) ns was associated with 97% of the fluorescence intensity, whereas ϕ long, E84C = 16.5 (+10.0, −4.5) ns was associated with 74% of the fluorescence intensity. The same was observed for uncertainties of the relative amplitudes (pj = βj/Σβj) of the long correlation times: at 25°C, plong, T121C = 0.724 (+0.021, −0.024), whereas plong, E84C = 0.400 (+0.061, −0.058).
The short correlation times recovered from analyses of the anisotropy data as double exponential were in the range of 1–2 ns for both mutants. The short correlation times and relative amplitudes were least well determined for T121C; these parameters were associated with less than 5% of the fluorescence intensity. By contrast, in the case of E84C-AEDANS from 5–35°C these parameters were associated with 25–50% of the fluorescence intensity. For example, at 25°C, ϕshort, T121C = 0.9 (+0.7, −0.5) ns and pshort, T121C = 0.276 (+0.166, −0.064), whereas ϕshort, E84C = 1.9 (+0.5, −0.5) ns and pshort, E84C = 0.600 (+0.042, −0.054).
Global analysis of anisotropy decays of sTF-E84C-AEDANS and sTF-T121C-AEDANS
The similarity in the long correlation times obtained from the unconstrained analyses of E84C-AEDANS and T121C-AEDANS suggest that the global harmonic mean rotational correlation times for the two sTF mutants are indistinguishable within the certainty of the data. In addition, the results indicate that the local motions of the AEDANS probe on T121C had an insignificant influence on determination of the long correlation times, which correspond to the global motions of the proteins in solution. This suggests that a global analysis of the fluorescence anisotropy decays of the IAEDANS-labeled mutants T121C and E84C, with the long correlation times fit in common as the global constraint and the short rotational correlation times fit as independent parameters, would provide better resolution of the segmental motions of the AEDANS probe on E84C. This resolution of segmental motions would help us determine how association with VIIa might affect the dynamics of the Pro79-Pro92 loop.
The results from global analyses of data obtained for temperatures from 5–35°C are given in Table 1; the results at each temperature are from a separate global analysis. In all cases, the global fits were statistically indistinguishable from the unconstrained fits for two correlation times. In the global analyses, the long correlation times remained essentially the same for T121C-AEDANS as those obtained from the unconstrained analyses, indicating that the time constants associated with the global motions of this mutant are well determined. By comparison, the long correlation times for E84C-AEDANS were 15–20% longer than those obtained from the unconstrained analyses, indicating that the time-constants associated with the global motions of this mutant are not as well determined in the unconstrained analyses. Depending on temperature, this is likely due to 75% to 50% of the fluorescence intensity being associated with the long correlation times of E84C-AEDANS as compared to 95%, or more, of the fluorescence intensity being associated with the long correlation times of T121C-AEDANS. Thus, the results of the global analyses of the IAEDANS-labeled proteins are consistent with the expectation that the global motions of the sTF mutants would be similar, if not indistinguishable.
Table 1.
Global analyses of the long correlation time of AEDANS-labeled mutants sTF-E84C and T121C
| Mutant | °C | η (cp) | χ2 | β1/Σβj∗ | ϕ1 (ns)† | β2/Σβj∗ | ϕ2 (ns)† | 〈r〉 | r0 |
|---|---|---|---|---|---|---|---|---|---|
| E84C | 5 | 1.543 | 1.23 | 0.50 (0.81) | 36.8 (−3.7, 4.3) | 0.50 (0.19) | 2.9 (−0.4, 0.5) | 0.121 | 0.245 |
| T121C | 1.17 | 0.80 (0.96) | 36.8 (−3.7, 4.3) | 0.20 (0.04) | 2.0 (−0.7, 0.9) | 0.146 | 0.255 | ||
| E84C | 15 | 1.157 | 1.26 | 0.43 (0.73) | 26.3 (−2.7, 3.6) | 0.57 (0.27) | 2.7 (−0.4, 0.5) | 0.098 | 0.228 |
| T121C | 1.25 | 0.81 (0.96) | 26.3 (−2.7, 3.6) | 0.19 (0.04) | 1.9 (−0.9, 1.3) | 0.129 | 0.248 | ||
| E84C | 25 | 0.905 | 1.27 | 0.39 (0.74) | 19.2 (−1.7, 2.6) | 0.61 (0.26) | 1.9 (−0.3, 0.4) | 0.081 | 0.219 |
| T121C | 1.12 | 0.72 (0.96) | 19.2 (−1.7, 2.6) | 0.28 (0.04) | 0.9 (−0.5, 0.9) | 0.109 | 0.262 | ||
| E84C | 35 | 0.731 | 1.26 | 0.35 (0.68) | 15.1 (−1.3, 1.7) | 0.65 (0.32) | 1.7 (−0.3, 0.3) | 0.064 | 0.203 |
| T121C | 1.24 | 0.75 (0.96) | 15.1 (−1.3, 1.7) | 0.25 (0.04) | 1.0 (−0.9, 0.8) | 0.097 | 0.252 |
Global analyses with the long correlation times, ϕ1, fit as the common parameter and the short correlation times, ϕ2, fit as independent parameters at a timing resolution of 70 ps/channel. The values for the viscosity, η, at each temperature were calculated from the values for water given in the 52nd edition of the CRC Handbook of Chemistry and Physics (1971–1972), using 1.016, the relative viscosity at 20°C of 0.155 M NaCl, as an approximate correction for the concentrative properties of the buffer.
The relative fluorescence intensity contribution of each anisotropy component is indicated within the parenthesis; β1 and β2 are the amplitudes of the correlation times, r0 is the limiting anisotropy, and 〈 r 〉 is the steady-state anisotropy calculated from the time-resolved parameters.
Upper and lower 95% confidence limits, calculated by the support-plane method (40), are indicated within the parenthesis, and χ2 is the reduced chi-squared statistic.
The useful time range for measuring rotational correlation times may extend from ∼0.1 to 10 〈τ〉, depending on the difference between the correlation times, their relative amplitudes, and the intensity average fluorescence lifetime of the probe (34,41,42). Considering that 〈τ〉 = 13–16 ns for E84C-AEDANS, the short rotational correlation times resolved from E84C-AEDANS anisotropy decay data are close to 0.1 〈τ〉, near the lower limit for resolution. Therefore, to assure that AEDANS was a satisfactory probe for assessing the impact of VIIa binding on the Pro79-Pro92 loop dynamics of sTF, we prepared the single-isomer F6IA conjugate E84C-F6A because fluorescein dianion has a fluorescence lifetime of ∼4 ns (43,44), which provides a closer match to the short correlation times observed for E84C. As expected, unconstrained fits of the anisotropy decay of E84C-F6A for two correlation times yielded anisotropy parameters close to those previously obtained for E84C labeled with Oregon Green 488 iodoacetamide, which had 〈τ〉 = 3.9 ns (34). When the E84C-F6A data were analyzed with the long correlation time fixed to the values obtained from the global analyses of the IAEDANS-labeled mutants, the χ2 values increased by <5%, indicating that the results were statistically indistinguishable. In addition, the short correlation times recovered were similar to those obtained for the IAEDANS conjugate. To test whether the two probes were also providing the same information about the relative amplitude of the short correlation times, the data sets from IAEDANS and F6IA-labeled E84C were evaluated by a global analysis in which the short rotational correlation times were fit as common parameters, the long correlation time at each temperature was fixed at the values obtained in the previous global analyses of T121C-AEDANS and E84C-AEDANS (Table 1), and the relative amplitudes, pshort = βshort/Σβj, were fit as independent parameters. The results of these analyses, given in Table 2, confirm that both probes report similar pshort values, which increase for both F6A and AEDANS as the temperature increases, consistent with increased probe mobility. Also consistent with increased probe mobility, the short correlation time decreases with increasing temperature.
Table 2.
Global analysis of the short correlation time of F6A- and AEDANS-labeled sTF-E84C
| Probe | °C | η (cp) | χ2 | β1/Σβj | ϕ1 (ns) | β2/Σβj | ϕ2 (ns) | 〈r〉 | r0 |
|---|---|---|---|---|---|---|---|---|---|
| AEDANS | 5 | 1.543 | 1.24 | 0.52 (−0.04, 0.03) | 36.8 | 0.48 (−0.03, 0.03) | 2.4 (−0.2, 0.3) | 0.120 | 0.252 |
| F6A | 1.14 | 0.53 (−0.02, 0.01) | 36.8 | 0.47 (−0.02, 0.02) | 2.4 (−0.2, 0.3) | 0.182 | 0.274 | ||
| AEDANS | 15 | 1.157 | 1.25 | 0.46 (−0.03, 0.03) | 26.3 | 0.54 (−0.03, 0.03) | 2.0 (−0.2, 0.2) | 0.099 | 0.242 |
| F6A | 1.12 | 0.50 (−0.02, 0.02) | 26.3 | 0.50 (−0.02, 0.02) | 2.0 (−0.2, 0.2) | 0.160 | 0.261 | ||
| AEDANS | 25 | 0.905 | 1.24 | 0.41 (0.77) | 19.2 | 0.59 (0.23) | 1.7 (−0.2, 0.2) | 0.080 | 0.225 |
| F6A | 1.12 | 0.46 (0.70) | 19.2 | 0.54 (0.30) | 1.7 (−0.2, 0.2) | 0.139 | 0.244 | ||
| AEDANS | 35 | 0.731 | 1.14 | 0.36 (−0.04, 0.04) | 15.1 | 0.64 (−0.05, 0.05) | 1.5 (−0.2, 0.2) | 0.065 | 0.212 |
| F6A | 1.10 | 0.44 (−0.02, 0.02) | 15.1 | 0.56 (−0.03, 0.03) | 1.5 (−0.2, 0.2) | 0.117 | 0.249 |
Global fit of the short correlation time, ϕ2, from the two probes with the long correlation time, ϕ1, fixed at the values reported in Table 1; the timing resolutions of the AEDANS and 6F data sets were 70 and 35 ps per channel, respectively. The lower and upper 95% confidence limits, calculated by the support-plane method (40), are reported within the parenthesis. The other parameters are defined in the legend for Table 1.
Anisotropy decays of IAEDANS-labeled sTF-E84C and sTF-T121C bound to VIIa
Unconstrained analyses of the time-resolved anisotropy data from the T121C-AEDANS:VIIa complex as a function of temperature were well described by single correlation times except at 35°C, where a small contribution of a short correlation time provided slightly better fits (<10% reduction in χ2). Unconstrained analyses of data for the E84C-AEDANS:VIIa complex required two correlation times at all temperatures to obtain statistically satisfactory fits; single correlation time fits yielded χ2 values >2. Similar to the fits for the mutants in the absence of VIIa, the single correlation time fits for the T121C-AEDANS:VIIa complex yielded longer times than did those for the E84C-AEDANS:VIIa complex. However, like the global fits for the two mutants in the absence of VIIa, as shown in Table 3, global analyses in which the long correlation time was fit as a common parameter for the two mutants at each temperature and a second correlation time was included as an independent parameter for the E84C-AEDANS:VIIa data, yielded statistically satisfactory fits. These analyses yielded short correlation times similar to those observed in the absence of VIIa (compare Tables 1 and 2), but with a 40–50% decrease in the relative amplitude, pshort. Table 3 also includes results for analysis of the 35°C data where a second correlation time was included as an independent parameter for the T121C-AEDANS:VIIa complex. Consistent with the <10% reduction obtained in χ2, error estimates from the 95% confidence limits show that the shorter correlation time is not well defined for the T121C-AEDANS complex with VIIa.
Table 3.
Time-resolved fluorescence anisotropy of AEDANS-labeled sTF mutants bound to VIIa
| Mutant | °C | η (cp) | χ2 | β1/Σβj | ϕ1 (ns) | β2/Σβj | ϕ2 (ns) | 〈r〉 | r0 |
|---|---|---|---|---|---|---|---|---|---|
| E84C | 5 | 1.543 | 1.17 | 0.76 (−0.07, 0.07) | 145 (−5, 6) | 0.24 (−0.02, 0.02) | 3.6 (−0.7, 0.9) | 0.199 | 0.271 |
| T121C | 1.10 | 1.00 | 145 (−5, 6) | 0.211 | 0.241 | ||||
| E84C | 15 | 1.157 | 1.16 | 0.75 (−0.01, 0.01) | 106 (−4, 4) | 0.25 (−0.03, 0.03) | 2.8 (−0.6, 0.6) | 0.175 | 0.260 |
| T121C | 1.10 | 1.00 | 106 (−4, 4) | 0.192 | 0.232 | ||||
| E84C | 25 | 0.905 | 1.14 | 0.68 (−0.01, 0.01) | 78 (−3, 3) | 0.32 (−0.04, 0.04) | 2.3 (−0.5, 0.5) | 0.155 | 0.257 |
| T121C | 1.08 | 1.00 | 78 (−3, 3) | 0.177 | 0.225 | ||||
| E84C | 35 | 0.731 | 1.15 | 0.61 (−0.01, 0.02) | 56 (−2, 2) | 0.39 (−0.04, 0.04) | 2.0 (−0.3, 0.4) | 0.127 | 0.243 |
| T121C | 1.18 | 1.00 | 56 (−2, 2) | 0.158 | 0.217 | ||||
| E84C | 35 | 0.731 | 1.14 | 0.60 (−0.01, 0.01) | 61 (−3, 2) | 0.40 (−0.03, 0.04) | 2.2 (−0.4, 0.4) | 0.128 | 0.243 |
| T121C | 1.08 | 0.87 (−0.02, 0.02) | 61 (−3, 2) | 0.13 (−0.03, 0.29) | 1.9 (−1.9, 1.7) | 0.157 | 0.240 |
Global motions of IAEDANS-labeled sTF-E84C and sTF-T121C free and bound to VIIa
Our previous studies by analytical ultracentrifugation and time-resolved anisotropy (9,20,34) have shown that in the presence of calcium ion, sTF and VIIa form a high-affinity heterodimer, that VIIa and its complex with sTF are both highly asymmetric, and that VIIa looses its segmental flexibility when it binds sTF. To our knowledge, there have been no reports about segmental motions in sTF, whether alone or in complex with VIIa.
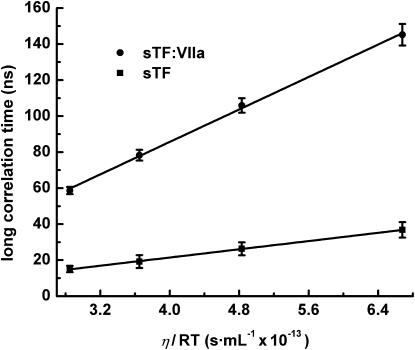
Fig. 2 shows the temperature and viscosity dependence of the long rotational correlation times obtained from the global fits of E84C-AEDANS and T121C-AEDANS alone and when bound to VIIa (data from Tables 1 and 3). The hydrodynamic behavior as a function of viscosity, η, and temperature, T, of both the free cofactor mutants and their complexes with VIIa is in accord with the Stokes-Einstein equation (45),
| (13) |
where V is the molecular volume of a spherical molecule and R is the gas constant. The values for the long correlation times, however, are about a factor of two longer than expected for a spherical, hydrated protein with Mr of 25 kDa, like sTF, or 75 kDa, like the complex with VIIa (9). For example, assuming a partial specific volume of 0.74 mL/g and average hydration of 0.28 g water/g protein, at 25°C spherical proteins with a molecular mass of 25 or 75 kDa would have rotational correlation times ϕsphere equal to 9.3 or 28 ns, respectively. However, the long correlation times from the global fits at 25°C are 19.2 and 75 ns, respectively, for the mutants and their complexes with VIIa. These long correlation times indicate greater surface to volume ratios than predicted for spherical, hydrated proteins of equivalent molecular mass.
Figure 2.
Long correlation times versus η /RT for sTF and the sTF:VIIa complex. Details are described in Results.
Using the relationships for hydrodynamics of ellipsoids developed by Small and Eisenberg (28), naive modeling of sTF and the complex with VIIa as prolate ellipsoids (a > b = c) yields axial ratios of ∼4:1 and 6:1, respectively. For example, these ratios predict that at 25°C, 〈ϕ〉 is equal to 17.8 ns and 74.6 ns for sTF and the sTF:VIIa complex, respectively. Considering the crystal structure models (10–13) as approximate prolates, the long axis of sTF would be ∼75 Å and the minor axes 20–25 Å yielding an axial ratio of ∼3:1, whereas the long axis of the sTF:VIIa complex would be ∼125 Å and the minor axes 30–35 Å yielding an axial ratio of ∼4:1. The hydrodynamic calculations are naive in part because they do not take into account the probe orientation relative to the molecular axes, which affect the preexponential factors of the rotational correlation times (Eq. 6). The calculations also tend to overestimate asymmetry because they do not take into account the contribution of the rugosity of protein surfaces, which contributes significantly to the friction coefficient (9,46). Taking these factors into account, in particular rugosity, the global hydrodynamics of the mutants and their complexes with VIIa obtained from the time-resolved anisotropy data are consistent with the overall asymmetries observed in the crystal structure models.
Segmental motions of sTF-E84C and sTF-T121C free and bound to VIIa
As seen in Fig. 1, A and B, sTF has two seven-stranded, β-sandwich, immunoglobulin-like domains—subdomains 1 and 2—joined by an extensive midriff region. Residue Glu84 is located in the Pro79-Pro92 loop of subdomain 1, which interacts with the EGF2 and catalytic domains of VIIa. Residue Thr121 is located in the turn between the first and second antiparallel β-strands of cofactor subdomain 2, which interacts primarily with the Gla and EGF1 domains of VIIa.
In the absence of VIIa, the anisotropy decays of E84C-AEDANS and T121C-AEDANS both were best fit by double exponentials, the shorter component having correlation times in the range 1–4 ns. This timescale is characteristic of correlation times that have been associated with segmental motions of the polypeptide backbone (47–50); correlation times in the range of 10 ns or longer have been associated with whole domain motions in multi-domain proteins (9,51–53). NMR studies indicate that local motions, which here would include the librational and rotational motions of the Cys side chain and the six-atom probe linker, would collectively result in an anisotropy decay component with a correlation time on the order of 100 ps or less (54). This is too fast for resolution in the fluorescence anisotropy decay data, and is consistent with the zero-time anisotropy values recovered for both mutants that were less (r0 = 0.20–0.26) than that determined for ME-AEDANS in glycerol (r0 = 0.318). The salient observation is that the relative amplitude of the short correlation time of E84C-AEDANS is about the same as the relative amplitude for the long correlation time (Table 1), indicating large-range segmental motion of the Pro79-Pro92 loop. The much smaller relative amplitude of the short correlation time of T121C-AEDANS is consistent with very little segmental motion of the polypeptide backbone of the antiparallel strands and the connecting turn containing Thr121.
When T121C-AEDANS is bound to VIIa, a single exponential provides an adequate fit of the anisotropy decay, indicating that VIIa binding essentially eliminates the intramolecular dynamics of the turn that joins the first and second antiparallel β-strands of subdomain 2. In the case of E84C, formation of the complex is accompanied by a twofold reduction in the relative amplitude of the short correlation time, indicating a decrease in the range of segmental motion of the Pro79-Pro92 loop, which is discussed below. There is also an increase in the time constant for the short correlation time; the time constants for the segmental motions of probe-labeled E84C calculated using Eq. 11 are 0.5–1 ns longer in the VIIa-bound state (Fig. 3). Although little change is observed in the time of the segmental motions in AEDANS-labeled E84C when bound to VIIa, the blue shift in the steady-state emission spectrum indicates that the probe's local environment is significantly perturbed, and the decrease in relative amplitude of the short correlation time indicates that the amount of segmental motion of the Pro79-Pro92 loop is reduced significantly.
Figure 3.
Segmental correlation times for sTF-E84-AEDANS free and bound to VIIa calculated from the short correlation times using Eq. 11. Details are described in Results.
Segmental flexibility can be considered in terms of restricted diffusion (55), and Kinosita et al. (56,57), have developed two ways for analyzing restricted diffusion using fluorescence anisotropy decay data. In one, a fluorophore wobbles uniformly within a cone (square-well potential). In the other, the probe distribution is Gaussian. Although restricted diffusion of a polypeptide loop in a protein is likely to be other than conical, the wobbling-cone model provides a simple but useful way to assess the impact of interactions with ligands on the segmental motions of domains and secondary structural elements (58–60). In the wobbling-cone model, the angle θ with respect to the symmetry axis of the cone can be estimated using the relationship
| (14) |
where r0 = rshort + rlong, the amplitudes of the short and long correlation times, respectively; alternatively, rlong/r0 = plong as defined in Eq. 10. Accordingly, from 5–35°C, the dynamics of E84C-AEDANS in the Pro79-Pro92 loop can be described by angles varying from 38–46° for the free state of the cofactor and by angles varying from 24–32° for the VIIa-bound state. This indicates first, that the cofactor interaction with the catalytic domain of VIIa reduces the conformational entropy of the Pro79-Pro92 loop, and second, that the loop entropy increases with temperature whether the cofactor is free or bound.
Discussion
Segmental flexibility, conformational change and VIIa activation
Kinetic studies by Nemerson and Gentry (7) showed that the enhancement of catalysis of VIIa was not solely due to assembly of the cofactor-enzyme-substrate ternary complex, TF:VIIa:X, on the membrane surface, but required TF as an essential enzyme activator for VIIa. This model implied that, as an essential cofactor, TF causes conformational changes in VIIa and these conformational changes make VIIa a substantially more efficient enzyme. Waxman et al. (9), determined by time-resolved fluorescence anisotropy and sedimentation velocity studies that when active-site-inhibited VIIa binds to sTF, there is loss of segmental flexibility in the enzyme's light chain and immobilization of the chymotrypsin-like catalytic heavy chain.
Higashi et al. (8), subsequently found that VIIa has greater affinity than VII for binding TF and that occupation of the active site of VIIa with inhibitors increased its affinity for TF, indicating allosteric interactions regulate activation of VIIa. In addition, inhibitors that caused larger increases in affinity provided greater protection from chemical modification for the α-amino group of the heavy chain, Ile16 (chymotrypsinogen numbering convention). The free α-amino group of Ile16 is generated by proteolytic cleavage between the EGF2 and protease domains, which converts VII to VIIa. The ability to form the salt bridge between the α-amino group of Ile16 and the carboxyl side chain of Asp194 is essential for stabilizing the active configuration of the catalytic triad (61). Based on these results, Higashi et al. (8), proposed that when VIIa is in the unbound state, the heavy chain retains zymogen-like features, and steric constraints prevent formation of the critical salt bridge between Ile16 and Asp194.
If free VIIa were to have a zymogen-like structure, the respective affinities of VII and uninhibited VIIa for TF should be very similar. In accord with this expectation, binding experiments with TF (62) and sTF (23), show that the affinity of VII for the cofactor is only slightly weaker than that of VIIa without an active-site inhibitor. Thus, the energetic cost associated with the structural and dynamic changes for conversion of zymogen VII to unbound enzyme VIIa is small; the energetic cost for essential activation is paid when VIIa binds to TF, and the protein-protein interactions in this complex favor a conformational state of VIIa that is catalytically efficient. According to the analysis of the zymogen structure by Eigenbrot et al. (16,63), many interactions at the binding interface between TF and VIIa may have allosteric influence on the active site of VIIa, including remodeling of the catalytic triad. This allosteric effect would be in accord with the finding that active-site transition-state inhibitors increase the affinity of VIIa for TF (8,64).
Allosteric linkage, cofactor loop dynamics, and VIIa activation
Reciprocity is required by the thermodynamics of allosteric linkage (15). However, the structural and dynamic changes affected by allosteric interactions between two proteins can occur in either protein or both. Because the crystal structures of sTF alone (10–12) and in complex with active-site-inhibited VIIa are similar and can be superimposed with a root mean-square deviation of 0.53 Å (13,14), it is possible that the conformational changes involved in essential activation of VIIa are confined to VIIa. However, as discussed by Eigenbrot et al. (16,63), in the zymogen conformation, if sTF were to maintain the same structure seen in the complex with active-site inhibited VIIa, VIIa residues Arg162 and Leu163 would have steric clash with TF residues Glu91, Pro92, and Tyr94 (Fig. 1 B). Furthermore, Kittur et al. (17) have shown that the sTF mutant E91A has an order of magnitude weaker affinity for VIIa than wild-type sTF, and its complex with VIIa has a twofold reduction in Km for conversion of IX to IXa and a modest reduction in both Km and kcat for conversion of X to Xa. Ruf et al. (18) have shown that the triple mutation of residues Glu84, Ser85, and Thr86 to Ala has little effect on the cofactor activity of sTF. Thus, the residues that have a direct role in the allosteric activation of VIIa are at the carboxyl terminal end of the Pro79-Pro92 loop. All of the loop residues are characterized by high thermal factors in the sTF structures by Harlos et al. (10), and Muller et al. (11,12); data for residues 87–90 are not reported in the latter structure. In the structure of the VIIa complex, residues 81–90 are absent because the sTF used for crystallization was subjected to limited subtilisin digestion. To our knowledge, there have been no previous reports concerning the structural dynamics of Pro79-Pro92 loop in the sTF:VIIa complex.
Summary
We report time-resolved anisotropy measurements of fluorescence probes covalently bound to the single Cys sTF mutants E84C and T121C, which show that interaction with uninhibited VIIa reduces segmental motions in the two subdomains of sTF: in subdomain 1, which interacts with the EGF2 and catalytic domains of VIIa, Glu84 resides in the Pro79-Pro92 loop; and in subdomain 2, which interacts with the Gla and EGF1 domains of VIIa, Thr121 is located in the turn between the first and second antiparallel β-strands (Fig. 1 A). Our results show that when sTF and VIIa associate, dynamic structural changes take place not only in enzyme VIIa but also in both subdomains of cofactor sTF. The results support the steric clash, proposed by Eigenbrot et al. (16,63), between subdomain 1 of sTF and zymogen-like VIIa, which would be resolved by conformational and dynamic changes during the allosteric, essential activation of VIIa. The anisotropy results show that the region that includes Thr121 has limited segmental flexibility in free sTF, which is eliminated on binding VIIa. The anisotropy of probes bound to E84C, however, show that the Pro79-Pro92 loop, which interacts with the catalytic domain of VIIa, has substantial segmental flexibility that is markedly reduced on binding to VIIa. The decrease in the motion of the Pro79-Pro92 loop that follows VIIa-sTF association indicates that entropy of the loop has a role in the thermodynamics of the protein-protein interactions that determine allosteric control of VIIa activation.
Acknowledgments
We thank Yale Nemerson, Thomas Laue, David Banner, William Konigsberg and Jolyon Jesty for discussions on the structural and functional interactions of TF, sTF and VIIa. We also thank William Konigsberg for providing the sTF encoding plasmid p6TF219 and William Laws for discussions on interpretation of time-resolved fluorescence anisotropy data. Finally, we thank Laurie Franklin for help with site-directed mutagenesis.
This research was supported by grant MCB-0517644 from the National Science Foundation and in part by grant RR-15583 (National Center for Research Resources, a component of the National Institutes of Health).
A preliminary account of this work was presented at the joint meeting of the Biophysical Society 52nd Annual Meeting and the 16th International Biophysics Congress of the International Union of Pure and Applied Biophysics (IUPAB), Feb. 2–6, 2008 (Long Beach, CA). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of National Science Foundation, National Center for Research Resources, National Institutes of Health, the Biophysical Society, or IUPAB.
References
- 1.Bach R.R. Initiation of coagulation by tissue factor. CRC Crit. Rev. Biochem. 1988;23:339–368. doi: 10.3109/10409238809082548. [DOI] [PubMed] [Google Scholar]
- 2.Bach R.R. Tissue factor encryption. Arterioscler. Thromb. Vasc. Biol. 2006;26:456–461. doi: 10.1161/01.ATV.0000202656.53964.04. [DOI] [PubMed] [Google Scholar]
- 3.Nemerson Y., Bach R. Tissue factor revisited. Prog. Hemost. Thromb. 1982;6:237–261. [PubMed] [Google Scholar]
- 4.Nemerson Y., Repke D. Tissue factor accelerates the activation of coagulation factor VII: the role of a bifunctional coagulation cofactor. Thromb. Res. 1985;40:351–358. doi: 10.1016/0049-3848(85)90270-1. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox J.N., Smith K.M., Schwartz S.M., Gordon D. Localization of tissue factor in the normal vessel wall and in the atherosclerotic plaque. Proc. Natl. Acad. Sci. USA. 1989;86:2839–2843. doi: 10.1073/pnas.86.8.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverberg S.A., Nemerson Y., Zur M. Kinetics of the activation of bovine coagulation factor X by components of the extrinsic pathway. Kinetic behavior of two-chain factor VII in the presence and absence of tissue factor. J. Biol. Chem. 1977;252:8481–8488. [PubMed] [Google Scholar]
- 7.Nemerson Y., Gentry R. An ordered addition, essential activation model of the tissue factor pathway of coagulation: evidence for a conformational cage. Biochemistry. 1986;25:4020–4033. doi: 10.1021/bi00362a006. [published erratum appears in Biochemistry 1987 Feb 10;26(3):974] [DOI] [PubMed] [Google Scholar]
- 8.Higashi S., Matsumoto N., Iwanaga S. Molecular mechanism of tissue factor-mediated acceleration of factor VIIa activity. J. Biol. Chem. 1996;271:26569–26574. doi: 10.1074/jbc.271.43.26569. [DOI] [PubMed] [Google Scholar]
- 9.Waxman E., Laws W.R., Laue T.M., Nemerson Y., Ross J.B.A. Human factor VIIa and its complex with soluble tissue factor: evaluation of asymmetry and conformational dynamics by ultracentrifugation and fluorescence anisotropy decay methods. Biochemistry. 1993;32:3005–3012. doi: 10.1021/bi00063a011. [DOI] [PubMed] [Google Scholar]
- 10.Harlos K., Martin D.M., O'Brien D.P., Jones E.Y., Stuart D.I. Crystal structure of the extracellular region of human tissue factor. Nature. 1994;370:662–666. doi: 10.1038/370662a0. [published erratum appears in Nature 1994 Oct 20;371(6499): 720] [DOI] [PubMed] [Google Scholar]
- 11.Muller Y.A., Ultsch M.H., de Vos A.M. The crystal structure of the extracellular domain of human tissue factor refined to 1.7 Å resolution. J. Mol. Biol. 1996;256:144–159. doi: 10.1006/jmbi.1996.0073. [DOI] [PubMed] [Google Scholar]
- 12.Muller Y.A., Ultsch M.H., Kelley R.F., de Vos A.M. Structure of the extracellular domain of human tissue factor: location of the factor VIIa binding site. Biochemistry. 1994;33:10864–10870. doi: 10.1021/bi00202a003. [DOI] [PubMed] [Google Scholar]
- 13.Banner D.W., D'Arcy A., Chene C., Winkler F.K., Guha A. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 14.Banner D.W. The factor VIIa /tissue factor complex. Thromb. Haemost. 1997;78:512–515. [PubMed] [Google Scholar]
- 15.Wyman J., Gill S.J. University Science Books; Mill Valley, CA: 1990. Binding and Linkage: Functional Chemistry of Biological Macromolecules. [Google Scholar]
- 16.Eigenbrot C., Kirchhofer D., Dennis M.S., Santell L., Lazarus R.A. The factor VII zymogen structure reveals reregistration of beta strands during activation. Structure. 2001;9:627–636. doi: 10.1016/s0969-2126(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 17.Kittur F.S., Manithody C., Morrissey J.H., Rezaie A.R. The cofactor function of the N-terminal domain of tissue factor. J. Biol. Chem. 2004;279:39745–39749. doi: 10.1074/jbc.M406628200. [DOI] [PubMed] [Google Scholar]
- 18.Ruf W., Schullek J.R., Stone M.J., Edgington T.S. Mutational mapping of functional residues in tissue factor: identification of factor VII recognition determinants in both structural modules of the predicted cytokine receptor homology domain. Biochemistry. 1994;33:1565–1572. doi: 10.1021/bi00172a037. [DOI] [PubMed] [Google Scholar]
- 19.Hasselbacher C.A., Rusinova E., Waxman E., Rusinova R., Kohanski R.A. Environments of the four tryptophans in the extracellular domain of human tissue factor: comparison of results from absorption and fluorescence difference spectra of tryptophan replacement mutants with the crystal structure of the wild-type protein. Biophys. J. 1995;69:20–29. doi: 10.1016/S0006-3495(95)79891-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waxman E., Ross J.B.A., Laue T.M., Guha A., Thiruvikraman S.V. Tissue factor and its extracellular soluble domain: the relationship between intermolecular association with factor VIIa and enzymatic activity of the complex. Biochemistry. 1992;31:3998–4003. doi: 10.1021/bi00131a015. [DOI] [PubMed] [Google Scholar]
- 21.Zemsky J., Rusinova E., Nemerson Y., Luck L.A., Ross J.B.A. Probing local environments of tryptophan residues in proteins: comparison of 19F nuclear magnetic resonance results with the intrinsic fluorescence of soluble human tissue factor. Proteins. 1999;37:709–716. [PubMed] [Google Scholar]
- 22.Hudson E.N., Weber G. Synthesis and characterization of two fluorescent sulfhydryl reagents. Biochemistry. 1973;12:4154–4161. doi: 10.1021/bi00745a019. [DOI] [PubMed] [Google Scholar]
- 23.Kelley R.F., Yang J., Eigenbrot C., Moran P., Peek M. Similar molecular interactions of factor VII and factor VIIa with the tissue factor region that allosterically regulates enzyme activity. Biochemistry. 2004;43:1223–1229. doi: 10.1021/bi035738i. [DOI] [PubMed] [Google Scholar]
- 24.Belford G.G., Belford R.L., Weber G. Dynamics of fluorescence polarization in macromolecules. Proc. Natl. Acad. Sci. USA. 1972;69:1392–1393. doi: 10.1073/pnas.69.6.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang T.J., Eisenthal K.B. Theory of fluorescence depolarization by anisotropic rotational diffusion. J. Chem. Phys. 1972;57:5094–5097. [Google Scholar]
- 26.Rigler R., Ehrenberg M. Fluorescence relaxation spectroscopy in the analysis of macromolecular structure and motion. Q. Rev. Biophys. 1976;9:1–19. doi: 10.1017/s0033583500002122. [DOI] [PubMed] [Google Scholar]
- 27.Barkley M.D., Kowalczyk A.A., Brand L. Fluorescence decay studies of anisotropic rotations of small molecules. J. Chem. Phys. 1981;75:3581–3593. [Google Scholar]
- 28.Small E.W., Isenberg I. Hydrodynamic properties of a rigid molecule: rotational and linear diffusion and fluorescence anisotropy. Biopolymers. 1977;16:1907–1928. doi: 10.1002/bip.1977.360160907. [DOI] [PubMed] [Google Scholar]
- 29.Lakowicz J.R. 3rd ed. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 30.Dale R.E., Chen L.A., Brand L. Rotational relaxation of the “microviscosity” probe diphenylhexatriene in paraffin oil and egg lecithin vesicles. J. Biol. Chem. 1977;252:7500–7510. [PubMed] [Google Scholar]
- 31.Förster T. Vandenhoeck and Ruprecht; Göttingen: 1951. Fluoreszenz Organischer Verbindungen. [Google Scholar]
- 32.Badea M.G., Brand L. Time-resolved fluorescence measurements. Methods Enzymol. 1979;61:378–425. doi: 10.1016/0076-6879(79)61019-4. [DOI] [PubMed] [Google Scholar]
- 33.Paoletti J., Le Pecq J.B. Corrections for instrumental errors in measurement of fluorescence and polarization of fluorescence. Anal. Biochem. 1969;31:33–41. doi: 10.1016/0003-2697(69)90238-3. [DOI] [PubMed] [Google Scholar]
- 34.Rusinova E., Tretyachenko-Ladokhina V., Vele O.E., Senear D.F., Ross J.B.A. Alexa and Oregon Green dyes as fluorescence anisotropy probes for measuring protein-protein and protein-nucleic acid interactions. Anal. Biochem. 2002;308:18–25. doi: 10.1016/s0003-2697(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 35.Waxman E. Mount Sinai School of Medicine of the City University of New York; New York: 1993. Thermodynamic and hydrodynamic behavior of the tissue factor:factor VII(a) complex. PhD dissertation. [Google Scholar]
- 36.Osterlund M., Owenius R., Carlsson K., Carlsson U., Persson E. Probing inhibitor-induced conformational changes along the interface between tissue factor and factor VIIa. Biochemistry. 2001;40:9324–9328. doi: 10.1021/bi010283n. [DOI] [PubMed] [Google Scholar]
- 37.Osterlund M., Owenius R., Persson E., Lindgren M., Carlsson U. Spectroscopic probing of the influence of calcium and the gla domain on the interaction between the first EGF domain in factor VIIa and tissue factor. Eur. J. Biochem. 2000;267:6204–6211. doi: 10.1046/j.1432-1327.2000.01693.x. [DOI] [PubMed] [Google Scholar]
- 38.Owenius R., Osterlund M., Lindgren M., Svensson M., Olsen O.H. Properties of spin and fluorescent labels at a receptor-ligand interface. Biophys. J. 1999;77:2237–2250. doi: 10.1016/S0006-3495(99)77064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owenius R., Osterlund M., Svensson M., Lindgren M., Persson E. Spin and fluorescent probing of the binding interface between tissue factor and factor VIIa at multiple sites. Biophys. J. 2001;81:2357–2369. doi: 10.1016/S0006-3495(01)75882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straume M., Frasier-Cadoret S.G., Johnson M.L. Least-squares analysis of fluorescence data. In: Lakowicz J.R., editor. Topics in Fluorescence Spectroscopy. Vol. 2. Plenum Press; New York: 1991. pp. 177–240. [Google Scholar]
- 41.Wahl P. Statistical accuracy of rotational correlation times determined by the photocounting pulse fluorimetry. Chem. Phys. 1977;22:245–256. [Google Scholar]
- 42.Wahl P. Measurement of Brownian correlation times by pulse fluorometry. NATO Adv. Sci. Inst. Ser., Ser. Life Sci. 1983;69:483–495. [Google Scholar]
- 43.Magde D., Rojas G.E., Seybold P.G. Solvent dependence of the fluorescence lifetimes of xanthene dyes. Photochem. Photobiol. 1999;70:737–744. [Google Scholar]
- 44.Martin M.M., Lindqvist L. Ph-dependence of fluorescein fluorescence. J. Lumin. 1975;10:381–390. [Google Scholar]
- 45.Einstein A. Theory of Brownian movement. Ann. Phys. 1906;19:371–381. [Google Scholar]
- 46.Teller D.C., Swanson E., de Haen C. The translational friction coefficient of proteins. Methods Enzymol. 1979;61:103–124. doi: 10.1016/0076-6879(79)61010-8. [DOI] [PubMed] [Google Scholar]
- 47.Berman H.A., Yguerabide J., Taylor P. Flexibility of the molecular forms of acetylcholinesterase measured with steady-state and time-correlated fluorescence polarization spectroscopy. Biochemistry. 1985;24:7140–7147. doi: 10.1021/bi00346a018. [DOI] [PubMed] [Google Scholar]
- 48.Fa M., Karolin J., Aleshkov S., Strandberg L., Johansson L.B. Time-resolved polarized fluorescence spectroscopy studies of plasminogen activator inhibitor type 1: conformational changes of the reactive center upon interactions with target proteases, vitronectin and heparin. Biochemistry. 1995;34:13833–13840. doi: 10.1021/bi00042a015. [DOI] [PubMed] [Google Scholar]
- 49.Hibbs R.E., Johnson D.A., Shi J., Taylor P. Structural dynamics of the acetylcholine binding protein: hydrodynamic and fluorescence anisotropy decay analyses. J. Mol. Neurosci. 2006;30:73–74. doi: 10.1385/JMN:30:1:73. [DOI] [PubMed] [Google Scholar]
- 50.Hibbs R.E., Radic Z., Taylor P., Johnson D.A. Influence of agonists and antagonists on the segmental motion of residues near the agonist binding pocket of the acetylcholine-binding protein. J. Biol. Chem. 2006;281:39708–39718. doi: 10.1074/jbc.M604752200. [DOI] [PubMed] [Google Scholar]
- 51.Hanson D.C., Yguerabide J., Schumaker V.N. Segmental flexibility of immunoglobulin G antibody molecules in solution: a new interpretation. Biochemistry. 1981;20:6842–6852. doi: 10.1021/bi00527a016. [DOI] [PubMed] [Google Scholar]
- 52.Hanson D.C., Yguerabide J., Schumaker V.N. Rotational dynamics of immunoglobulin G antibodies anchored in protein A soluble complexes. Mol. Immunol. 1985;22:237–244. doi: 10.1016/0161-5890(85)90156-7. [DOI] [PubMed] [Google Scholar]
- 53.Holowka D., Wensel T., Baird B. A nanosecond fluorescence depolarization study on the segmental flexibility of receptor-bound immunoglobulin E. Biochemistry. 1990;29:4607–4612. doi: 10.1021/bi00471a015. [DOI] [PubMed] [Google Scholar]
- 54.Li Z., Raychaudhuri S., Wand A.J. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yguerabide J. Nanosecond fluorescence spectroscopy of macromolecules. Methods Enzymol. 1972;26:498–578. doi: 10.1016/s0076-6879(72)26026-8. [DOI] [PubMed] [Google Scholar]
- 56.Kinosita K., Ikegami A., Kawato S. On the wobbling-in-cone analysis of fluorescence anisotropy decay. Biophys. J. 1982;37:461–464. doi: 10.1016/S0006-3495(82)84692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kinosita K., Kawato S., Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys. J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bayley P., Martin S., Browne P., Royer C. Time-resolved fluorescence anisotropy studies show domain-specific interactions of calmodulin with IQ target sequences of myosin V. Eur. Biophys. J. 2003;32:122–127. doi: 10.1007/s00249-002-0274-7. [DOI] [PubMed] [Google Scholar]
- 59.Kim S.J., Beard W.A., Harvey J., Shock D.D., Knutson J.R. Rapid segmental and subdomain motions of DNA polymerase beta. J. Biol. Chem. 2003;278:5072–5081. doi: 10.1074/jbc.M208472200. [DOI] [PubMed] [Google Scholar]
- 60.Schroder G.F., Alexiev U., Grubmuller H. Simulation of fluorescence anisotropy experiments: probing protein dynamics. Biophys. J. 2005;89:3757–3770. doi: 10.1529/biophysj.105.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oppenheimer H.L., Labouesse B., Hess G.P. Implication of an ionizing group in the control of conformation and activity of chymotrypsin. J. Biol. Chem. 1966;241:2720–2730. [PubMed] [Google Scholar]
- 62.Dickinson C.D., Ruf W. Active site modification of factor VIIa affects interactions of the protease domain with tissue factor. J. Biol. Chem. 1997;272:19875–19879. doi: 10.1074/jbc.272.32.19875. [DOI] [PubMed] [Google Scholar]
- 63.Eigenbrot C., Kirchhofer D. New insight into how tissue factor allosterically regulates factor VIIa. Trends Cardiovasc. Med. 2002;12:19–26. doi: 10.1016/s1050-1738(01)00139-6. [DOI] [PubMed] [Google Scholar]
- 64.Sorensen B.B., Persson E., Freskgard P.-O., Kjalke M., Ezban M. Incorporation of an active site inhibitor in factor VIIa alters the affinity for tissue factor. J. Biol. Chem. 1997;272:11863–11868. doi: 10.1074/jbc.272.18.11863. [DOI] [PubMed] [Google Scholar]
- 65.DeLano, W. L. 2007. MacPyMOL: A PyMOL-based molecular graphics application for MacOS X. DeLano Scientific, Palo Alto, CA. http://www.pymol.org).