Abstract
The p7 protein from hepatitis C virus is critical for the assembly and secretion of infectious virus, making it an attractive drug target. It is thought to be a viroporin with a demonstrated ion channel activity when reconstituted into planar lipid bilayers. Electron microscopy experiments suggest that p7 oligomers coexist as hexamers and heptamers. Proposed models of p7 oligomers assume the N-terminal helix to be the pore lining helix. Here, we demonstrate, via electrophysiology, that Cu2+ has an inhibitory effect on the p7 ion channel and that the amino acid responsible for this inhibition is one histidine in each monomer. This information coupled with the p7 sequence data suggests that the N-terminal helix of p7 does indeed form the transmembrane pore and that this histidine is pore-lining. The information will aid in the construction of oligomeric pore-models and the interpretation of electron microscopy data.
Hepatitis C virus (HCV) infects more than 170 million people worldwide and is one of the major causes of liver disease including chronic hepatitis, cirrhosis, and heptacellular carcinoma (1). No vaccine is available and the current treatment, pegylated interferon-α and ribavirin is expensive and ineffective in ∼50% of patients (2). Therefore, there is great desire to identify alternative drug targets. The virally encoded p7 protein, shown to be crucial for the secretion of infectious particles, is one such target (3,4).
The HCV p7 monomer is a hydrophobic protein of 63 amino acids (5) located at the junction of the structural and nonstructural (replicative) proteins of the HCV polyprotein. Two topologies for monomeric p7 have been proposed: a double membrane-spanning hairpin topology in the endoplasmic reticulum with the N- and C-termini facing the endoplasmic reticulum lumen (6), and an L-shaped form with the C-terminus facing the cytoplasm (7,8).
HCV p7 is capable of forming cation selective ion channels in planar lipid bilayers (9–12). In vitro HCV p7 ion channel activity can be inhibited by a range of compounds including amantadine, which also inhibits the viroporin M2 of the influenza A virus, and long alkyl chain iminosugar derivatives, such as N-nonyl-deoxynojirimycin (NN-DNJ), which are known to inhibit endoplasmic reticulum α glucosidase I and II as well as HCV p7 (11,13).
The oligomerization state of the channel has been under debate. Using electron microscopy, bacterially expressed GST-his-tagged and GST-FLAG-p7 monomers have been suggested to assemble into hexameric and heptameric ion channels, respectively (9,10). Molecular dynamics studies of the channel suggested a potential oligomerization of p7 monomers into a hexameric bundle, though other oligomerization states were not ruled out (14). Both the heptameric models of p7 proposed by Patargias et al. and Clarke et al. assume that the N-terminal helix of p7 lines the pore (9,14). In the absence of high resolution structural data, it is desirable to establish which residues may line the pore to help interpret low resolution electron microscopy data.
Gandhi et al. showed that M2 can be inhibited by Cu2+ but not by Mg2+ (15). This inhibition is thought to be due to the chelation of Cu2+ by histidine side chains which line the pore of M2, thus causing ion-channel block. Analysis of the p7 sequence from 26 representative strains (Fig. S1 in the Supporting Material) indicates that histidines are found in the N-terminal helix of the p7 protein. One strain, H77, possesses only one histidine at position 17 (Fig. 1 A). We thus postulated that if H77-p7 channels were inhibited by Cu2+, it would lend support to the hypothesis that the N-terminal helix lines the pore. Such information will be invaluable in the construction of future models.
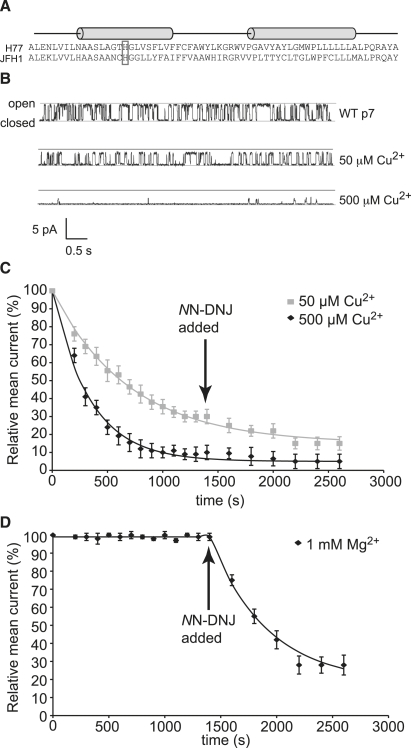
Figure 1.
(A) Sequences of HCV p7 protein from the H77 and JFH-1 strains. The gray box indicates H-17. Cylinders indicate predicted helical regions. (B) Typical channel recording traces. (C) Time course of inhibition of the wild-type H77 p7 ion channel protein by 50 μM (gray squares) and 500 μM (black diamonds) Cu2+. The symbols in this and the following figures show the recorded currents (mean ± SE for n = 10), and the lines show the currents fit with an optimal exponential decay constant using MATLAB (The MathWorks, Natick, MA). (D) Time course of inhibition by 1mM Mg2+. At t = 1400 s, 500 μM of NN-DNJ was added to confirm the presence of p7 ion channels.
Using chemically synthesized H77 p7 in black lipid membranes comprised of phosphatidylcholine and phosphatidylethanolamine lipids, we determined whether p7 ion channel activity could be inhibited by Cu2+ (see the Supporting Material for methods). Addition of 50 μM Cu2+ to the bathing solution reduces the relative mean current to ∼20% (Fig. 1, B and C. The mean conductance drops from 25.5 ± 1.8 pS to 13.9 ± 1.2 pS over this time period, and the open channel probability, Po, shifts from 0.33 ± 0.08 to 0.22 ± 0.05). When the concentration of Cu2+ was increased to 500 μM, the inhibition was even more pronounced (mean conductance = 7.2 ± 0.5 pS, Po = 0.13 ± 0.04).
Control experiments with 1mM Mg2+ were performed (Fig. 1 D) where the channels were confirmed to be insensitive to Mg2+ (mean conductance 24.4 ± 1.6 pS, Po = 0.31 ± 0.08). Evidence that the channels were comprised of p7 protein was provided by the addition of 500 μM NN-DNJ at t = 1400 s, where Mg2+ treated channels were inhibited substantially (Fig. 1 D). NN-DNJ was also added to the experiments with Cu2+ at t = 1400 s, but in this case no further inhibition was observed because by this point the maximum level of inhibition had already been obtained.
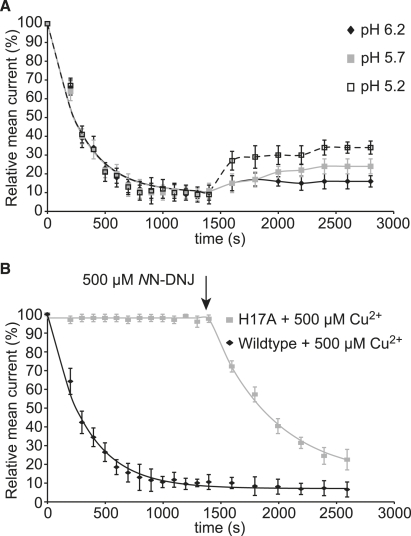
Gandhi et al. showed that Cu2+ binds less strongly to His-37 of influenza M2 at low pH (5.2-6.2). Histidine residues in solution have a pKa of ∼ 6.5. If H-17 is solvent exposed, we reasoned that it might show sensitivity to pH in terms of its protonation state and consequently its ability to coordinate Cu2+. We examined the recovery from the Cu2+ induced block by addition of 0.05 M HCl into the cis chamber until the desired pH was reached. The results are summarized in Fig. 2 A and show that decreasing pH leads to an increase in the extent of recovery. We do not know the orientation of the channels within the black lipid membranes. One explanation of why the recovery is incomplete may be due to a proportion of the channels being oriented such that the H-17 is less accessible to protons in the cis chamber.
Figure 2.
(A) Addition of 0.05 M HCl at t = 1400 s to pH 6.2 (black diamonds), pH 5.7 (gray squares), and pH 5.2 (open squares). (B) Comparison of the H-17A mutant peptide (gray squares) with wild-type p7 (black diamonds). 500 μM Cu2+ is added at t = 0. The addition of 500 μM NN-DNJ at t = 1400 s has little additional effect on the wild-type channel but inhibits the mutant channels.
To test our hypothesis further, the mutant H-17A was synthesized and when reconstituted into a planar lipid bilayer, exhibited wild-type channel activity (mean conductance = 24.8 ± 1.7 ps, Po = 0.31 ± 0.08). In this case, however, the addition of 500 μM of Cu2+ to the bathing solution did not inhibit p7 mutant ion channel recordings but did inhibit wild-type p7, as seen in Fig. 2 B. As expected, the addition of 500 μM NN-DNJ at 1400 s, inhibited both the wild-type and the mutant ion channel (Fig. 2 B).
Comparison of the H77 strain with the JFH-1 strain (Fig. 1 A) revealed that there is a histidine at an equivalent position. We reasoned that peptides from this strain should also exhibit Cu2+ inhibition, and the experiments described above were repeated for the JFH-1 strain. Results were very similar (Supporting Material), supporting our conclusions for the H77 strain.
We have demonstrated that Cu2+ is capable of inhibiting the p7 ion channel of HCV in a planar lipid bilayer. The inhibition is concentration-dependent. In addition, the effect of the inhibitor activity can also be recovered by decreasing the pH of the bathing solution. The lower the pH, the quicker and greater the reversal of inhibition appears to be. These studies lead us to conclude that the N-terminal helix of p7 lines the pore through which ions are conducted and that the H-17 residue faces the pore. This information should provide a useful constraint in the development of oligomeric models of the pore.
Supporting Material
Methods, results, figures, and references are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(08)00048-9.
Supporting Material
Acknowledgments
The authors thank P. Stansfeld, S. Woodhouse, J. O'Leary, P. Luik, T. Whitfield, S. Pollock, and C. Wee for comments, G. Boldt and P. Wentworth Jr. for chemical synthesis and R. Antrobus for mass spectrometry. We also thank the Wellcome Trust and Overseas Research Students Awards Scheme.
P.C.B. is a Research Councils United Kingdom Fellow. N.Z. is a Glycobiology Career Development Fellow and Senior Research Fellow of Linacre College.
References and Footnotes
- 1.Ahsan N., Rao K.V. Hepatobiliary diseases after kidney transplantation unrelated to classic hepatitis virus. Semin. Dial. 2002;15:358–365. doi: 10.1046/j.1525-139x.2002.00087.x. [DOI] [PubMed] [Google Scholar]
- 2.Feld J.J., Hoofnagle J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 3.Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C., Lindenbach B.D., Pragai B.M., McCourt D.W., Rice C.M. Processing in the hepatitis C virus E2–NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrere-Kremer S., Montpellier-Pala C., Cocquerel L., Wychowski C., Penin F. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin S., Clarke D., McCormick C., Rowlands D., Harris M. Signal peptide cleavage and internal targeting signals direct the hepatitis C virus p7 protein to distinct intracellular membranes. J. Virol. 2005;79:15525–15536. doi: 10.1128/JVI.79.24.15525-15536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isherwood B.J., Patel A.H. Analysis of the processing and transmembrane topology of the E2p7 protein of hepatitis C virus. J. Gen. Virol. 2005;86:667–676. doi: 10.1099/vir.0.80737-0. [DOI] [PubMed] [Google Scholar]
- 9.Clarke D., Griffin S., Beales L., Gelais C.S., Burgess S. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin S.D., Beales L.P., Clarke D.S., Worsfold O., Evans S.D. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 11.Pavlovic D., Neville D.C., Argaud O., Blumberg B., Dwek R.A. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 13.Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 14.Patargias G., Zitzmann N., Dwek R., Fischer W.B. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 15.Gandhi C.S., Shuck K., Lear J.D., Dieckmann G.R., DeGrado W.F. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J. Biol. Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.