Abstract
Mutations of PCDH15, encoding protocadherin 15, can cause either combined hearing and vision impairment (type 1 Usher syndrome; USH1F) or nonsyndromic deafness (DFNB23). Human PCDH15 is reported to be comprised of 35 exons and encodes a variety of isoforms with 3 to 11 ectodomains (EC), a transmembrane domain and a carboxy-terminal cytoplasmic domain (CD). Building on these observations we describe an updated gene structure that has four additional exons of PCDH15 and isoforms that can be subdivided into four classes. Human PCDH15 encodes three alternative, evolutionarily conserved unique cytoplasmic domains (CD1, CD2 or CD3). Families ascertained on the basis of prelingual hearing loss were screened for linkage of this phenotype to markers for PCDH15 on chromosome 10q21.1. In seven of twelve families segregating USH1 we identified homozygous mutant alleles (1 missense, 1 splice site, 3 nonsense and 2 deletion mutations) of which six are novel. One family was segregating nonsyndromic deafness DFNB23 due to a homozygous missense mutation. To date in our cohort of 557 Pakistani families, we have found 11 different PCDH15 mutations that account for deafness in 13 families. Molecular modeling provided mechanistic insight into the phenotypic variation in severity of the PCDH15 missense mutations. We did not find pathogenic mutations in five of the twelve USH1 families linked to markers for USH1F, which suggest either the presence of mutations of yet additional undiscovered exons of PCDH15, mutations in the introns or regulatory elements of PCDH15, or an additional locus for type I USH at chromosome 10q21.1.
Keywords: DFNB23, Usher syndrome, protocadherin 15, PCDH15, deafness, retinitis pigmentosa
Introduction
Causes of profound childhood deafness include environmental and genetic factors. In North American and European populations hereditary deafness is estimated to occur in 1 in every 2,000 newborns (Morton 1991). Approximately 30% of this hearing impairment co-occurs with other clinical features such as loss of vision (Bergstrom et al. 1971). Usher syndrome (USH) is characterized by a loss of vision due to retinitis pigmentosa (RP) and bilateral sensorineural deafness (Smith et al. 1994). From studies in Scandinavia, Colombia, United Kingdom and the United States the prevalence of USH is between 1/16,000 and 1/50,000 (Petit 2001).
USH is classified into three clinical subtypes. Type 1 USH (USH1) is the most genetically heterogeneous. To date, there are seven USH1 loci (USH1B, USH1C, USH1D, USH1E, USH1F, USH1G, and USH1H) and five USH1 genes have been identified (Ahmed et al. 2008; Petit 2001; Weil et al. 2003). Particular mutations of four of these USH1 genes, MYO7A, USH1C, CDH23, and PCDH15 can also cause nonsydromic hearing loss, DFNB2, DFNB18, DFNB12 and DFNB23, respectively (Ahmed et al. 2003; Ahmed et al. 2002; Bork et al. 2001; Riazuddin et al. 2008).
Mouse models have been instrumental in identifying the genes for human USH1 (Ahmed et al. 2001; Alagramam et al. 2001b; Bolz et al. 2001; Weil et al. 1995; Weil et al. 2003). For example, the Ames waltzer (av) phenotype is due to recessive mutations of Pcdh15 (Alagramam et al. 2001a). Homozygous av mice show degeneration of inner ear neuroepithelia associated with deafness and vestibular dysfunction but no RP (Ball et al. 2003; Haywood-Watson et al. 2006), and are only a model for DFNB23 nonsyndromic deafness (Ahmed et al. 2003).
We previously reported a large number of wild type alternatively spliced transcripts of mouse Pcdh15, which utilize a subset of the 39 exons, and can encode a signal sequence, an extracellular domain with 3 to 11 ECs, a single-pass transmembrane domain and one of three different carboxy-terminus cytoplasmic domains (Ahmed et al. 2006) referred to as CD1, CD2 or CD3. Here we report mutations of PCDH15 in seven families segregating USH1F, and one family segregating nonsyndromic deafness DFNB23. We found six novel mutant alleles of PCDH15. We also provide a more complete gene structure for PCDH15 that includes four additional exons encoding three alternative cytoplasmic domains.
Materials and methods
Subject enrollment
This study was approved by the Institutional Review Board (IRB) at the National Centre of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758), and the Central NeuroScience IRB at the National Institutes of Health, USA (OH-93-N-016). Subjects were ascertained in rural areas of the Punjab and Sindh provinces of Pakistan. Written informed consent was obtained from all adults and parents of minors under the age of 18 years.
Clinical evaluation
We performed medical history interviews to identify possible clinical features of syndromic hearing loss and rule out potential environmental causes. Affected and some unaffected subjects underwent a general otologic examination. Hearing was evaluated in some affected and unaffected subjects by pure-tone air- and bone-conduction audiometry with or without tympanometry. No air-bone gaps were observed in any tested individuals. Vestibular function was assessed by tandem gait, Romberg testing and electronystagmography (ENG) with caloric stimulation. Funduscopic and electroretinography (ERG) examinations were performed by an ophthalmologist to confirm the absence or presence of RP. The ages of the affected individuals at the time of examination ranged from 8 to 27 years.
DNA isolation, genotyping and mutational analyses
Genomic DNA was extracted from peripheral blood samples using a standard protocol. We first screened for linkage of the deafness phenotype to STR (short tandem repeat) markers for all of the reported DFNB/USH loci using genomic DNA from affected and unaffected members of 557 families segregating autosomal recessive hearing loss. The USH1F/DFNB23 linked markers used are D10S1643, D10S546, and D10S2522.
All of the exons of PCDH15 (accession numbers AY029237; EU718480; EU718481; EU718482) and approximately 100 base pairs of intronic sequences flanking each exon (Ahmed et al. 2003; Ahmed et al. 2001) were PCR-amplified and sequenced using genomic DNA from an affected member of each USH1F/DFNB23 family. In the eight families where mutations in PCDH15 were found, DNA from all affected and unaffected members were then examined for the mutation.
cDNA cloning and sequence analysis of PCDH15
Downstream of the exon encoding the reported carboxy-terminus of the cytoplasmic domain (CD1) of human PCDH15 (Ahmed et al. 2003) there are regions of sequence conservation with predicted open reading frames. RT-PCR analyses revealed transcripts of PCDH15 in human retina cDNA (GEtRare™, Genemed) that have a subset of additional alternatively spliced exons that encode two novel cytoplasmic domains designated CD2 and CD3. All PCR products were subcloned and both strands were sequenced. PCR primers used for the amplification of PCDH15 are reported in Ahmed et al. 2001. The primers used to PCR amplify the 4 novel exons of PCDH15 described here are provided in Supplemental Table 1.
Molecular Modeling
Molecular modeling was performed using the C-cadherin crystal structure as a template (PDB-identifier 1q55) (He et al. 2003). Sequence identity between the EC repeat 2 and 3 of protocadherin 15 and C-cadherin was 26% over 252 amino acids. Modeling was done with the WHAT IF server (http://swift.cmbi.ru.nl) and YASARA (www.yasara.org) using standard parameter settings (Krieger et al. 2002; Vriend 1990).
Results
Linkage and Mutational Analyses
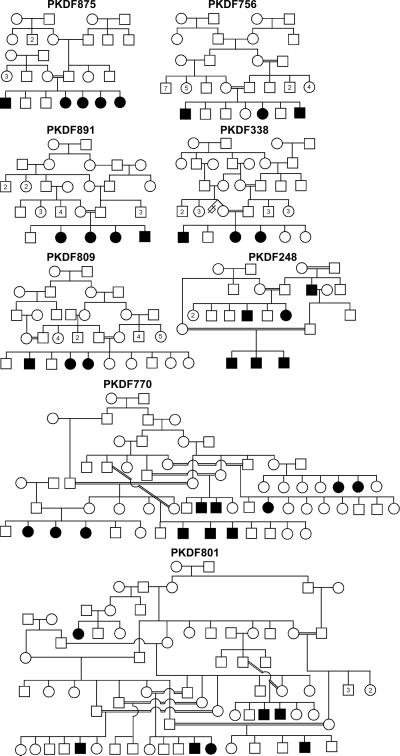
In 13 unreported Pakistani families (Fig. 1 and data not shown), we observed co-segregation of deafness with homozygosity for USH1F/DFNB23-linked STR marker genotypes. We identified homozygous mutant alleles of PCDH15 in affected individuals from eight of these 13 families (Fig. 1 and Table 1). These variants co-segregated with deafness in each of the eight families (Fig. 1) and were not found in 100 ethnically matched hearing control individuals (Table 1).
Fig. 1.
Pedigrees of eight Pakistani families segregating recessive sensorineural hearing loss linked to markers for PCDH15.
Table 1.
Mutations and clinical data of affected members of eight families segregating mutations of PCDH15
| Nucleotide changea |
Exon | Effect on Protein |
Domain | Allele frequency |
Family | Ethnicity | Auditory phenotype (sensorineural)d |
Visual phenotypee | Age at ambulation (months) |
Vestibular functionf |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Missense mutations: | |||||||||||
| c.400C→G | 5 | p.R134G | EC1 | 0/200 | PKDF756 | Punjabi | severe to profound | +ERG at age 12 years, +FE +ERG at age 16 years, +FE |
12-13 | +TG, +RM +TG, +RM |
|
| c.533A→G | 6 | p.D178G | EC2 | 0/200 | PKDF875 | Punjabi | profound | −FE at age 24 years −FE at age 27 years |
12-15 | −TG, −RM −TG, −RM |
|
| Splicing variants: | |||||||||||
| c.3717+1G→T | 27 | splicing error | b | 0/200 | PKDF248 | Sindhi | profound | ±FE at age 15 years ±FE at age 16 years |
12-15 | −TG, −RM −TG, −RM |
|
| Nonsense mutations: | |||||||||||
| c.7C→T | 2 | p.R3X | SP | 0/200 | PKDF338 | Punjabi | profound | ±FE at age 14 years ±FE at age 16 years |
24-28 | −TG, −RM −TG, −RM |
|
| c.1940C→G | 16 | p.S647X | EC6 | 0/200 | PKDF809 | Punjabi | profound | ±FE at age 17 years −FE at age 27 years |
12-15 | −TG, −RM −TG, −RM |
|
| c.2052C→A | 17 | p.Y684X | EC6 | 0/200 | PKDF801 | Punjabi | profound | ±FE at age 12 years −FE at age 21 years |
12-15 | −TG, −RM −TG, −RM |
|
| Deletions: | |||||||||||
| c.2483delT | 19 | p.E829KfsX12 | c | 0/200 | PKDF891 | Punjabi | profound | ±FE at age 8 years −FE at age 10 years −FE at age 12 years |
12-15 | −TG, −RM −TG, −RM −TG, −RM |
|
| c.4257delA | 32 | p.L1419FfsX99 | CD1 | 0/200 | PKDF770 | Saraiki | profound | −FE at age 19 years −FE at age 23 years |
12-15 | −TG, −RM −TG, −RM |
|
Codon numbering is based on cDNA and starts with the first in-frame methionine (accession no. AY029237). DNA numbering based on +1 as the A of the initiation codon.
Located in sequence between EC11 and transmembrane domain.
Located in sequence between EC7 and EC8.
evaluated by pure-tone audiometry.
eletroretinography (ERG) response: −, extinguished; ±, subnormal; +, normal; for fundus examination (FE) results: −, typical RP; ±, subnormal RP-like findings; +, normal.
+, normal vestibular function; −, vestibular dysfunction as determined by Tandem Gait (TG) and Romberg tests (RM).
SP, signal peptide; EC, ectodomain; CD1, cytoplasmic domain 1.
Missense mutations
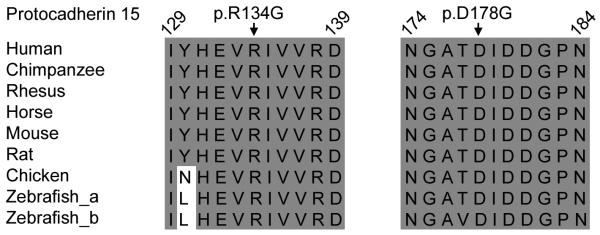
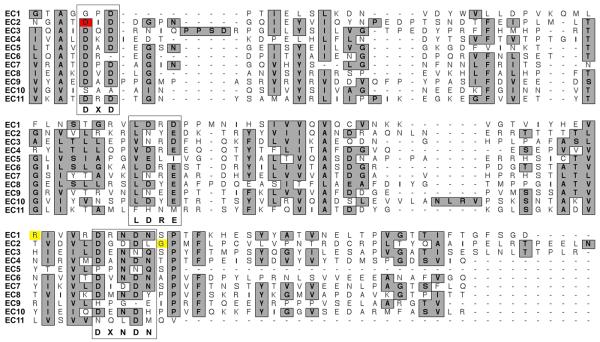
In family PKDF756 we found a transversion mutation (c.400C→G) in exon 5, resulting in a substitution of glycine for arginine at position 134 (p.R134G). This residue is located in the first ectodomain (EC1; Table 1). Amino acid alignments of several protocadherin 15 orthologues indicate conservation of arginine-134 (Fig. 2). A ClustalW amino acid sequence alignment of the eleven EC domains of protocadherin 15 reveals that the p.R134G mutation does not disrupt a conserved calcium-binding motif (Fig. 3).
Fig. 2.
ClustalW multiple amino acid sequence alignment of protocadherin 15 orthologs in two noncontiguous regions show that p.R134 and p.D178 are conserved across species (shaded background, same amino acids; light background, non-conserved amino acids).
Fig. 3.
Alignment of the eleven protocadherin 15 EC domains with missense mutations causing USH1F (red) and DFNB23 (yellow). A gray background indicates highly conserved or similar residues. The DXD, LDRE and DXNDN calcium-binding motifs are boxed.
All affected individuals of family PKDF756 were said to have begun walking at or before 12 months of age, and subsequent examinations show normal vestibular function (Table 1). Fundus examinations of two older individuals (12 and 16 years) show no evidence of RP, which was further confirmed by normal ERG waves amplitudes. However, these affected individuals are not old enough to make a conclusive diagnosis of nonsyndromic hearing loss DFNB23. The onset of RP may be delayed and appear in the second or third decade of life. However, the oldest affected individual in family PKDF70 segregating p.R134G was 44 years old at the time of examination and had normal ERG wave amplitudes (Ahmed et al. 2003). These data indicate that p.R134G is associated with nonsyndromic hearing loss DFNB23 (Ahmed et al. 2003).
The subjects from the remaining seven Pakistani families segregating PCDH15 mutations are profoundly deaf and started walking independently between 12-28 months (Table 1). Balance function of the 13 affected individuals from these seven families was evaluated by tandem gait and Romberg test. None of these affected individuals had a normal tandem gait or a normal Romberg test (Table 1). Fundus examination of the 13 affected individuals revealed a varied occular phenotype (Table 1) that ranged from slight pigmentary changes to complete retinal degeneration.
Sequence analysis of all PCDH15 exons (Table 1) in affected members of USH1 family PKDF875 demonstrated homozygosity for a missense change at a conserved residue in EC2 (c.533A→G, p.D178G; Fig. 2) encoded by exon 6. Alignments of protocadherin 15 EC domains shows that p.D178G does disrupt a conserved DXD calcium-binding motif of EC2 (Fig. 3).
Splice-site mutation
Family PKDF248 is segregating a splice-donor-site mutation (c.3717+1G→T) in intron 27 that alters the consensus “G” of the “GT” donor splice site of exon 27 (Table 1). The GENSCAN (http://genes.mit.edu/GENSCAN.html) predicted effect of c.3717+1G→T, when included in the otherwise wild type genomic sequence, is the use of a cryptic splice donor site in intron 27, a shift in the translation reading frame followed by a premature stop codon (p.V1240LfsX2). If mutant mRNA is not entirely degraded by the nonsense mediated decay pathway, the predicted protein could encode all of the 11 EC domains, but would lack the transmembrane and the cytoplasmic domain.
Protein truncating mutations
Three mutations causing premature stop codons were identified (Table 1). Affected individuals of family PKDF338 are homozygous for a transition mutation (c.7C→T) resulting in a stop codon at position 3 (p.R3X). This mutation was previously reported segregating with USH1 in two families (Ahmed et al. 2001; Alagramam et al. 2001b). In family PKDF809, we found a transversion mutation (c.1940C→G), converting a serine codon at position 647 to a stop codon (p.S647X). This mutation is predicted to truncate the protein in EC6 domain. Another nonsense mutation (c.2052C→A) causes a truncation in exon 17 encoding the EC6 domain (p.Y684X; Table 1) in family PKDF801. Affected members of families PKDF891 and PKDF770 (Fig. 1) are segregating a homozygous c.2483delT, which is predicted to truncate the protein before EC8, and c.4257delA located in exon 32 encoding cytoplasmic domain 1 (CD1), respectively (Table 1).
Novel exons of PCDH15
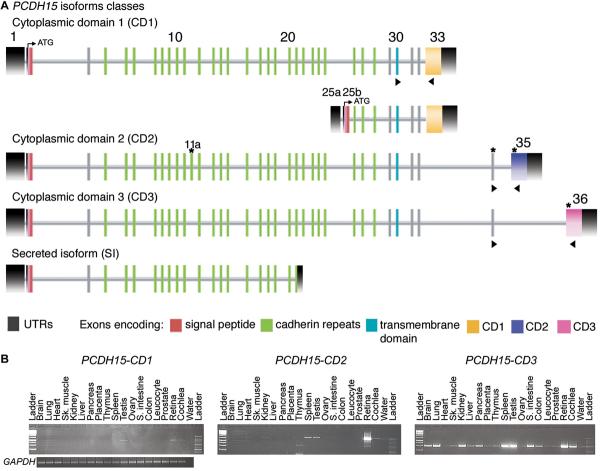
For five families segregating deafness linked to markers for PCDH15 we did not find pathogenic variants of PCDH15 after sequencing the 35 reported exons of this gene. Four of these five families have significant simulated LOD scores of 3.8, 5.2, 3.3 and 3.6. These data suggest either the presence of mutations of yet additional undiscovered exons of PCDH15, mutations in the introns or regulatory elements of PCDH15 (Alagramam et al. 2007), or an additional locus for type I USH at chromosome 10q21.1. Conserved synteny between mouse and human allowed us to identify four new exons (previously annotated as ESTs in the NCBI database), which encode two novel, conserved, cytoplasmic domains (CD2 and CD3) of protocadherin 15 (Fig. 4A). The amino acid sequences of CD1, CD2 and CD3 are entirely different from one another. Consistent with the nomenclature for the isoforms of Pcdh15 we reported for the mouse (Ahmed et al. 2006), there are also four isoform classes of human isoforms of PCDH15 (Fig. 4A). In human NK/T lymphoma cells, Rouget-Quermalet and co-authors reported a secreted isoform of protocadherin 15 (SI), which contains the first 21 exons of PCDH15 encoding the signal peptide and six EC (Rouget-Quermalet et al. 2006).
Fig. 4.
Structure and expression profile of PCDH15 isoforms. a Four isoform classes of PCDH15 defined by the presence or absence of one of three alternative, unique cytoplasmic domains (CD1, CD2, or CD3). Additional exons discovered after the initial report of PCDH15 structure (Ahmed et al. 2001) are designated with a letter-suffix (11a, 25a, 25b) since they are located among the previously reported 33 exons (Ahmed et al. 2003). Black shaded boxes designate the 5′UTR and 3′UTR. A signal peptide (red) is encoded by exon 2 and a transmembrane domain (blue) is encoded by exon 30. Four newly discovered exons identified in this study are marked with asterisks. Exons encoding the cadherin repeats are represented by green boxes and the exons 33 (yellow), 35 (purple) and 36 (pink) encode three alternative cytoplasmic domains (CD1, CD2, CD3), respectively. b Expression profiles of the three isoform classes of PCDH15 that have different cytoplasmic domains (CD1, CD2 and CD3) are shown. PCR primers (arrowheads in panel a) were designed to amplify CD1, CD2 and CD3 from cDNA prepared from different human tissues. Left expression profile, PCDH15-CD1 mRNA is present in testis, retina, and cochlear cDNA. Middle profile, PCDH15-CD2 is found in human heart, kidney, thymus, spleen, testis, retina, and cochlea. Right profile, PCDH15-CD3 is widely expressed. PCR amplification of GAPDH cDNA was used as a control for the quality and quantity of RT-PCR template.
The amino acid sequences of human and mouse CD1, CD2 and CD3 cytoplasmic domains of PCDH15 have 55% (67%), 83% (91%) and 77% (84%) identity (similarity), respectively (Supplemental Figs. 1-3). In addition to characterizing the isoforms of human PCDH15, we also determined the expression pattern of CD1, CD2 and CD3 isoforms in various tissues. PCDH15-CD1 has a limited pattern of expression and was detected in human testis, retina and cochlear cDNAs (Fig. 4B). PCDH15-CD2 expression was present in the human heart, kidney, thymus, spleen, testis, retina and cochlear cDNAs (Fig. 4B), whereas PCDH15-CD3 is a widely transcribed isoform and was detected in almost all of the cDNAs from 18 tissues that we tested (Fig. 4B). However, mutational screening of the novel exons of PCDH15 encoding CD2 and CD3 did not reveal additional pathogenic variants.
The USH1 linkage interval for the five presumably PCDH15-mutation-negative families include about 42 genes. After excluding putative regulatory elements and genomic deletions encompassing PCDH15, the 42 genes would then be screened for mutations, one of which may prove to be a third USH1 gene on chromosome 10q11.2-q21.1.
Molecular Modeling
The aspartic acid (p.D178) mutated in family PKDF875 segregating USH1 resides in the calcium binding motif in the loops between EC2 and EC3 (http://www.cmbi.ru.nl/~hvensela/PCDH15/). Glycine does not have a side chain in contrast to aspartic acid and therefore cannot interact with the calcium atoms. As a result the p.D178G allele is predicted to show decreased calcium affinity. The previously reported p.G262D mutation associated with DFNB23 is located close to the calcium-binding residues in the loops between EC2 and EC3 (Ahmed et al. 2003). Even though glycine-262 is not directly involved in binding calcium, it is predicted to help maintain the correct shape of the calcium-binding domain. Aspartic acid has a large side chain, while glycine does not have a side chain, so the aspartic acid at this position will disturb the local structure that is needed to bind calcium. R134G is also associated with nonsyndrome deafness DFNB23 (Table 1). This mutation is located in the middle of EC1 with its side chain pointing outwards. This domain is known to be important for interaction with a partnering cadherin or protocadherin monomer (Prakasam et al. 2006). Mutation from the hydrophilic and positively charged arginine into a glycine might disturb these interactions. Indeed p.R134G was shown in vitro to result in the loss of interaction between protocadherin 15 and cadherin 23 (Kazmierczak et al. 2007).
Discussion
To date, we have identified 18 families co-segregating deafness linked to markers for USH1F/DFNB23 (Ahmed et al. 2003; Ahmed et al. 2001). Mutations of PCDH15 were found in 13 of these 18 families. We did not find a pathogenic mutation in five of the twelve USH1 families. CDH23 in humans is located approximately 17.8 cM from PCDH15 on chromosome 10q22.1 (Bolz et al. 2001; Bork et al. 2001). Mutations of CDH23 can cause either type 1 USH or DFNB12 nonsyndromic hearing loss (Astuto et al. 2002; Bork et al. 2001). On the basis of meiotic recombinations in affected individuals, we can exclude mutations of CDH23 as the cause of USH1 segregating in four of these five PCDH15-linked but PCDH15 mutation-negative families. However, our genetic analyses do not exclude PCDH15.
To date, 21 recessive alleles of PCDH15 at the USH1F locus and two mutant DFNB23 alleles have been reported (Ahmed et al. 2003; Cremers et al. 2007). In addition, digenic inheritance was suggested as an explanation for three USH1 singeltons (Zheng et al. 2005). Each affected individual was found to be carrying one recessive mutant allele of PCDH15 and one recessive mutant allele of CDH23 (Zheng et al. 2005), and it is the combination of these two non-allelic mutations that was presumed to cause type 1 USH (Zheng et al. 2005). However, not all the exons of PCDH15 could have been checked for mutations and mutations were screened by SSCP analyses, not by direct sequencing of genomic DNA. Moreover, one of the three USH1 “digenic subjects” (family 1677) was reported to be homozygous for a pathogenic mutation T1209A of CDH23, while also a carrier of 5601delAAC in PCDH15 (Zheng et al. 2005) and is thus not an example of digenic inheritance. In our opinion, the evidence for digenic inheritance responsible for type 1 Usher syndrome is not robust.
In our cohort of 557 Pakistani families ascertained on the basis of severe to profound deafness, 2.3% have mutations of PCDH15. The majority of mutations of PCDH15 occur within the region encoding the EC domains or in CD1 (Supplementary Table 2). Mutations associated with nonsyndromic deafness DFNB23 or type 1 USH have not been found in the exons encoding the transmembrane domain or in exons encoding the alternative cytoplasmic domains CD2 and CD3 (Supplementary Table 2). Protocadherin-15-CD2 and protocadherin-15-CD3 are expressed in many cell types outside of the auditory system, and perhaps have other necessary functions. Although our sample size of mutations of PCDH15 is still small, it is tempting to speculate that mutations of the exons encoding CD2 and CD3 may not result in Usher syndrome but some other pleiotropic disorder.
The three missense mutations (p.R134G, p.D178G, p.G262D) of PCDH15 are located within exons 5 to 8 encoding the extracellular N-terminus of protocadherin 15. R134G and G262D are associated with nonsyndromic deafness DFNB23 and do not changes the conserved sequence of the DXD, LDRE and DXNDN motifs of the EC domains. These motifs are responsible for calcium-binding and rigidification of the EC domain (Alattia et al. 1997). So far only one homozygous missense mutation (p.D178G) is associated with USH1F and it does alter the canonical DXD motif of EC2 (Fig. 3), which is predicted to result in a reduced affinity for calcium. By comparison, all truncating mutant alleles of PCDH15 are associated with type 1 USH. Similarly, all of the 31 reported truncating mutant alleles of CDH23 cause USH1D (Astuto et al. 2002; Bolz et al. 2001; Bork et al. 2001; Ouyang et al. 2005; Roux et al. 2006). A better understanding as to how the retina is spared in deaf individuals homozygous for presumably hypomorphic mutations of PCDH15, CDH23, MYO7A or USH1C (Ahmed et al. 2003; Ahmed et al. 2002; Bork et al. 2001; Riazuddin et al. 2008) should be helpful in developing interventions that delay the age of onset and/or slow the progression of the RP component of Usher syndrome.
Supplementary Material
Acknowledgements
The authors are grateful to the families who made this research possible. We thank Barbara Ploplis and Nicholas Kusnezov for their technical support. We also thank Julie Schultz, Karen Friderici, and Anne Madeo for suggestions regarding this manuscript. This study was supported by the Higher Education Commission, Islamabad, Pakistan; Ministry of Science and Technology, Islamabad, Pakistan and by intramural funds to TBF from the National Institute on Deafness and Other Communication Disorders, NIH (1 ZO1 DC000039-11).
Footnotes
Electronic database information
GENSCAN, http://genes.mit.edu/GENSCAN.html
Protocadherin 15 Molecular Modeling data, http://www.cmbi.ru.nl/~hvensela/PCDH15
WHAT IF server, http://swift.cmbi.ru.nl
YASARA, www.yasara.org
REFERENCES
- Ahmed Z, Riazuddin S, Khan S, Friedman P, Riazuddin S, Friedman T. USH1H, a novel locus for type I Usher syndrome, maps to chromosome 15q22-23. Clin Genet. 2008 doi: 10.1111/j.1399-0004.2008.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, Griffith AJ, Frolenkov GI, Belyantseva IA, Richardson GP, Friedman TB. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci. 2006;26:7022–34. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, Sieving P, Riazuddin S, Griffith AJ, Friedman TB, Belyantseva IA, Wilcox ER. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–23. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, Menon PS, Deshmukh D, Griffith AJ, Riazuddin S, Friedman TB, Wilcox ER. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum Genet. 2002;110:527–31. doi: 10.1007/s00439-002-0732-4. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Miller ND, Adappa ND, Pitts DR, Heaphy JC, Yuan H, Smith RJ. Promoter, alternative splice forms, and genomic structure of protocadherin 15. Genomics. 2007;90:482–92. doi: 10.1016/j.ygeno.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001a;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Van Camp G, Hageman GS, Woychik RP, Smith RJ, Hagemen GS. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001b;10:1709–18. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- Alattia JR, Ames JB, Porumb T, Tong KI, Heng YM, Ottensmeyer P, Kay CM, Ikura M. Lateral self-assembly of E-cadherin directed by cooperative calcium binding. FEBS Lett. 1997;417:405–8. doi: 10.1016/s0014-5793(97)01333-1. [DOI] [PubMed] [Google Scholar]
- Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, Ohliger SJ, Riazuddin S, Morell RJ, Khan S, Riazuddin S, Kremer H, van Hauwe P, Moller CG, Cremers CW, Ayuso C, Heckenlively JR, Rohrschneider K, Spandau U, Greenberg J, Ramesar R, Reardon W, Bitoun P, Millan J, Legge R, Friedman TB, Kimberling WJ. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet. 2002;71:262–75. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SL, Bardenstein D, Alagramam KN. Assessment of retinal structure and function in Ames waltzer mice. Invest Ophthalmol Vis Sci. 2003;44:3986–92. doi: 10.1167/iovs.02-1009. [DOI] [PubMed] [Google Scholar]
- Ben-Yosef T, Ness SL, Madeo AC, Bar-Lev A, Wolfman JH, Ahmed ZM, Desnick RJ, Willner JP, Avraham KB, Ostrer H, Oddoux C, Griffith AJ, Friedman TB. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N Engl J Med. 2003;348:1664–70. doi: 10.1056/NEJMoa021502. [DOI] [PubMed] [Google Scholar]
- Bergstrom L, Hemenway WG, Downs MP. A high risk registry to find congenital deafness. Otolaryngol Clin North Am. 1971;4:369–99. [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del CSCM, Vila MC, Molina OP, Gal A, Kubisch C. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–12. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, Kimberling WJ, Kulm M, de Brouwer AP, van Wijk E, te Brinke H, Cremers CW, Hoefsloot LH, Banfi S, Simonelli F, Fleischhauer JC, Berger W, Kelley PM, Haralambous E, Bitner-Glindzicz M, Webster AR, Saihan Z, De Baere E, Leroy BP, Silvestri G, McKay GJ, Koenekoop RK, Millan JM, Rosenberg T, Joensuu T, Sankila EM, Weil D, Weston MD, Wissinger B, Kremer H. Development of a genotyping microarray for Usher syndrome. J Med Genet. 2007;44:153–60. doi: 10.1136/jmg.2006.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood-Watson RJ, 2nd, Ahmed ZM, Kjellstrom S, Bush RA, Takada Y, Hampton LL, Battey JF, Sieving PA, Friedman TB. Ames Waltzer deaf mice have reduced electroretinogram amplitudes and complex alternative splicing of Pcdh15 transcripts. Invest Ophthalmol Vis Sci. 2006;47:3074–84. doi: 10.1167/iovs.06-0108. [DOI] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–13. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- Hutchin T, Coy NN, Conlon H, Telford E, Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, Liu XZ, Bitner-Glindzicz M, Mueller R. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK - implications for genetic testing. Clin Genet. 2005;68:506–12. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, Kachar B. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Le Guedard S, Faugere V, Malcolm S, Claustres M, Roux AF. Large genomic rearrangements within the PCDH15 gene are a significant cause of USH1F syndrome. Mol Vis. 2007;13:102–7. [PMC free article] [PubMed] [Google Scholar]
- Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- Ouyang XM, Yan D, Du LL, Hejtmancik JF, Jacobson SG, Nance WE, Li AR, Angeli S, Kaiser M, Newton V, Brown SD, Balkany T, Liu XZ. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum Genet. 2005;116:292–9. doi: 10.1007/s00439-004-1227-2. [DOI] [PubMed] [Google Scholar]
- Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–97. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. Proc Natl Acad Sci U S A. 2006;103:15434–9. doi: 10.1073/pnas.0606701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, Zafar AU, Khan SN, Sabar F, Javid FT, Wilcox ER, Tsilou E, Boger ET, Sellers JR, Belyantseva IA, Riazuddin S, Friedman TB. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29:502–11. doi: 10.1002/humu.20677. [DOI] [PubMed] [Google Scholar]
- Rouget-Quermalet V, Giustiniani J, Marie-Cardine A, Beaud G, Besnard F, Loyaux D, Ferrara P, Leroy K, Shimizu N, Gaulard P, Bensussan A, Schmitt C. Protocadherin 15 (PCDH15): a new secreted isoform and a potential marker for NK/T cell lymphomas. Oncogene. 2006;25:2807–11. doi: 10.1038/sj.onc.1209301. [DOI] [PubMed] [Google Scholar]
- Roux AF, Faugere V, Le Guedard S, Pallares-Ruiz N, Vielle A, Chambert S, Marlin S, Hamel C, Gilbert B, Malcolm S, Claustres M. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90% J Med Genet. 2006;43:763–8. doi: 10.1136/jmg.2006.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Berlin CI, Hejtmancik JF, Keats BJ, Kimberling WJ, Lewis RA, Moller CG, Pelias MZ, Tranebjaerg L. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet. 1994;50:32–8. doi: 10.1002/ajmg.1320500107. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–6. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–1. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12:463–71. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, Johnson KR, Liu XZ. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14:103–11. doi: 10.1093/hmg/ddi010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.