Abstract
Receptors of bacterial chemotaxis form clusters at the cell poles, where clusters act as “antennas” to amplify small changes in ligand concentration. It is worthy of note that chemoreceptors cluster at multiple length scales. At the smallest scale, receptors form dimers, which assemble into stable timers of dimers. At a large scale, trimers form large polar clusters composed of thousands of receptors. Although much is known about the signaling properties emerging from receptor clusters, it is unknown how receptors localize at the cell poles and what the determining factors are for cluster size. Here, we present a model of polar receptor clustering based on coupled trimers of dimers, where cluster size is determined as a minimum of the cluster-membrane free energy. This energy has contributions from the cluster-membrane elastic energy, penalizing large clusters due to their high intrinsic curvature, and receptor-receptor coupling that favors large clusters. We find that the reduced cluster-membrane curvature mismatch at the curved cell poles leads to large and robust polar clusters, in line with experimental observation, whereas lateral clusters are efficiently suppressed.
Introduction
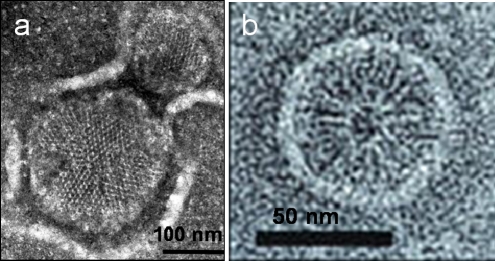
Chemoreceptor clustering is widely conserved among bacteria and archaea (1), allowing cells to detect chemicals in the environment with high sensitivity over a wide range of background concentrations. In the bacteria Escherichia coli, Salmonella enterica, and Caulobacter crescentus, receptor clustering is well documented and occurs at multiple length scales. At a small scale, chemotaxis receptors form stable homodimers, which then assemble into larger complexes with receptors of different chemical specificities intermixed (2). Three homodimers, connected at their signaling tip, form a trimer of dimers (named “trimer” from here on) (2–4), believed to be the smallest stable signaling unit (5,6). At a larger scale, thousands of receptors (7) form ∼200-nm large polar clusters (cf. Fig. 1 a) (3,12–16). Despite the excellent characterization of much of the bacterial chemotaxis network, it is unknown how receptors localize at the cell poles and how they assemble into large polar clusters.
Figure 1.
Electron micrographs of chemoreceptor clusters. (a) Extended clusters in membrane preparations (8–11). Clusters of similar size are observed at the poles of living cells (12,13). Image is courtesy of Michael Manson. (b) Self-assembled round micelle at receptor-dimer resolution (23). Image printed with written permission from publisher. All experiments are based on Tsr-receptor overexpression, as well as membrane extraction, negative staining, and freezing for imaging.
Polar localization appears to be an intrinsic property of chemoreceptors (17,18). It hardly depends on the presence or absence of the receptor-bound kinase CheA and adaptor protein CheW (19), and is unaffected by removal of the periplasmic ligand-binding domain of the receptors (19). It is also a passive process, since newly synthesized receptors, initially inserted at random positions in the membrane, diffuse and ultimately become trapped at the cell poles (20,21). Most important, polar localization appears to depend on membrane curvature. First, inhibition of actin-homolog MreB in growing cells leads to cell swelling and a diffuse receptor distribution, with remaining receptor localization in areas of increased cell curvature (22). Second, receptor-membrane extracts self-assemble into round micelles after receptor overexpression and cell lysis (23) (Fig. 1 b). From electron micrographs, the intrinsic curvature of the trimer structure can be estimated (24). Third, other two-component receptor dimers, e.g., the receptor LuxQ of the quorum-sensing pathway, dimerize without forming trimers of dimers and are evenly distributed over the cell surface (25). Taken together, these observations suggest that the distinct trimer structure, with its increased intrinsic curvature, is responsible for polar receptor localization.
Although trimers may have a tendency to localize at the cell poles and areas of high membrane curvature, tight clustering requires an attractive coupling among the trimers. The conventional view is that CheA and CheW mediate interactions among receptors. Alternative models include swapping of the cytoplasmic receptor domains (26) and membrane-mediated coupling (27) (see Results and Discussion section). The high sensitivity and cooperativity obtained from in vivo fluorescence resonance energy transfer (FRET) (28,29) and in vitro (30) data demonstrate that the functional units of receptor signaling are indeed larger than trimers. These observations are supported by recent quantitative models (29,31–35).
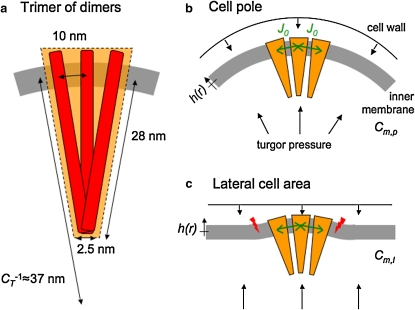
Based on these observations, we propose a model for polar receptor localization and clustering due to the high intrinsic curvature of trimers and an attractive trimer-trimer coupling. Specifically, we consider a membrane-embedded cluster composed of trimers. For a spherocylindrical cell, we assume that the average membrane curvature at the poles is twice as large as average curvature at the lateral surface area, and that trimers have a high intrinsic curvature (Fig. 2 a). The intrinsic curvature of a trimer tends to deform the membrane (Fig. 2 c), penalizing large clusters of trimers. However, attractive coupling between trimers favors cluster formation, leading to a competition between these two opposing energy contributions. Using continuum elastic theory, we derive an analytical expression for the total cluster-membrane energy. We find that due to the reduced cluster-membrane curvature mismatch at the poles, trimers favorably cluster at the poles and not at the lateral cell area. Furthermore, the cluster-size distribution is determined by the cluster-membrane energetics, as well as the trimer density in the cell membrane. Our predicted average cluster size is in line with experimental observation.
Figure 2.
Schematic of membrane-inserted receptors. (a) Trimer (orange shaded area) of dimers (red bars) with geometric parameters. CT is the intrinsic curvature of a trimer. (b) Cluster of three trimers at the cell pole. Trimer-trimer coupling strength J0 is indicated by green arrows. Also shown are the cell wall and the inner membrane (gray) with curvature Cm,p. The height profile h(r) describes the cluster-membrane deformation as measured relative to the preferred height due to cell wall and turgor pressure. (c) Same cluster at a lateral position with membrane curvature Cm,l. Red arrows indicate energetically unfavorable membrane deformations due to the cluster-membrane curvature mismatch.
Model
Receptor geometry
In our model, receptor dimers are assumed to always be associated in trimers, the smallest stable signaling unit (5,6). In Fig. 2 a, the receptor dimer length and width, as well as the distance between neighboring dimers within a trimer, are taken from partial crystal structures (4) and electron microscopy (23). Very similar parameters were used to model the physical response of trimers to osmolytes measured by homo-FRET (36). It is important to note that the estimated value for the intrinsic curvature of a trimer corresponds closely to the inverse radius of self-assembled micelles (Fig. 1 b (23)). The value for the trimer cross section A (Table 1) is consistent with a three-dimensional model of the receptor cluster (37,38) and estimates from cryoelectron microscopy (8,13,23).
Table 1.
Summary of standard parameters
| Parameter | Value | Meaning |
|---|---|---|
| λ (kBT/nm4) | 0.25 (40) | Pinning modulus |
| κc (kBT) | 120∗ | Bending stiffness of cluster |
| κm (kBT) | 25 (40) | Bending stiffness of membrane |
| C−1m,p (nm) | 400 (39) | Inverse of polar membrane curvature |
| C−1m,l (nm) | 800† | Inverse of lateral membrane curvature |
| C−1T (nm) | 37 (23) | Inverse of trimer-of-dimer curvature |
| J0 (kBT) | 3∗ | Trimer-trimer coupling strength |
| A (nm2) | 200 (37) | Trimer cross section |
Standard parameters are used throughout calculations unless specified otherwise.
Cluster bending stiffness and trimer-trimer coupling strength are varied in Fig. 5c to check for robustness.
Value for lateral membrane curvature inverse is based on a spherocylindrical cell.
Elastic cluster-membrane energy
The elastic energy of a membrane-embedded cluster is determined by cluster and membrane bending energies and a pinning potential
| (1) |
The first term, proportional to the bending stiffness, κc, of the receptor cluster, penalizes deviations between the total cluster curvature, , and the preferred cluster curvature, which is equal to the intrinsic trimer curvature, CT. The total cluster curvature is defined by the two principal curvatures C1 and C2 (39). The second term in Eq. 1, proportional to the pinning modulus, λ (40,41), penalizes deviations of the cluster height, , from the preferred height h0, determined by the shape of the curved cell wall. The third and fourth terms in Eq. 1 mirror the first and second, respectively, and describe the cluster-surrounding membrane with total curvature , preferred curvature Cm, and height . Bending stiffnesses κc and κm arise from optimal packing of receptors and lipids, respectively, aiming to protect hydrophobic residues from polar water. The pinning modulus arises due to the turgor pressure, which pushes the membrane and cluster outward, whereas the rigid cell wall pushes them inward. The net effect is a penalty for deformations away from the preferred cell shape (see Fig. 2) (40,41).
Let us define the relative height perturbation, , as measured relative to the preferred height, , in the direction of the normal, pointing in the radial direction outward from surface (Fig. 2, b and c). This allows us to perform the following calculations in the so-called normal gauge, where the total curvatures for the cluster (and membrane), , are now given by Cm + (Cm2 − 2CG)hc(m) + ∇2hc(m) to the lowest order, following from the first-order variation of the geometry (42). Here, CG is the Gaussian curvature. We use two further approximations: 1), ∇2hc(m) >> (Cm2 − 2CG)hc(m), justified for small amplitude and large bending ripples (hence, the contribution proportional to hc(m) is neglected); and 2), the flat Laplacian, ∇2 ≈ ∂2/∂x2 + ∂2/∂y2 (replacing the curvilinear Laplacian), which is valid for sufficiently small clusters. Introducing and ΔC = CT − Cm, Eq. 1 can thus be written as
| (2) |
Parameter ΔC is the important curvature mismatch, i.e., the difference between the cluster and membrane curvatures. The elastic energy model in Eq. 2 neglects surface tension, as well as a Gaussian curvature effect due to the different cluster and membrane elastic properties (see Appendix A for a justification). In this article, the model is applied to both polar and lateral clusters using standard parameter values given in Table 1. Very similar models were previously applied to describe mixtures of lipids with different curvatures (40,41,43–45). For a general review and alternative elastic-energy models, see (46,47).
Considering a circular cluster of radius R, the total height profile is composed of for the cluster (r < R) and for the membrane (r > R), and is determined by minimizing the total elastic cluster-membrane energy with respect to variation of h(x, y). According to Nielsen et al. (48), minimizing an elastic energy of the generic form
| (3) |
leads to the Euler-Lagrange equation
| (4) |
| (5) |
Replacing Ψ by the expressions in Eq. 2 leads to the fourth-order linear differential equation (49–51) for the cluster (membrane):
| (6) |
which is independent of ΔC in the small deformation approximation. Since we consider a circular cluster located at the origin of the coordinate system, we apply cylindrical symmetry from now on. To solve Eq. 6 for cluster and membrane, one needs four boundary conditions for each equation. We require ∂hc/∂r = 0 at the origin and that the membrane deformation vanishes far away from the cluster, i.e., and . We further impose that the solutions for cluster and membrane match at the cluster-membrane interface, i.e., hc(R) = hm(R) and ∂hc/∂r|R = ∂hm/∂r|R.
Equation 6 can be solved by applying the Kelvin differential equation
| (7) |
leading to for the cluster (membrane). In Eq. 7, the second-order derivative is now calculated using ∇2 = ∂2/∂r2 + 1/r ∂/∂r. The solution to Eq. 7 is given by (52)
| (8) |
and
| (9) |
where ber0, bei0, ker0, and kei0 are the zeroth-order Kelvin function, and I0 and K0 are the zeroth-order modified Bessel functions of the first and second kind, respectively.
To construct the solution for the cluster (r < R), only the modified Bessel function of the first kind has zero slope at r = 0, whereas to construct the solution for the membrane (r > R), only the modified Bessel function of the second kind has a vanishing real part at infinity. To obtain real solutions, we need to add their complex conjugates
| (10) |
and
| (11) |
where a–d are real parameters to be determined by matching the boundary conditions.
Solution coefficients
After boundary conditions are matched, the solution coefficients in Eqs. 10 and 11 are given by
| (12) |
| (13) |
| (14) |
| (15) |
where S = 2ℜ[(a + ib)βc,2I1(βc,2R)] is the slope at the cluster-membrane interface, H0 is the cluster height at r = 0, and H1 is the height at r = R, i.e., at the cluster-membrane interface. We further used ∂I0(βr)/∂(βr) = βI1(βr) and ∂K0(βr)/∂(βr) = − βK1(βr). The remaining unknown parameters, such as H0 and H1, are determined by numerically minimizing the elastic energy.
Analytic expression for elastic energy
The integrals in Eq. 2 can be solved analytically using integration by parts twice (49,53) and exploiting Eq. 6, leading to
| (16) |
The higher-order derivatives are calculated from the solutions Eqs. 10 and 11 using (52)
| (17) |
| (18) |
| (19) |
and
| (20) |
| (21) |
| (22) |
In the limit of very large clusters, the elastic energy of Eq. 16 reduces to
| (23) |
since the other contributions to the elastic energy grow more slowly with size (cf. Fig. 5 a). The number of trimers in a cluster of radius R is given by N ≈ R2π/A.
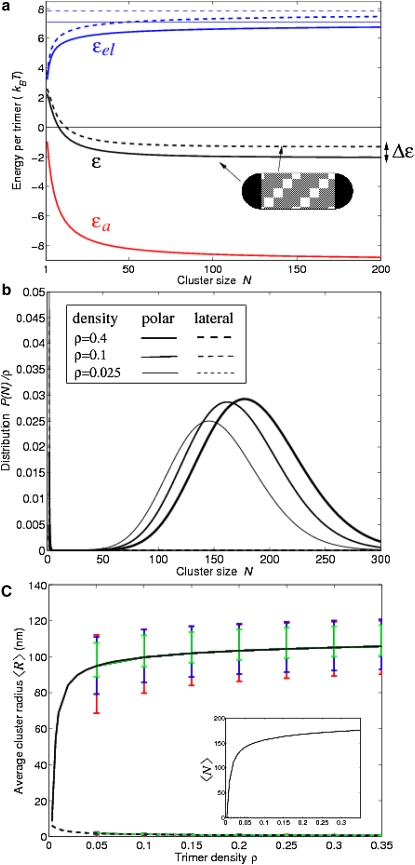
Figure 5.
Quantifying polar receptor clustering. (a) Cluster-membrane energy density, i.e., energy per trimer as a function of cluster size N. Total energy density ɛ (black) is the sum of repulsive elastic energy density ɛel (blue) and attractive cluster energy density ɛa (red). Solid lines correspond to cell poles and dashed lines to lateral positions. Thin solid and dashed blue lines indicate asymptotic limits of the elastic energy density for at the poles and lateral positions, respectively. Polar clusters are stabilized by energy density Δɛ. (b) Distribution of cluster sizes for three values of the trimer density (occupancy fraction), ρ = 0.4, 0.1, and 0.025 at the poles (solid lines) and lateral positions (dashed lines). (c) Average cluster radius 〈R〉 at the poles (black solid line) and lateral area (black dashed line) as a function of trimer density, ρ, based on standard parameters, including trimer-trimer coupling, J0, trimer curvature, CT, and cluster bending stiffness, κc from Table 1. Error bars indicate robustness to parameter changes: upper error bars for poles and lateral area are 1.1 for J0 (red), 0.98 for CT (dimer-dimer distance reduced by 1 nm (green)), and 0.92 for κc (blue); lower error bars for poles and lateral area are 0.9 for J0 (red), 1.03 for CT (dimer-dimer distance increased by 1 nm (green)), and 1.09 for κc (blue). (Inset) Average cluster size (number of trimers) 〈N〉 as a function of trimer density for standard parameters.
Attractive trimer-trimer coupling
In our model, trimers interact favorably when in close contact, driving cluster formation. The total coupling energy of a cluster of radius R is described by
| (24) |
For a triangular lattice like that used in Fig. 3, we consider two expressions for the average coupling energy per trimer. Assuming a cluster made of concentric rings formed around a central trimer, the coupling energy is given by (41)
| (25) |
where J0 is the coupling energy between two neighboring trimers (Table 1). This expression is exact in the limit of large clusters. As an alternative, we fit
| (26) |
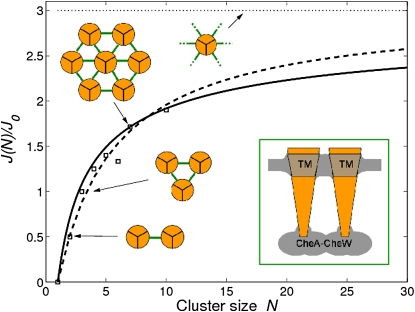
to exact interaction energies of small compact clusters and obtain parameter N0.5 = 4.72. According to Fig. 3, Eq. 25 overestimates the interaction energy for small clusters, whereas Eq. 26 overestimates the interaction energy for large clusters. In the limit , both models produce . In the following, we use Eq. 26, since it helps to stabilize large clusters.
Figure 3.
Average interaction energy J(N) per trimer (in units of coupling strength J0) as a function of cluster size (number of trimers, N). Trimers are shown by orange disks, and trimer-trimer coupling of strength J0 by green bars. Clusters are assumed to have a triangular lattice structure. Symbols represent small compact clusters of minimal circumference. Solid and dashed lines represent the “ring model” and “fit-to-symbols model”, respectively (see Model section). The horizontal dotted line shows the average interaction energy for an infinitely large cluster. (Inset) Possible mechanisms of trimer-trimer coupling including that mediated by receptor-bound CheA-CheW and membrane deformations based on large hydrophobic transmembrane domains.
The total energy is the sum of elastic energy and the attractive energy
| (27) |
where we explicitly included the dependence on the cluster size N.
Distribution of cluster sizes
For a spherocylindrical cell, the preferred membrane curvature at the poles Cm = Cm,p is twice as large as the preferred membrane curvature at the lateral area Cm = Cm,l = Cm,p/2, but still smaller than the trimer curvature, CT. This leads to a smaller cluster-membrane curvature mismatch at the poles, ΔCp = CT − Cm,p, than at the lateral area, ΔCl = CT − Cm,l = CT − Cm,p/2. Consequently, the total energy (Eq. 27) of a cluster is also smaller at the poles than at the lateral area Ep(N) < El(N), favoring polar clustering.
Based on the total energies, statistical mechanics is used to calculate the cluster-size distribution at the poles/lateral area (p/l) (41,54)
| (28) |
where the exponential Boltzmann factor describes the probability of observing a cluster of N trimers at the poles/lateral area for chemical potential μ. The chemical potential represents the energy required or released by inserting a trimer into the membrane, and is adjusted to fulfill an overall target trimer density on the cell surface (occupancy fraction) ρ via
| (29) |
Using the cluster-size distributions, the average cluster sizes at the poles and lateral area are given by
| (30) |
Conditions for large polar clusters
To find the conditions that favor polar clustering, we consider the total energy density ɛ = Ep/l/N, i.e., the total energy of the membrane-embedded cluster per trimer. Generally, minimization with respect to N determines the energetically preferred cluster size. We note the following. First, the energy density is generally a monotonically decreasing function of N, which eventually saturates for large N (cf. Fig. 5 a). This indicates that maximal cluster sizes are energetically favorable. Second, the elastic energy density is always smaller at the poles than at the lateral area, demonstrating that polar clustering is energetically predominant.
Let us consider the total energy densities in the limit . In this limit, the total cluster-membrane energy density at the poles/lateral area is given by
| (31) |
Consequently, the energy-density difference between the poles and lateral area is provided by
| (32) |
where we used Cm,l = Cm,p/2. Increasing CT beyond 3/4 Cm,p favors polar over lateral clusters. Specifically, for NΔɛ > 1kBT, a cluster of N trimers is significantly more favorable at the poles than at the lateral area at temperature T.
Results and Discussion
Based on experimental observations outlined in the Introduction, we propose a model for polar receptor localization and clustering (Fig. 2). The ingredients and model assumptions are:
-
1.
An individual trimer of dimers (trimer), believed to be the smallest stable signaling unit (5,6), has a high intrinsic curvature CT (Fig. 2 a). The cell membrane has a higher curvature at the cell poles Cm,p than at the lateral area Cm,l. For a spherocylindrical cell, we have specifically Cm,l = Cm,p/2. Since CT > Cm,p > Cm,l, individual trimers favor the cell poles energetically, although this effect is very small by itself (fraction of thermal energy kBT).
-
2.
Trimers are coupled with strength J0 when in close proximity (Fig. 2, b and c), driving cluster formation at the poles and lateral area (Fig. 3).
-
3.
Due to the cluster-membrane curvature mismatch, growing clusters deform the membrane and are energetically penalized. However, since the cluster-membrane curvature mismatch at the poles, ΔCp = CT − Cm,p, is smaller than the corresponding mismatch at the lateral area, ΔCl = CT − Cm,p/2, this energy penalty is reduced at the poles (Fig. 2 b).
As outlined in the Model section, the model is implemented by considering a membrane-embedded cluster of radius R. The height profile of the cluster and the membrane minimizes the elastic energy, which is determined by the cluster and membrane preferred curvatures (respective CT and Cm,p/l) and their bending stiffnesses (respective κc and κm). Furthermore, a pinning modulus λ (40,41) pushes the membrane and the cluster against the rigid cell wall (Fig. 2, b and c). This penalizes large deformations of the cluster and the membrane. The main findings are:
-
1.
Considered separately, poles and lateral area favor maximal cluster sizes energetically.
-
2.
Actual cluster size is determined by trimer density (entropy), where increasing trimer density pushes distribution of cluster sizes to larger values.
-
3.
Polar-only clustering is a result of the reduced curvature mismatch at the poles, energetically stabilizing polar clusters and suppressing lateral ones.
Our results are in line with the experimental observation of large polar clusters, which were found to be robust (17–19) and only slightly affected by attractant binding (55,56), expression level variation (15,19,57), and receptor methylation (11,58–60).
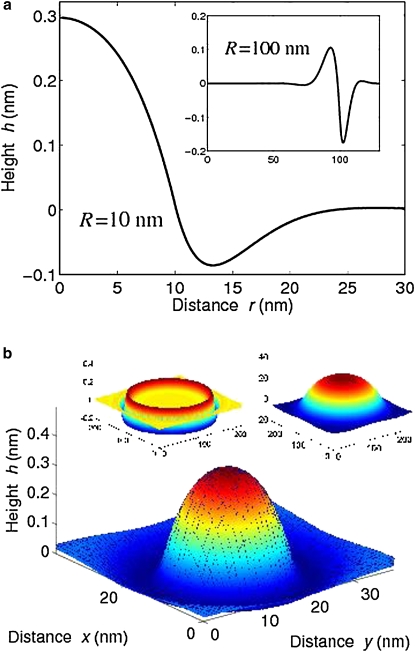
Fig. 4 shows typical cluster-membrane height profiles for two different cluster radii, R = 10 nm and R = 100 nm. The height profile minimizes the cluster-membrane elastic energy given in Eq. 2. The profile of the small cluster bulges out into the periplasmic space in a convex manner, whereas the large cluster of physiological size is flattened. The latter effect appears consistent with images from cryoelectron microscopy (12,13). In our model, large deformations are suppressed by the pinning potential (Fig. 4 b, left inset). Significant reduction of the pinning modulus, λ, leads to strongly curved clusters (Fig. 4 b, right inset). Note that the maximal deformation of the large curved cluster can easily exceed the width of the periplasmic space (20 nm (61)), emphasizing the importance of the pinning modulus.
Figure 4.
Cluster-membrane height profiles. (a) Profile h(r) as a function of distance r from the center for a small cluster of radius R = 10 nm. (Inset) Profile for a large cluster of radius R = 100 nm. (b) Three-dimensional profile for a small cluster. (Left inset) Three-dimensional profile for a large cluster. For parameters κc, κm, and λ, see Table 1. (Right inset) Large cluster for a very small pinning modulus (λ = 10−5kBT/nm4).
How large are clusters and how do their sizes differ between cell poles and lateral positions? Consider a membrane-embedded cluster of radius R or number of trimers N ≈ R2π/A, where A is the trimer cross section. The total trimer energy E is equal to the sum of the unfavorable elastic energy, Eel, and the favorable attractive energy, Ea. Dividing by the number of trimers, N, results in the corresponding energy densities ɛ, ɛel, and ɛa. Generally, the minimum of the total energy density ɛ as a function of cluster size N provides the preferred cluster size. Fig. 5 a shows that key requirements for stable polar clusters are fulfilled: (1) The energy density reaches its lowest value in the limit , energetically favoring maximal clusters. (2) Although the energy-density difference Δɛ between the poles and lateral cell area can be smaller than the thermal energy kBT, a cluster of N trimers is stabilized at the poles and suppressed at the lateral area when NΔɛ > kBT (see Model section).
Due to the finite trimer density, cluster sizes are always finite. To obtain the predicted distribution of cluster sizes, we consider the combined system of cell poles and lateral area. The distribution of cluster sizes can be calculated using Boltzmann statistics of equilibrium statistical mechanics (see Model section). A chemical potential is further adjusted to obtain a certain target trimer density. Fig. 5 b shows the size distributions of polar and lateral clusters for three different trimer densities. Even at low trimer densities, very few residual trimers, stabilized by entropy, remain unclustered at the poles and lateral area. The average radii of polar clusters shown in Fig. 5 c correspond well with the observed cluster diameters of ∼200 nm, whereas lateral clusters are significantly suppressed.
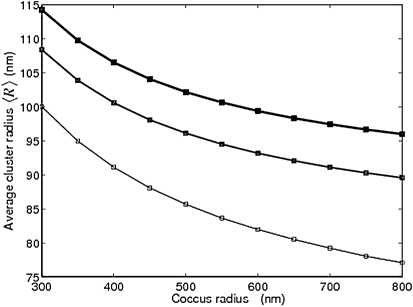
As shown in Fig. 5 c, our model predicts the average cluster size as a function of trimer density. This prediction can be tested experimentally through imaging. The fluorescence intensity of a cluster, e.g., measured using receptor-GFP fusion proteins (20), is proportional to the number of receptors in the cluster. As an alternative, imaging by cryoelectron microscopy can provide spatial cluster dimensions (12,13). The dependence on trimer density can be studied by expressing receptors from an inducible plasmid. A recent experiment using the fusion protein CheY-YFP as a fluorescent marker indeed indicated a strong correlation between receptor expression level and polar fluorescence intensity (62). Furthermore, the predicted link between membrane curvature and clustering can be tested by quantifying receptor fluorescence intensities for cell-shape phenotypes, e.g., when inhibiting actin-homolog MreB (20), which is responsible for the rod shape in bacteria. As an alternative, cocci cells or round membrane vesicles can be used, allowing the study of receptor clustering in the presence of only a single membrane curvature. In Fig. 6, we show the predicted average cluster size as a function of coccus radius and receptor density (expression level). Increasing the coccus radius decreases the coccus curvature and leads to a larger cluster-membrane curvature mismatch, which reduces cluster size. In contrast, increasing the receptor density shifts cluster size distribution toward larger clusters.
Figure 6.
Predicted receptor clustering in cocci cells. Average cluster radius, 〈R〉, as a function of coccus radius. Solid lines of varying thickness correspond to the three densities from Fig. 5b.
In recent experiments the physical responses of dimers were measured by homo-FRET using receptor-YFP fusions (36,63). These data indicate that the dimer-dimer distance in a trimer (distance between receptor C-termini) shrinks by 10% upon osmolyte stimulation. Osmolytes act as repellents and are presumably sensed through receptor-membrane coupling. To see if polar clustering is robust against such perturbations, Fig. 5 c shows that a 10% increase of the dimer-dimer distance (CT−1 = 36.2 nm (lower green error bars)) destabilizes clusters very little, whereas a 10% decrease of the dimer-dimer distance (CT−1 = 38.8 nm (upper green error bars)) stabilizes clusters even further. To illustrate the robustness of polar clustering with respect to other model parameters, we also varied cluster bending stiffness κc and trimer-trimer coupling strength J0. Fig. 5 c shows that these parameter variations only lead to modest changes in cluster stability (blue and red error bars, respectively).
Although our model provides a robust mechanism for the formation of large polar clusters, it has limitations.
-
1.
A recent study showed that, in addition to large polar clusters, there are also small lateral clusters at future division sites, such as 1/2, as well as 1/4 and 3/4 cell length (14). However, these lateral clusters appear immobile, presumably due to anchoring, and hence may form through a different mechanism. In Rhodobacter sphaeroides, immobile lateral clusters of chemotaxis receptor homologs were even found in the cytoplasm (64). Our model does suggest that clusters form at the new poles once cell division occurs and the membrane pinches off. Newly synthesized receptors, inserted by the Sec-machinery throughout the cell surface (20,65), would begin to cluster at the new cell poles. However, if equilibration is too slow to grow new polar clusters from scratch after cell division, lateral clusters may be useful by serving as nucleation sites.
-
2.
Structural work on receptors and receptor-bound proteins in Thermotoga maritima suggests that, at least for this bacterium, receptor dimers may assemble into linear oligomers and not trimer-based clusters (66). A bioinformatics study on chemotaxis receptors across many species also addresses this issue, but does not favor one model over the other (67). On the other hand, cryoelectron microscopy of Caulobacter crescentus strongly supports trimer-based clusters (13).
-
3.
Our model does not include interactions between multiple clusters. Such interactions may largely be unimportant, since fluorescence images generally indicate only one, rarely more, cluster per cell pole (14).
-
4.
It can further not be ruled out that receptors do not localize to the poles themselves, but that certain polar lipids, e.g. cardiolipin (68), provide favorable sites for receptor localization and clustering.
-
5.
Although some elastic properties are included in our model, others are not, e.g., membrane thickness deformation due to large hydrophobic transmembrane domains (49). Thickness deformation leads to a line tension, i.e., an elastic energy proportional to the cluster circumference, 2πR, affecting polar and lateral clusters equally. Although this effect does not change the stability of polar clusters, the line tension may provide a mechanism for trimer-trimer coupling (69) (see next paragraph) and have influence on the cluster size distribution.
What are the possible mechanisms responsible for trimer-trimer coupling?
-
1.
Coupling mediated by CheA and CheW (Fig. 3, inset). The presence of CheA and CheW increases polar clustering only modestly (15,16,59). However, overexpression of CheA decreases the receptor cooperativity measured by FRET, whereas overexpression of CheW increases it (29).
-
2.
Coupling mediated by elastic membrane deformations. Receptor activity has been shown to depend on receptor-membrane interactions (36,63,70), which, we speculate, may provide a mechanism for trimer-trimer coupling (69,71). In support of this mechanism, receptor transmembrane regions are unusually large (24–30 residues) compared to the membrane thickness (30 Å or ∼20 residues) (72). Such large regions possibly lead to significant membrane deformations to protect the hydrophobic receptor residues from water (Fig. 3, inset). If transmembrane regions change with activity, e.g., within the receptor piston model (70,73–75), trimer-trimer coupling may even depend on the receptor activity state, as proposed for the approximate two-state osmolarity-sensing MscL pore (27).
-
3.
Coupling mediated by swapping of cytoplasmic domains of neighboring dimers (26).
Many other sensory receptors cluster as well, including B-cell (76), T-cell (77), Fcγ (78), synaptic (79), and ryanodine (80) receptors. This indicates that receptor clustering serves important regulatory functions in the cell, e.g., adjusting signaling properties, recruiting auxiliary proteins, or kinetically proofreading unexpected stimuli. Unlike bacteria, eukaryotic receptor clustering appears much more dynamic, including receptor diffusion and internalization, and the underlying physical mechanism remains little understood. Our work adds additional support to the idea that the elastic properties of receptors and membrane may be a general design principle to regulate receptor localization and clustering (47,81).
Appendix A: Surface Tension and Gaussian Curvature
The elastic energy in Eq. 2 neglects surface tension and Gaussian curvature terms. Here, we show that these two elastic energy contributions are indeed very small.
Surface tension
In the small deformation approximation (Monge representation) for cluster and membrane, the contribution from the surface tension to the elastic energy (Eq. 2) is given by
| (A1) |
where σc(m) is the surface tension of the cluster (membrane) and . Surface tension arises in part from the attractive receptor-receptor and lipid-lipid interactions. Minimization of the total elastic energy (Eq. 2 plus Eq. A1) leads to the Euler-Lagrange equation
| (A2) |
for the cluster (membrane). Using integration by parts twice (49,53), and applying Eq. A2, the elastic-energy contribution due to the surface tension of the cluster and membrane is given by
| (A3) |
where R is the cluster radius, H1 is the cluster-membrane height at the interface (r = R), and S is the slope of the cluster-membrane at the interface. This energy describes a line tension (∼2πR) and vanishes for σc = σm, since cluster and membrane contributions point in opposite radial directions.
We apply perturbation theory to estimate the significance of the surface-tension contribution. For this purpose, we use our previously calculated height profile, Eqs. 10 and 11, obtained without the surface-tension term. Using σm = σc/4 = 1 kBT/nm2 (49) and a physiological cluster radius of R = 100 nm, we find that the estimated energy contribution from the surface tensions is much smaller than the elastic energy (Eq. 2), i.e., Eelσ/Eel ≈ 0.004, justifying the neglect of this term.
Gaussian curvature
The contribution to the elastic energy from the Gaussian curvature can be neglected for homogeneous membranes that do not change their topology (Gauss-Bonnet theorem). However, this contribution is technically nonzero for our cluster-membrane system. In the small deformation approximation, this contribution is given by (39)
| (A4) |
where KG,c(m) is the Gaussian curvature modulus for the cluster (membrane). In Wiggins and Phillips (49), Eq. 129 shows that the Gaussian energy contribution has two parts, one topological, which is just a constant since our membrane contains a single receptor cluster, and the second a contour integral along the cluster boundary. Based on Eq. 131 of Wiggins and Phillips (49), we obtain
| (A5) |
For our height profile (Eqs. 10 and 11), as well as KG,c(m) = −κc(m)/2 (39,49), this energy contribution is significantly smaller than the elastic energy in Eq. 2, i.e., |EelG|/Eel ≈ 0.0006, justifying the neglect of this term. As expected, this energy contribution vanishes when the elastic properties of cluster and membrane become identical for KG,c = KG,m.
Acknowledgments
We thank Kerwyn Huang, Michael Manson, Ranjan Mukhopadhyay, Samuel Safran, Victor Sourjik, Sriram Subramaniam, and Ned Wingreen for helpful discussions and two anonymous referees for valuable suggestions.
We acknowledge funding from the Biotechnology and Biological Sciences Research Council grant BB/G000131/1 and from the Centre for Integrated Systems Biology at Imperial College.
References
- 1.Gestwicki J.E., Lamanna A.C., Harshey R.M., McCarter L.L., Kiessling L.L. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J. Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Studdert C.A., Parkinson J.S. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames P., Studdert C.A., Reiser R.H., Parkinson J.S. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K.K., Yokota H., Kim S.H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 5.Studdert C.A., Parkinson J.S. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldog T., Grimme S., Li M., Sligar S.G., Hazelbauer G.L. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M., Hazelbauer G.L. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAndrew R.S., Ellis E.A., Manson M.D., Holzenburg A. TEM analysis of chemoreceptor arrays in native membranes of E. coli. Microsc. Microanal. 2004;10:416–417. [Google Scholar]
- 9.McAndrew R.S., Ellis E.A., Lai R.Z., Manson M.D., Holzenburg A. Identification of Tsr and Tar chemoreceptor arrays in E. coli inner membranes. Microsc. Microanal. 2005;11:1190–1191. [Google Scholar]
- 10.Lai R.Z., Manson J.M., Bormans A.F., Draheim R.R., Nguyen N.T. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 11.McAndrew R.S., Ellis E.A., Lai R.Z., Manson M.D., Holzenburg A. Effects of chemoreceptor modification on the structures of Tsr arrays. Microsc. Microanal. 2006;12:378–379. [Google Scholar]
- 12.Zhang P., Khursigara C.M., Hartnell L.M., Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briegel A., Ding H.J., Li Z., Werner J., Gitai Z. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol. Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiem S., Kentner D., Sourjik V. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 2007;26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddock J.R., Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 16.Sourjik V., Berg H.C. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 17.Kentner D., Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Schulmeister S., Ruttorf M., Thiem S., Kentner D., Lebiedz D. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kentner D., Thiem S., Hildenbeutel M., Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 20.Shiomi D., Yoshimoto M., Homma M., Kawagishi I. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 2006;60:894–906. doi: 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J., Xiao J., Ren X., Lao K., Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 22.Shih Y.-L., Kawagishi I., Rothfield L. The MreB and Min cytoskeletal-like systems play independent roles in prokaryotic polar differentiation. Mol. Microbiol. 2005;58:917–928. doi: 10.1111/j.1365-2958.2005.04841.x. [DOI] [PubMed] [Google Scholar]
- 23.Weis R.M., Hirai T., Chalah A., Kessel M., Peters P.J. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefman J., Zhang P., Hirai T., Weis R.M., Juliani J. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 2004;186:5052–5061. doi: 10.1128/JB.186.15.5052-5061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neiditch M.B., Federle M.J., Pompeani A.J., Kelly R.C., Swem D.L. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolanin P.M., Stock J.B. Bacterial chemosensing: cooperative molecular logic. Curr. Biol. 2004;14:R486–R487. doi: 10.1016/j.cub.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Ursell T., Huang K.C., Peterson E., Phillips R. Cooperative gating and spatial organization of membrane proteins through elastic interactions. PLoS Comput. Biol. 2007;3:e81. doi: 10.1371/journal.pcbi.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sourjik V., Berg H.C. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sourjik V., Berg H.C. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 30.Li G., Weis R.M. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of. Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 31.Bray D., Levin M.D., Morton-Firth C.J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 32.Mello B.A., Tu Y. An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc. Natl. Acad. Sci. USA. 2005;102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keymer J.E., Endres R.G., Skoge M., Meir Y., Wingreen N.S. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc. Natl. Acad. Sci. USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endres R.G., Wingreen N.S. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc. Natl. Acad. Sci. USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skoge M.L., Endres R.G., Wingreen N.S. Receptor-receptor coupling in bacterial chemotaxis: evidence for strongly coupled clusters. Biophys. J. 2006;90:4317–4326. doi: 10.1529/biophysj.105.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaknin A., Berg H.C. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2006;103:592–596. doi: 10.1073/pnas.0510047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu T.S., Le Novère N., Levin M.D., Beavil A.J., Sutton B.J. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.H., Wang W., Kim K.K. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boal D. Cambridge University Press; Cambridge, UK: 2002. Mechanics of the Cell. [Google Scholar]
- 40.Huang K.C., Mukhopadhyay R., Wingreen N.S. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput. Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukhopadhyay R., Huang K.C., Wingreen N.S. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys. J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capovilla R., Guven J., Santiago J.A. Deformations of the geometry of lipid vesicles. J. Phys. Math. Gen. 2003;36:6281–6295. [Google Scholar]
- 43.Lipowsky R. Budding of membranes induced by intermembrane domains. J. Phys. II (France) 1992;2:1825–1840. [Google Scholar]
- 44.Seifert U. Curvature-induced lateral phase segregation in two-component vesicles. Phys. Rev. Lett. 1993;70:1335–1338. doi: 10.1103/PhysRevLett.70.1335. [DOI] [PubMed] [Google Scholar]
- 45.Komura S., Shimokawa N. Tension-induced morphological transition in mixed lipid bilayers. Langmuir. 2006;22:6771–6774. doi: 10.1021/la053135x. [DOI] [PubMed] [Google Scholar]
- 46.Kozlov M.M., Andelman D. Theory and phenomenology of mixed amphiphilic aggregates. Curr. Opin. Colloid Interface Sci. 1996;1:362–366. [Google Scholar]
- 47.Brown F.L.H. Elastic modeling of biomembranes and lipid bilayers. Annu. Rev. Phys. Chem. 2008;59:685–712. doi: 10.1146/annurev.physchem.59.032607.093550. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen C., Goulian M., Andersen O.S. Energetics of inclusion-induced bilayer deformations. Biophys. J. 1998;74:1966–1983. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiggins P., Phillips R. Membrane-protein interactions in mechanosensitive channels. Biophys. J. 2005;88:880–902. doi: 10.1529/biophysj.104.047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H.W. Deformation free energy of bilayer membrane and its effect on Gramicidin channel lifetime. Biophys. J. 1986;50:1061–1070. doi: 10.1016/S0006-3495(86)83550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dan N., Safran S.A. Effects of lipid characteristics on the structure of transmembrane proteins. Biophys. J. 1998;75:1410–1414. doi: 10.1016/S0006-3495(98)74059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abramowitz M., Stegun I. Dover; New York: 1968. Handbook of Mathematical Functions. [Google Scholar]
- 53.Landau L.D., Lifshitz E.M. 3rd ed. Butterworth-Heinemann; Oxford, UK: 1986. Theory of Elasticity. [Google Scholar]
- 54.Endres R.G., Falke J.J., Wingreen N.S. Chemotaxis receptor complexes: from signaling to assembly. PLoS Compl. Biol. 2007;3:e150. doi: 10.1371/journal.pcbi.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamanna A.C., Ordal G.W., Kiessling L.L. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol. Microbiol. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- 56.Homma M., Shiomi D., Homma M., Kawagishi I. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at a cell pole. Proc. Natl. Acad. Sci. USA. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skidmore J.M., Ellefson D.D., McNamara B.P., Couto M.M., Wolfe A.J. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J. Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiomi D., Banno S., Homma M., Kawagishi I. Stabilization of polar localization of a chemoreceptor via its covalent modifications and its communication with a different chemoreceptor. J. Bacteriol. 2005;187:7647–7654. doi: 10.1128/JB.187.22.7647-7654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liberman L., Berg H.C., Sourjik V. Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J. Bacteriol. 2004;186:6643–6646. doi: 10.1128/JB.186.19.6643-6646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lybarger S.R., Maddock J.R. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J. Bacteriol. 1999;181:5527–5529. doi: 10.1128/jb.181.17.5527-5529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matias V.R., Al-Amoudi A., Dubochet J., Beveridge T.J. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiem S., Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol. Microbiol. 2008;68:1228–1236. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- 63.Vaknin A., Berg H.C. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson S.R., Wadhams G.H., Armitage J.P. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:8209–8214. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gebert J.F., Overhoff B., Manson M.D., Boos W. The Tsr chemosensory transducer of Escherichia coli assembles into the cytoplasmic membrane via a SecA-dependent process. J. Biol. Chem. 1988;263:16652–16660. [PubMed] [Google Scholar]
- 66.Park S.Y., Borbat P.P., Gonzalez-Bonet G., Bhatnagar J., Freed J.H. Reconstruction of the chemotaxis receptor:kinase assembly. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 67.Alexander R.P., Zhulin I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mileykovskaya E., Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dan N., Pincus P., Safran S.A. Membrane-induced interactions among inclusions. Langmuir. 1993;9:2768–2771. [Google Scholar]
- 70.Draheim R.R., Bormans A.F., Lai R.Z., Manson M.D. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2005;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 71.Aranda-Espinoza H., Berman A., Dan N., Pincus P., Safran S. Interaction between inclusions embedded in membranes. Biophys. J. 1996;71:648–656. doi: 10.1016/S0006-3495(96)79265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boldog T., Hazelbauer G.L. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 2004;13:1466–1475. doi: 10.1110/ps.04648604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ottemann K.M., Xiao W., Shin Y.K., Koshland D.E. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 74.Peach M.L., Hazelbauer G.L., Lybrand T.P. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 2002;11:912–923. doi: 10.1110/ps.4640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller A.S., Falke J.J. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schamel W.W., Reth M. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 2000;13:5–14. doi: 10.1016/s1074-7613(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 77.Germain R.N., Stefanova I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu. Rev. Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 78.Chacko G.W., Duchemin A.M., Coggeshall K.M., Osborne J.M., Brandt J.T. Clustering of the platelet Fcγ receptor induces noncovalent association with the tyrosine kinase p72syk. J. Biol. Chem. 1994;269:32435–32440. [PubMed] [Google Scholar]
- 79.Griffith L.C. Receptor clustering: nothing succeeds like success. Curr. Biol. 2004;14:R413–R415. doi: 10.1016/j.cub.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 80.Yin C.C., Blayney L.M., Lai F.A. Physical coupling between ryanodine receptor-calcium release channels. J. Mol. Biol. 2005;349:538–546. doi: 10.1016/j.jmb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 81.Groves J.T. Bending mechanics and molecular organization in biological membranes. Annu. Rev. Phys. Chem. 2007;58:697–717. doi: 10.1146/annurev.physchem.56.092503.141216. [DOI] [PubMed] [Google Scholar]