Abstract
Genetic variants in embryonic lethal, abnormal vision, Drosophila-like 4 (ELAVL4) have been reported to be associated with onset age of Parkinson disease (PD) or risk for PD affection in Caucasian populations. In the current study we genotyped three single nucleotide polymorphisms in ELAVL4 in a Caucasian study sample consisting of 712 PD patients and 312 unrelated controls from the GenePD study. The minor allele of rs967582 was associated with increased risk of PD (odds ratio = 1.46, nominal P value = 0.011) in the GenePD population. The minor allele of rs967582 was also the risk allele for PD affection or earlier onset age in the previously studied populations. This replication of association with rs967582 in a third cohort further implicates ELAVL4 as a PD susceptibility gene.
Introduction
Parkinson disease (PD) is a complex neurodegenerative disorder with both environmental and genetic factors contributing to risk. Specific mutations in several genes have been identified which cause rare familial forms of PD including SNCA (Park1) (Polymeropoulos et al. 1997), Parkin (Park2) (Kitada et al. 1998), PINK1 (Park6) (Valente et al. 2004), DJ1 (Park7) (Bonifati et al. 2003), and LRRK2 (Park8) (Paisan-Ruiz et al. 2004). The role that these genes play in idiopathic PD is an area of active investigation, with LRRK2 being the most common.
Embryonic lethal, abnormal vision, Drosophila-like 4 (ELAVL4) (aliases HUD, PNEM) is located on chromosome 1p in a region [PARK10] linked to late onset PD in an Icelandic population (Hicks et al. 2002) and to age at onset of PD in a set of families ascertained from the US and Australia (Li et al. 2002). Follow-up studies in the US and Australian cohorts revealed evidence of association between single nucleotide polymorphism (SNP) markers in ELAVL4 and age at onset of PD (Noureddine et al. 2005). Subsequent examination in distinct cohorts identified association between ELAVL4 and risk for PD in an Irish case-control cohort, but not in Norwegian or US samples (Haugarvoll et al. 2007). Given Celtic origins in the Icelandic population this may suggest an Irish founder effect of the ELAVL4 association (Haugarvoll et al. 2007).
ELAVL4 is a human homologue of the Drosophilia gene elav, which is implicated in neuronal differentiation and maintenance. The ELAVL proteins are mRNA binding proteins and all members of the family contain three RNA-recognition motifs. Among other targets ELAVL4 binds to tau (Aranda-Abreu et al. 1999), which has been previously implicated in PD (see Healy et al. (2004) for meta analysis).
We examined association between ELAVL4 and the outcomes PD affection and onset age in the GenePD cohort.
Subjects and methods
Subjects
PD cases from families with multiple affected individuals were ascertained from 30 clinical sites as part of the GenePD Study. Neurologists from the participating sites examined and confirmed the diagnosis of each PD affected individual included in the study as previously described (Maher et al. 2002). Diagnostic criteria for PD were based on the United Kingdom PD Society Brain Bank Criteria (Gibb and Lees 1988) with slight modification. For example, multiple affected family members and head trauma were not considered exclusionary criteria. PD patients who were known to carry two parkin mutations (Sun et al. 2006) or a LRRK2 mutation were not included in the current analysis. The controls include Caucasian individuals recruited by the GenePD sites, which include some spouses, as well Caucasian controls recruited independently of the GenePD families. A set of 712 Caucasian PD patients from 404 families (55.5% male; mean onset age 60.8 ± 11.8) and 312 Caucasian unrelated controls (52.6% male; mean age at DNA collection 62.6 ± 11.0) were available for association testing. All participants signed informed consent forms and the study was approved by the Institutional Review Board at each of the participating clinical sites.
Genotype data
Tag SNPs with minor allele frequency >0.05 were selected to cover the region where previous association had been observed (Noureddine et al. 2005). Tag SNPs were replaced by previously published SNPs when there was substantial linkage disequilibrium between the Tag SNP and previously published SNP. Three SNPs in ELAVL4 (see Table 1) were genotyped in all individuals using TaqMan technology on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems Inc., Foster City, CA). Call rates were 0.95, 0.93, and 0.93 for rs967582, rs3902720, and rs12024093, respectively and all three SNPs were in Hardy–Weinberg equilibrium in controls subjects (P > 0.05).
Table 1.
Association results [odds ratio (OR), nominal P values (P), and minor allele frequencies (MAF)] for PD affection status and age at onset for ELAVL4 SNPs
| SNP | bp locationa |
MAF cases (%) |
MAF controls (%) |
Affection |
Age at onset |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Additive |
Dominant |
Recessive |
Additive |
Dominant |
Recessive |
||||||||||
| OR | P | OR | P | OR | P | Beta | P | Beta | P | Beta | P | ||||
| rs967582 | 50294192 | 36.6 | 30.8 | 1.303 | 0.021 | 1.460 | 0.011 | 1.272 | 0.294 | −0.208 | 0.764 | −0.058 | 0.951 | −0.644 | 0.616 |
| rs3902720 | 50317357 | 31.7 | 30.0 | 1.080 | 0.559 | 1.137 | 0.454 | 1.016 | 0.955 | −0.670 | 0.390 | −0.667 | 0.499 | −1.244 | 0.472 |
| rs12024093 | 50340871 | 18.9 | 17.3 | 1.114 | 0.502 | 1.147 | 0.460 | 1.056 | 0.909 | −1.212 | 0.223 | −1.591 | 0.144 | 0.001 | 1.000 |
Results are from analyses with no covariate adjustment
Chromosome 1 NCBI build 35; HapMap pairwise LD r2 (rs967582, rs3902720) = 0.03, r2 (rs967582, rs12024093) = 0.11, r2 (rs3902720, rs12024093) = 0.55
ELAVL4 association analysis
Linear and logistic models were used to assess the association between the genotype data in the ELAVL4 SNPs and age at onset of PD and PD affection status, respectively. For all analyses generalized estimating equations were employed to account for the non-independence of observations from within the same family. Analyses of age at onset included PD affected individuals only. Additive, dominant and recessive genetic models were considered. Analyses for PD affection and onset age were performed both with and without covariates in the model. Covariates included in the adjusted model for affection status were sex, age, and smoking defined as “ever smoked” (individuals who reported having, prior to the onset of PD symptoms, smoked regularly for at least 1 year) or “never smoked” (individuals who did not meet the ever smoked criteria). Covariate information was missing for some subjects and there were 662 PD and 285 control individuals included in the adjusted analysis for affection status. For the age at onset outcome, a single covariate indicating whether or not the subject was a heterozygote for a parkin mutation (Sun et al. 2006), was utilized and 676 PD subjects were included in the adjusted analysis.
Meta analysis of previously published results (Haugarvoll et al. 2007; Noureddine et al. 2005) and the current study examining SNP rs967582 and PD affection status was performed by combining P values as implemented in the program Metal (http://www.sph.umich.edu/csg/abecasis/metal/). Specifically, for each SNP a Z statistic was computed for each study based on the study specific P value and direction of the estimated effect. An overall Z statistic (and then corresponding P value) was computed as a weighted average of the study specific Z statistics, with the weights proportional to the square root of the number of individuals within each study. Weights were based on sample size defined as the number of subjects included in case control studies or the number of families used in the pedigree disequilibrium test. This method of meta analysis does not provide a summary effect measure but allows combination of association measures from different study designs such as the case-control (Haugarvoll et al. 2007) and family based designs (Noureddine et al. 2005) used in prior analysis of this region. No information about the direction of effect was provided for one population (Noureddine et al. 2005). Meta analysis was performed first assuming the direction of the effect was the same as the other studies and again, conservatively assuming the opposite direction from the other studies. Age at onset results were available for only two of the five populations and meta analysis was not performed for that outcome.
Results
Association results for the three ELAVL4 SNPs are shown in Table 1. Significant association was observed with either a dominant (P = 0.011) or additive model (P = 0.021) for PD affection status and SNP rs967582. Significant association remained after covariate adjustment (P = 0.029 for additive model and P = 0.020 for dominant model). No association was observed for age at onset of PD.
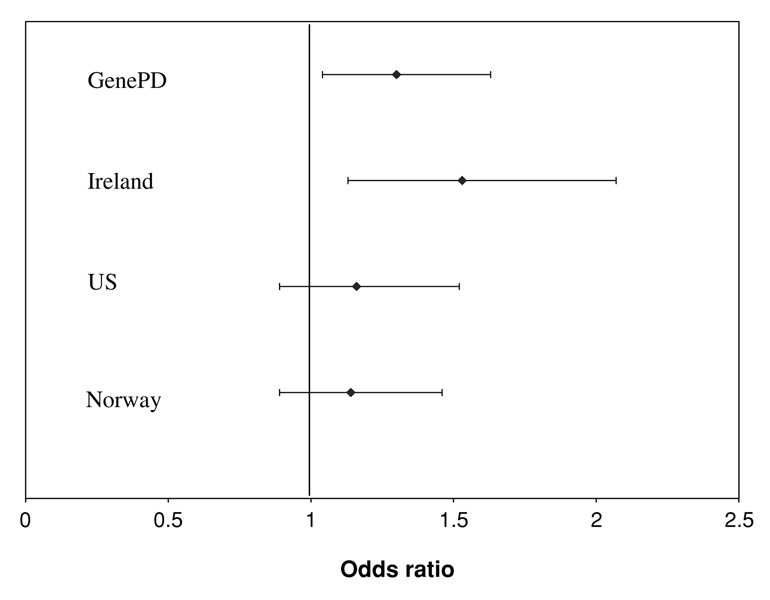
Meta analysis across five populations indicated overall association between SNP rs967582 and risk of PD affection status. In a conservative analysis, which assumed that the observed direction of effect that was unreported in (Noureddine et al. 2005) was opposite that of the other studies the meta analysis P value equaled 0.0063. Figure 1 shows the odds ratio and 95% confidence interval for the four populations for which estimates are available.
Fig. 1.
Odds ratio and 95% confidence interval for risk of PD affection status with increasing number of minor alleles of rs967582 from the current analysis (GenePD) and previously published populations (Haugarvoll et al. 2007). Another study in a US population found no association using a pedigree disequilbrium test. This study is not included in the figure as no effect measure was available (Noureddine et al. 2005)
Discussion
We examined association between risk of PD or age at onset of PD and SNPs in ELAVL4 in the GenePD study cohort. Significant association between the rs967582 SNP, and PD affection status was observed in this internationally recruited sample of familial PD cases, when examining nominal P values. A total of nine tests (three SNPs and three genetic models) were performed. Using a conservative Bonferroni correction the adjusted P value for rs967582 is 0.099 for the dominant model.
The minor allele of this SNP had previously been identified as associated with earlier age at onset of PD using family based association testing (Noureddine et al. 2005). In the current study, family relationships were accounted for analytically, and a population based association test was implemented. SNP rs967582 was recently associated with risk for PD in an Irish population but no significant association was observed for this SNP or others in ELAVL4 in an independent US or Norwegian cohort (Haugarvoll et al. 2007). Although it was suggested that the pattern of significance was due to a Celtic founder effect, the ORs for rs967582 in the US and Norwegian samples are in the same direction as those seen for the Irish population (1.14 and 1.16, respectively) (Haugarvoll et al. 2007). The current study, which represents an international sample of familial PD cases, suggests that the Haugarvoll et al. (2007) study may have had insufficient power to detect small increased odds of PD associated with the minor allele in their non-Celtic samples. In the Haugarvoll et al. (2007) study, as with the current study, the C minor allele is the risk allele, and the two studies report similar allele frequencies and odds ratio estimates. Meta analysis provides evidence of association across five populations.
The current meta analysis included all published association analyses between PD and SNPs in ELAVL4. Publication bias may lead to the dissemination of positive association findings rather than negative. This is a limitation of meta analyses performed using published results.
Replication of results is an important, but not final, step in implicating a gene as a risk factor for a complex disease. The minor allele in SNP rs967582 has now been implicated in three Caucasian PD populations for ELAVL4. Association studies in additional populations as well as biological studies to clarify the role of this gene in PD risk are justified.
Acknowledgments
This study was supported by PHS grant R01 NS36711-05 “Genetic Linkage Study in PD” and the Bumpus Foundation. The DNA samples contributed by the Parkinson Institute—Istituti Clinici di Perfezionamento, Milan, Italy, were from the “Human genetic bank of patients affected by PD and parkinsonisms,” supported by Italian Telethon grant n. GTF04007 and by the “Fondazione Grigioni per il Morbo di Parkinson.”
Contributor Information
Anita L. DeStefano, Email: adestef@bu.edu, Department of Biostatistics, Boston University School of Public Health, 715 Albany Street, Crosstown Center, 3rd floor, Boston, MA 02118, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
Jeanne Latourelle, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
Mark F. Lew, Department of Neurology, University Southern California School of Medicine, Los Angeles, CA 90033, USA
Oksana Suchowersky, Department of Clinical Neuroscience, University of Calgary, Calgary, AB, Canada T2N-4N1.
Christine Klein, Department of Neurology, University of Lübeck, 23538 Lübeck, Germany.
Lawrence I. Golbe, Department of Neurology, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08903, USA
Margery H. Mark, Department of Neurology, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08903, USA
John H. Growdon, Department of Neurology, Massachusetts General Hospital, Boston, MA 02114, USA
G. Fredrick Wooten, Department of Neurology, University of Virginia Health System, Charlottesville, VA 22908, USA.
Ray Watts, Department of Neurology, University of Alabama at Birmingham, Birmingham, AL 35233, USA.
Mark Guttman, Department of Medicine, University of Toronto, Toronto, Canada L6B-1C9.
Brad A. Racette, Department of Neurology, Washington University School of Medicine, Saint Louis, MO 63110, USA
Joel S. Perlmutter, Department of Neurology, Washington University School of Medicine, Saint Louis, MO 63110, USA
Lynn Marlor, Barrow Neurological Institute, Phoenix, AZ 85013, USA.
Holly A. Shill, Sun Health Research Institute, Sun City, AZ 85351, USA
Carlos Singer, Department of Neurology, University of Miami, Miami, FL 33136, USA.
Stefano Goldwurm, Parkinson Institute, Istituti Clinici di Perfezionamento, 39100 Milan, Italy.
Gianni Pezzoli, Parkinson Institute, Istituti Clinici di Perfezionamento, 39100 Milan, Italy.
Marie H. Saint-Hilaire, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA
Audrey E. Hendricks, Department of Biostatistics, Boston University School of Public Health, 715 Albany Street, Crosstown Center, 3rd floor, Boston, MA 02118, USA
Adam Gower, Department of Bioinformatics, Boston University, Boston, MA 02215, USA.
Sally Williamson, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
Michael W. Nagle, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA
Jemma B. Wilk, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA
Tiffany Massood, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
Karen W. Huskey, Department of Biostatistics, Boston University School of Public Health, 715 Albany Street, Crosstown Center, 3rd floor, Boston, MA 02118, USA Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
Kenneth B. Baker, Departments of Neurology and Neuroscience, Cleveland Clinic Foundation, Cleveland, OH 44195, USA
Ilia Itin, Departments of Neurology and Neuroscience, Cleveland Clinic Foundation, Cleveland, OH 44195, USA.
Irene Litvan, Department of Neurology, University of Louisville School of Medicine, Louisville, KY 40202, USA.
Garth Nicholson, Neurology Department, University of Sydney ANZAC Research Institute, Concord Hospital, Sydney, Australia.
Alastair Corbett, Neurology Department, University of Sydney ANZAC Research Institute, Concord Hospital, Sydney, Australia.
Martha Nance, Struthers Parkinson’s Center, Minneapolis, MN 55427, USA.
Edward Drasby, Port City Neurology, Scarborough, ME 04074, USA.
Stuart Isaacson, Parkinson’s Disease and Movement Disorder Center of Boca Raton, Boca Raton, FL 33486, USA.
David J. Burn, Institute for Ageing and Health/Regional Neurosciences Centre, Newcastle University, NE4 6BE Newcastle upon Tyne, UK
Patrick F. Chinnery, Institute for Ageing and Health/Regional Neurosciences Centre, Newcastle University, NE4 6BE Newcastle upon Tyne, UK
Peter P. Pramstaller, Department of Neurology, General Regional Hospital Bolzano, 39100 Bolzano, Italy
Jomana Al-hinti, Department of Neurology, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Anette T. Moller, Department of Neurology, Aarhus University Hospital, 8000 Aarhus, Denmark
Karen Ostergaard, Department of Neurology, Aarhus University Hospital, 8000 Aarhus, Denmark.
Scott J. Sherman, Department of Neurology, University of Arizona, Tucson, AZ 85724, USA
Richard Roxburgh, Department of Neurology, Auckland City Hospital, Auckland, New Zealand.
Barry Snow, Department of Neurology, Auckland City Hospital, Auckland, New Zealand.
John T. Slevin, Department of Neurology, University of Kentucky College of Medicine, Lexington, KY 40536, USA
Franca Cambi, Department of Neurology, University of Kentucky College of Medicine, Lexington, KY 40536, USA.
James F. Gusella, Molecular Neurogenetics Unit, Center for Human Genetic Research, Massachusetts General Hospital, Harvard Medical School Boston, MA 02114 Boston, USA
Richard H. Myers, Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA
References
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, Squitieri F, Krieger E, Vanacore N, van Swieten JC, Brice A, van Duijn CM, Oostra B, Meco G, Heutink P. DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003;24:159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugarvoll K, Toft M, Ross OA, Stone JT, Heckman MG, White LR, Lynch T, Gibson JM, Wszolek ZK, Uitti RJ, Aasly JO, Farrer MJ. ELAVL4, PARK10, and the Celts. Mov Disord. 2007;22:585–587. doi: 10.1002/mds.21336. [DOI] [PubMed] [Google Scholar]
- Healy DG, Abou-Sleiman PM, Lees AJ, Casas JP, Quinn N, Bhatia K, Hingorani AD, Wood NW. Tau gene and Parkinson’s disease: a case-control study and meta-analysis. J Neurol Neurosurg Psychiatry. 2004;75:962–965. doi: 10.1136/jnnp.2003.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks AA, Petursson H, Jonsson T, Stefansson H, Johannsdottir HS, Sainz J, Frigge ML, Kong A, Gulcher JR, Stefansson K, Sveinbjornsdottir S. A susceptibility gene for late-onset idiopathic Parkinson’s disease. Ann Neurol. 2002;52:549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher NE, Golbe LI, Lazzarini AM, Mark MH, Currie LJ, Wooten GF, Saint-Hilaire M, Wilk JB, Volcjak J, Maher JE, Feldman RG, Guttman M, Lew M, Waters CH, Schuman S, Suchowersky O, Lafontaine AL, Labelle N, Vieregge P, Pramstaller PP, Klein C, Hubble J, Reider C, Growdon J, Watts R, Montgomery E, Baker K, Singer C, Stacy M, Myers RH. Epidemiologic study of 203 sibling pairs with Parkinson’s disease: the GenePD study. Neurology. 2002;58:79–84. doi: 10.1212/wnl.58.1.79. [DOI] [PubMed] [Google Scholar]
- Noureddine MA, Qin XJ, Oliveira SA, Skelly TJ, van der Walt J, Hauser MA, Pericak-Vance MA, Vance JM, Li YJ. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum Genet. 2005;117:27–33. doi: 10.1007/s00439-005-1259-2. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Sun M, Latourelle JC, Wooten GF, Lew MF, Klein C, Shill HA, Golbe LI, Mark MH, Racette BA, Perlmutter JS, Parsian A, Guttman M, Nicholson G, Xu G, Wilk JB, Saint-Hilaire MH, DeStefano AL, Prakash R, Williamson S, Suchowersky O, Labelle N, Growdon JH, Singer C, Watts RL, Goldwurm S, Pezzoli G, Baker KB, Pramstaller PP, Burn DJ, Chinnery PF, Sherman S, Vieregge P, Litvan I, Gillis T, MacDonald ME, Myers RH, Gusella JF. Influence of heterozygosity for parkin mutation on onset age in familial Parkinson disease: the GenePD study. Arch Neurol. 2006;63:826–832. doi: 10.1001/archneur.63.6.826. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]