Abstract
Type IV pili are major bacterial virulence factors supporting adhesion, surface motility, and gene transfer. The polymeric pilus fiber is a highly dynamic molecular machine that switches between elongation and retraction. We used laser tweezers to investigate the dynamics of individual pili of Neisseria gonorrheae at clamped forces between 8 pN and 100 pN and at varying concentration of the retraction ATPase PilT. The elongation probability of individual pili increased with increasing mechanical force. Directional switching occurred on two distinct timescales, and regular stepping was absent on a scale > 3 nm. We found that the retraction velocity is bimodal and that the bimodality depends on force and on the concentration of PilT proteins. We conclude that the pilus motor is a multistate system with at least one polymerization mode and two depolymerization modes with the dynamics fine-tuned by force and PilT concentration.
Introduction
Type IV pili are major bacterial virulence factors that mediate adhesion to host cells and abiotic surfaces. The length of the polymeric cell appendages is dynamic, and the dynamics depend on the expression of the pilus retraction protein PilT (1,2) or its homologs. Dynamic properties and mechanical force generation via pili control a variety of functions such as host cell response (3,4), twitching motility (5), horizontal gene transfer (6,7), and biofilm formation (8). Infectivity is decreased in enteropathogenic Escherichia coli strains with nondynamic pili (9). During infection of endothelial cells by Neisseria meningiditis, the PilT level has been reported to be upregulated (10), suggesting that the PilT concentrations are altered during infection. Therefore, it is important to understand the interplay between PilT concentration, external mechanical force, and pilus dynamics.
Type IV pili form helical filaments with a pitch of 4 nm in which the effective length of a single subunit is on the order of 1 nm (11). The length of a pilus is controlled by two ATPases: PilF supports pilus elongation by polymerization of pilin subunits stored in the cell inner membrane, and PilT is required for pilus retraction, most likely in conjunction with organelle depolymerization. PilT has ATPase activity (12) and can form a hexamer in vitro (13). Both ATP binding and hydrolysis are necessary to support twitching motility (14).
Laser tweezers experiments demonstrated that pili generate remarkably high force by retraction (1). Variation of the pilus concentration did not alter the stalling force of 100 pN, indicating that individual pili generate force on the order of 100 pN (15); therefore, to our knowledge, the pilus motor is the strongest linear motor reported in the literature to date. Pilus bundling supports force generation in the range of 1 nN (16). Mechanical force near the stalling force induces switching from pilus retraction to pilus elongation at reduced PilT concentration (17). The velocity versus force relationship shows a characteristic profile in which the retraction velocity is constant at 1 μm/s at low forces and decreases exponentially at forces > 50 pN. Theoretical models using small ensembles of Brownian motors, but not isolated motors, can retrieve these characteristics in agreement with a hexameric structure of the PilT complex (18). Different molecular models have been proposed to explain the pilus motor mechanism (19–22). To elucidate the working mechanism of the pilus machine, it is essential to characterize the influence of force and the PilT concentration of the kinetic scheme to generate the characteristic shape of the velocity versus force relationship for both pilus retraction and pilus elongation.
In this study, we investigated directional switching of pilus dynamics while the force was clamped at different values between 8 pN and 100 pN for several seconds. We found that directional switching occurred at two distinct timescales and that the probability for pilus elongation increased with force and decreased with PilT concentration. Unexpectedly, we found that pilus retraction velocity was bimodal and that the existence of the high velocity mode as a function of increasing force was dependent on PilT concentration. These findings demonstrate that the type IV pilus system is a multistate system in which the free energy states are controlled by both PilT concentration and mechanical force. We hypothesize that this multistate system enables the bacterium to rapidly adjust its dynamics, i.e., directionality and velocity, to adjust the tension applied by pili. This finding is likely to be important for coordination of twitching motility and the interaction with mammalian host cells.
Materials and Methods
Bacterial strains and media
We used strains N. gonorrheae MS11 (WT), an isogenic derepressible strain MS11pilT ind (15,29), and an isogenic pilT-overexpressing MS11pilT oe strain (10). Bacteria were maintained on agar containing 5 g/L NaCl (Roth, Darmstadt, Germany); 4 g/L K2HPO4 (Roth); 1 g/L KH2PO4 (Roth); 15 g/L protease peptone (no. 3; BD Biosciences, Bedford, MA); 0.5 g/L starch (Sigma Aldrich, St. Louis, MO); 10 mL/L IsoVitaleX enrichment and 10 mL/L VCN (both purchased at BD Biosciences); and 0 or 0.25 mM isopropyl β-D-thiogalactoside (IPTG; Biomol, Hamburg, Germany) and were grown overnight at 37°C and 5% CO2.
Experimental setup
Retraction experiments were performed in phenol red-free Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) with 2 mM L-glutamine (Gibco), 8 mM sodium pyruvate (Gibco), 5 mM ascorbic acid (Roth), 30 mM HEPES (Roth), and 0 or 0.25 mM IPTG (Biomol) at 37°C. The bacteria were immobilized by attachment to a polystyrene-coated cover slide. For retraction experiments, a suspension of gonococci and 2 μm carboxylated bovine serum albumin–treated latex beads (Polysciences, Warrington, PA) was mounted on a microscope slide and sealed.
Performance of the setup
The detector assembly was calibrated by moving it by a linear actuator (M-403.12S; Physik Instrumente (PI), Karlsruhe, Germany) at a constant speed and holding a bead in the laser trap without applying any force to it. Using the drag force method, the linear range of detector and trap was measured to be ± 450 nm (data not shown).
The total performance of the apparatus was determined by the following procedure: A bead was bound to a pilus of an MS11pilT− bacterium (a mutant that does not perform pilus retraction), and the sample was moved with the motorized microscope table. The force clamp was thereby triggered, and the track was recorded. We determined the randomness from the average of 20 tracks with an average duration of ∼1 s for a velocity of 2.5 μm/s to be 0.2127 ± 0.0006 for an assumed step size of 1 nm. No peak in the histogram of pairwise distances was observed down to 1 nm for a track of 1 s duration with the same speed. In the range from 0.2 nm to 1 nm, variations with a peak-to-peak amplitude of ±12% were measured. A line fit to 4 tracks with durations in the second range resulted in residuals with a standard deviation (SD) of 3.5 ± 0.7 nm. Please note that this value depends on the length of the selected tracks. To determine the step size, the performance of the apparatus on short timescales is more relevant. From 10 track parts of 100 ms duration each, we determined the SD of the residuals to be 2.1 ± 0.5 nm. The algorithm for the dissection of the tracks provided single data pieces for each motor movement. A measurement with a stuck bead on a cover glass and a motor velocity of ∼200 nm/s resulted in a randomness of 0.06 ± 0.02. In these data sets, no peak in the histogram of pairwise distances was observed (see Fig. 2).
Figure 2.
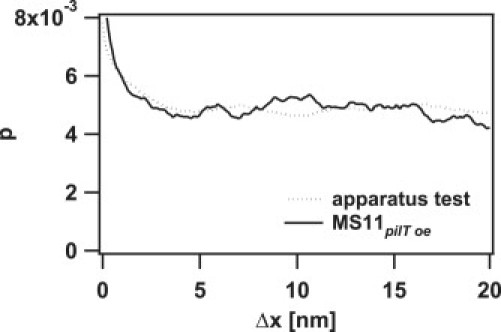
Pairwise displacements of pilus retraction. This example shows the normalized probability p of pairwise displacements Δx from a pilus retraction event of MS11pilT oe at 100 pN with an average velocity of 251 nm/s (solid line) and a test in which the microscope stage moved a stuck bead on a cover glass with a velocity of 283 nm/s (dotted line).
Randomness calculation
The randomness was calculated according to Svoboda et al. (23). A line fit was subtracted from the track. The track and an in-time shifted copy were subtracted from each other, and the squared values were averaged. Data from different retraction events were averaged with a weight corresponding to the root of the number of datapoints contributed to each point by each event. A linear fit weighted by the SD obtained from averaging was then applied between 5 and 100 ms. The slope represented the randomness divided by the step size. The error was obtained from the linear fit.
Results
Type IV pili switch between retraction and elongation
We improved a previously established assay for characterizing retraction kinetics and force generation by single pili (15) by implementing a force clamp (Fig. 1 a) to laser tweezers. Individual N. gonorrheae bacteria were immobilized on a polystyrene-coated coverslip. Using laser tweezers, a 2 μm polystyrene bead was approached to the bacterium. Eventually, a pilus bound to the bead and, upon pilus retraction, displaced it by a distance d from the center of the laser trap. After reaching a preset force threshold, a computer-controlled feedback was triggered that displaced the bacterium by a distance x(t) to keep the force (i.e., the distance d between the center of the laser trap and the bead) constant. The assay enabled us to follow the dynamics of a single pilus for up to several seconds while applying a constant force. Due to the force clamp, no elastic effects interfered with the determination of velocities and length.
Figure 1.
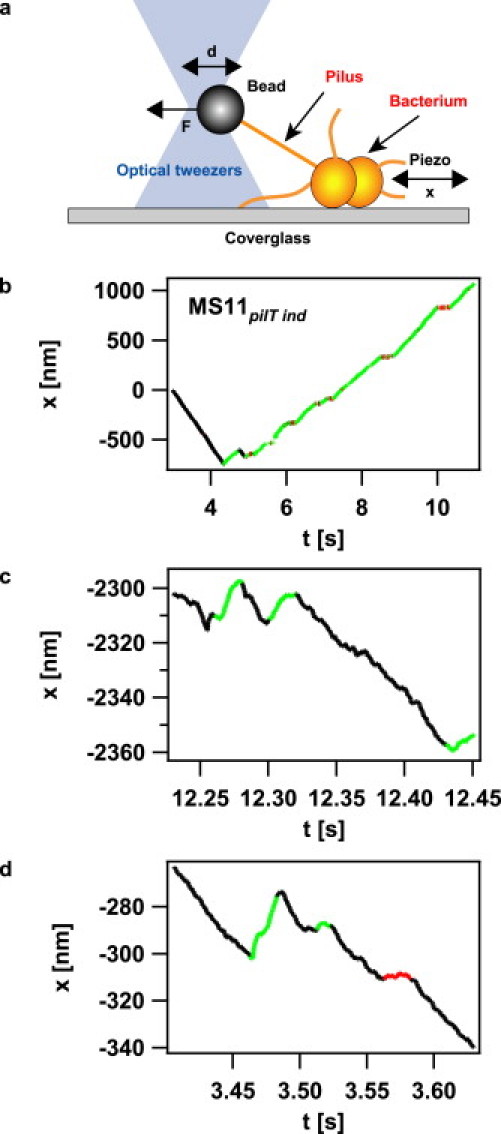
Experimental setup. (a) Sketch of the setup. A bacterium was immobilized on a polystyrene-coated cover glass. When a pilus was bound to the bead in the laser trap and retracted, the bead was displaced by a distance d from the center of the trap. At a predefined distance d corresponding to a force F, a feedback was triggered that moved the piezo table to keep d constant. (b) Typical length change x of a single pilus of MS11pilT ind as a function of time at 60 pN; (c and d) MS11pilT oe at 100 pN. The curves b to d were broken down into retraction intervals (black), elongation intervals (green), and pauses (red).
The movement of the bacterium with respect to the bead is a measure for the length change of the pilus, i.e., negative displacement (xt − xt − τ < 0) indicated pilus retraction and positive displacement indicated pilus elongation (xt − xt −τ > 0) (Fig. 1, b–d). The data were dissected into periods of pilus retraction, elongation, and pausing using parameters explained in detail in the Supporting Material (see Fig. S1 in the Supporting Material).
Analysis of stepping
We calculated the pairwise displacement histogram (Fig. 2) of pilus retraction at 100, 80, and 60 pN, and we found no peaks that were significantly different from control measurements in which a bead stuck to the cover glass was moved using an electric motor. Because the resolution of our apparatus was determined to be 2.1 ± 0.5 nm for a 100 ms window (Materials and Methods), our data show no regular stepping on a scale > 3 nm.
Unresolved switching may occur at a shorter lengthscale. To circumvent these limitations, we analyzed the data statistically using the randomness parameter r = (〈(x(t) − 〈x(t)〉)2〉 − 2〈η2〉)/δ〈x(t)〉, where x(t) is the displacement at the time t, δ is the step size, and 〈η2〉 is the Brownian displacement of the bead (23). As expected, we found that the randomness increased with external force and decreasing PilT concentration as the probability for directional switching increased (Fig. 3). At 8 pN, the randomness was independent of PilT concentration in the range of r = 3. Because we found that the elementary step size is < 3 nm, this observation suggests additional dynamics (back steps and pauses) at the millisecond timescale or shorter.
Figure 3.
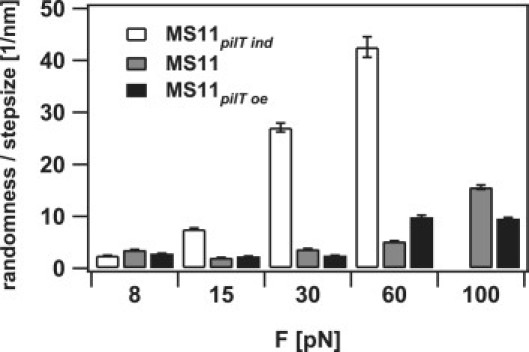
Randomness parameter r normalized to step size: (gray) MS11; (white) MS11pilT ind; and (black) MS11pilT oe. Error bars represent SD.
Probability for pilus elongation increases with increasing force and decreasing PilT concentration
We analyzed how external force affected the probabilities for pilus retraction, elongation, and pausing after pilus retraction triggered the measurement. The probability for a pilus to retract, elongate, or pause was defined as the contribution of the state to the total duration of all tracks under the specified conditions. At low forces, pilus retraction was clearly dominant; the probability for elongation and pausing was low but nonzero, i.e., 0.5% elongation and 2.5% pausing (Fig. 4, a and c). At high forces (i.e., in the range of 100 pN), the probability for elongation and pausing increased strongly to 17% and 8%, respectively (Fig. 4, b and c).
Figure 4.
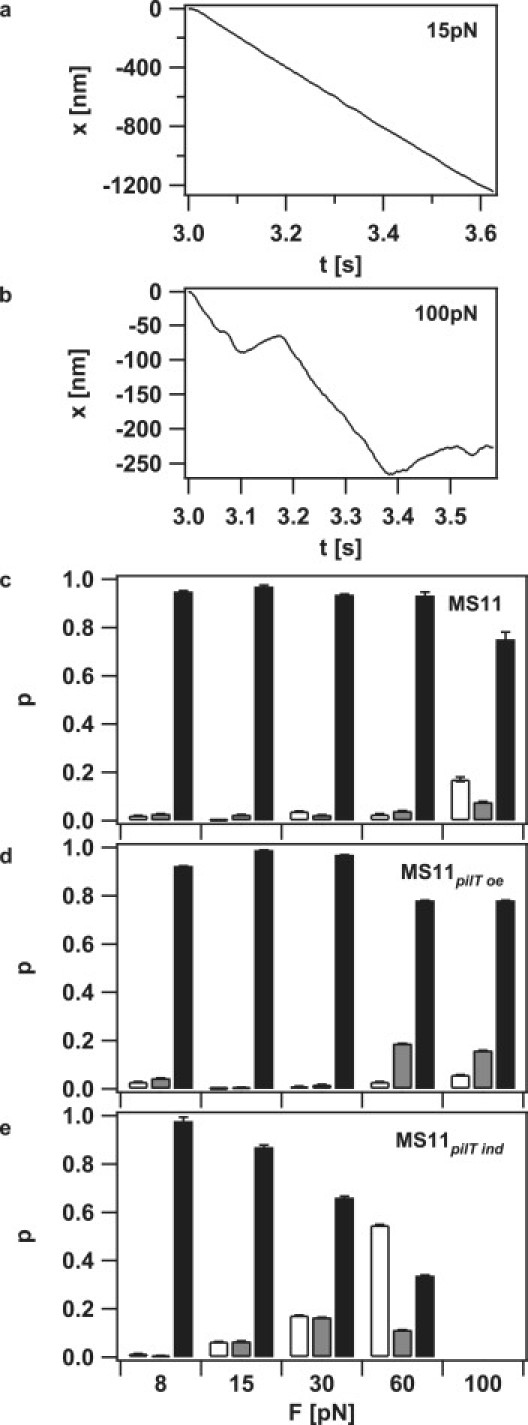
External force enhances the probability for pilus elongation. (a) Length change of a single pilus at 15 and (b) at 100 pN. Negative length change corresponds to pilus retraction. (c–e) Probability for pilus elongation (white), pausing (gray), and pilus retraction (black) for (c) MS11, (d) MS11pilT oe (increased PilT), and (e) MS11pilT ind (decreased PilT). The corresponding data set contains 465 retraction curves. Error bars represent SD obtained from statistical resampling.
We next investigated the influence of PilT concentration on pilus dynamics at varying forces. To this end, we used a PilT-overproducing strain in which the PilT concentration was increased and a pilT-inducible strain with decreased PilT concentration compared to the wild-type strain. Relative PilT concentrations were determined by Western blotting and demonstrated a ∼25-fold variation of PilT concentration between the different mutant strains (Fig. S4). When pilT was overexpressed, retraction probability was largely unaffected; however, the elongation probability decreased to 6% at 100 pN, whereas the pausing probability increased to 16% (Fig. 4 d). Decreasing the PilT concentration strongly affected the retraction/elongation probability ratio. Although the values are comparable with the wild-type at 8 pN, the elongation probability increased dramatically to 55% at 60 pN compared to 3% for the wild-type (Fig. 4 e). Due to the decreased frequency of retraction events, we did not extend the measurements to greater forces.
Switching between retraction and elongation occurs at two distinct timescales
The individual tracks show two time intervals in which the pilus motor changes direction. In the example shown in Fig. 1, c and d, a stretch of retraction was disrupted by short stretches of elongation at a timescale of several milliseconds and a lengthscale of several nanometers, whereas switching also occurred at a much larger timescale of hundreds of milliseconds and hundreds of nanometers after 4 s (Fig. 1 b). To quantify the timescales at which switching occurred, we plotted the cumulative distribution of retraction durations and elongation durations (Fig. 5, a and b). The underlying time resolution was 10 ms. The distribution could be fitted by a double exponential function, whereas the fit to a single exponential function was poor. We restricted the analysis to the derepressible mutant (low PilT) because the elongation was best characterized in this mutant. The average track length was ∼1 s. At reduced PilT concentration, the best fit by the double exponential function showed a timescale of tens of milliseconds and a distinct timescale of hundreds of milliseconds, with a tendency for decreasing values with increasing force for pilus retraction (Fig. 5, c and d). Note that the absolute values of the timescales depend on the time resolution set to dissect the data and that τ1 is likely to be overestimated since our temporal resolution is 10 ms (Figs. S1 and S2). The fact that switching occurs at distinct timescales and the orders of magnitude are independent of data dissection. For high forces, during which switching occurs most frequently, the pilus retraction velocity is 700 nm/s; thus, the different scales correspond to a few pilin monomers or several hundreds of monomers, respectively.
Figure 5.
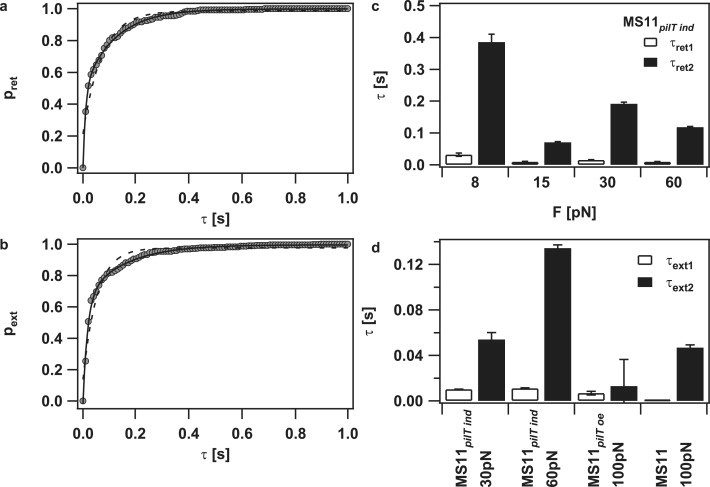
Different timescales of pilus elongation and retraction intervals. (a) Cumulative histogram of pilus retraction intervals (MS11pilT ind, 60 pN; circles). (dashed line) Best fit to a single exponential function and (solid line) best fit to a double exponential function with τret1 = 9 ± 1 ms, τret2 = 118 ± 2 ms. (b) Cumulative histogram of pilus elongation intervals (MS11pilT ind, 60 pN; circles). (dashed line) Best fit to a single exponential function and (solid line) best fit to a double exponential function with τext1 = 11.5 ± 0.5 ms, τext2 = 134 ± 3 ms. (c) Pilus retraction intervals obtained from the double exponential fit of cumulative histogram (MS11pilT ind) of pilus retraction intervals. (d) Pilus elongation intervals obtained from the double exponential fit of cumulative histogram of pilus elongation intervals.
Pilus retraction velocity is bimodal
To investigate how the velocity of pilus retraction was affected by external force, we plotted the velocity histograms (Fig. 6 a). The distribution was bimodal, with average velocities of 745 ± 42 nm/s and 2165 ± 253 nm/s up to forces of 15 pN; at higher forces, the high velocity mode disappeared. The position of the low velocity peak did not shift as a function of external force up to forces of 100 pN, indicating that the retraction velocity of this mode was independent of force (Fig. 6 c). In the strain with reduced PilT concentration, the high velocity mode was only detectable at 8 pN but not at higher forces (Fig. 6 b). In the PilT-overproducing strain, the high velocity mode was detectable at F < 100 pN, and the average velocity of both modes decreased with increasing force (Fig. 6, c and d). We rarely observed switching of retraction speed within one curve (Fig. 7), indicating that the average switching frequency between different velocity modes is < 0.1 Hz. Because switching between the two modes occurred within one retraction event, cellular individuality (e.g., ATP levels) cannot exclusively account for the bimodality of pilus retraction.
Figure 6.
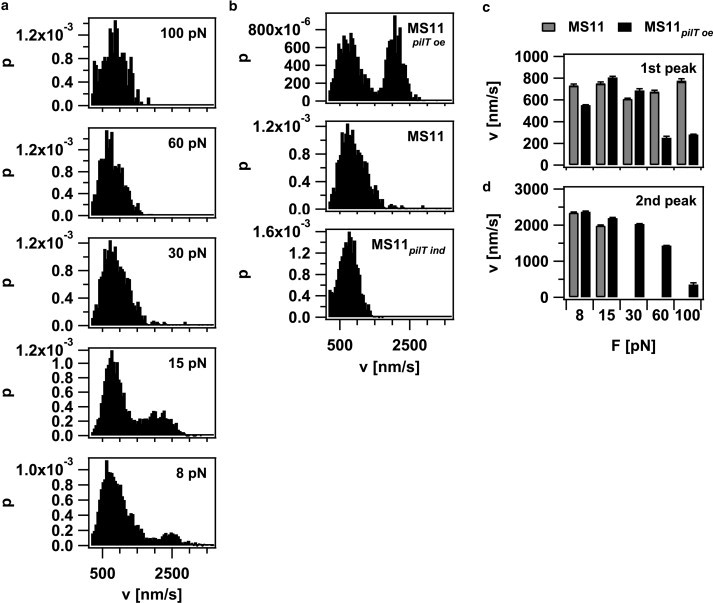
Velocity of pilus retraction is bimodal. (a) Histogram of retraction velocities at varying force (MS11) of in total 149 events from 29 bacteria (b) Histogram of retraction speeds at varying PilT concentrations at 30 pN. (c) Average velocity of the low velocity mode: (gray) MS11 and (black) MS11pilT oe. (d) Average velocity of the high velocity mode: (gray) MS11 and (black) MS11pilT oe.
Figure 7.
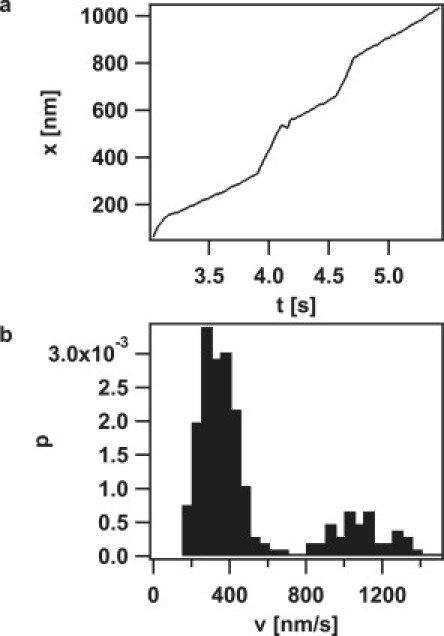
Velocity switching events during a single pilus retraction event. (a) Example of length change of the pilus of a MS11pilT oe bacterium at 100 pN over time with distinct velocities modes. (b) Corresponding histogram of the retraction speed of a.
Discussion
Our results lead to a better understanding of the control and molecular mechanism of type IV pilus dynamics and narrow down potential molecular motor mechanisms. First, the elementary step length is < 3 nm. Second, we found that directional switching of pilus length change occurred at forces between 8 pN and 100 pN and at 5-fold increased or 5-fold decreased PilT concentration as compared to the wild-type. Elongation and retraction intervals showed two distinct timescales—one timescale corresponding to a few monomers assembled into or disassembled from the pilus within 10 ms or less, and another timescale of more than ∼100 ms corresponding to hundreds of monomers—suggesting different underlying processes. Third, we found that pilus retraction velocity is bimodal, suggesting that two different compositions of the motor complex support pilus retraction. These observations strongly suggest that the type IV pilus system is a multistate system in which the dynamics can be fine-tuned by switching between different states.
Velocity versus force relationship of pilus retraction is determined by directional switching probability
In preliminary experiments, we characterized the velocity versus force relationship of type IV pilus retraction at varying force and low temporal and spatial resolution. We found that the velocity was constant at forces < 50 pN and, when the force was increased above this level, the velocity decayed exponentially with a stalling force of 110 pN (15). At the stalling force, we observed that pili often elongated at reduced PilT concentration (17). In this study, we improved the assay in two respects: We increased the spatial and temporal resolution, and we clamped the force to distinguish between force-induced effects and events occurring on a timescale > 100 ms. For these reasons, we could detect directional switching at timescales of 10 ms and 100 ms with the wild-type and at decreased or increased PilT concentrations. Our refined data cannot be reconciled with a simple Arrhenius-like scheme in which backward steps are neglected; two new aspects must be taken into account. First, the average velocity at low force is determined by an average value of the high velocity and the low velocity mode. At forces >30 pN, the high velocity mode disappears in a wild-type background, and the location of the low velocity mode is independent of force up to forces of 100 pN. In contrast, the probability for elongation increases continuously with increasing force; therefore, the average velocity of pilus retraction decreases. Please note that it is not possible in this setup to determine the stalling force because “stalled” pili would not trigger the force clamp.
Why does the probability for pilus elongation depend on force? The fact that the probability for pilus elongation depends both on force and on PilT concentration may be explained by force-dependent PilT binding constant to a complex at the base of the pilus. Diminished PilT function under tension may be compensated by elevated PilT levels, leading to a stabilization of the pilus retraction process at high PilT levels. In contrast, the elongation intervals increased with increasing force. Because we did not observe force dependence of the low velocity mode for retraction, the probabilities cannot be governed by a lowered energy barrier for the addition of a subunit due to the applied force. At increased PilT concentration and at high forces, the probability of pausing increased sharply, and the low velocity mode shifted toward lower values. This finding might indicate that, at high forces, the pilus retraction machinery often gets trapped or locked in a state that cannot support pilus dynamics. At wild-type levels of PilT, the pilus system may escape, releasing tension to enable PilT and, eventually, associated proteins to regain their working position and restart retraction.
The elementary steps of pilus retraction and elongation are < 3 nm
Different models have been proposed to explain the molecular mechanism of pilus elongation and retraction. It has been suggested that PilT directly binds to the pilus and acts as a cyclic molecular motor that displaces the pilus fiber with respect to the membrane by a powerstroke mechanism (19). In this scenario, the elementary step may be the translocation of either one subunit (∼1 nm) or one helical turn (∼4 nm). Studies based on structural and yeast two-hybrid experiments (21,24) have proposed a piston-like mechanism with an elementary step length of 1 nm in which the PilG protein would act as a scaffold for binding of either PilT or PilF. To address the question of whether the elementary step size was 1 helical turn or 4 nm, we carried out a pairwise subtraction of displacements and found no indication of steps on this order (i.e., 4 nm or higher).
Analysis of retraction curves at high forces and low velocities provided no evidence for steps larger than the resolution of our setup of 3 nm (i.e., in the range of 3 subunits). We conclude, therefore, that the elementary step is <1 helical turn.
Even at very low forces and high PilT concentration, the randomness r was still significantly higher than r = 1, the value expected for a motor with a Poisson waiting time distribution and without back steps. The high randomness value thus indicates unresolved steps at or below the millisecond timescale, potentially through significant backstepping.
Pilus dynamics is controlled by switching between multiple states
Assuming that a single process induced directional switching, we would expect an exponential distribution of elongation and retraction intervals as observed for run lengths of kinesin and myosin V (25,26). Our experiments, however, are clearly at odds with single exponential behavior; yet, they are in good agreement with a double exponential fit, indicating that directional switching occurred on two different timescales that are separated by 1 order of magnitude. Furthermore, switching probability increased with decreasing PilT concentration.
In summary, we tentatively suggest that the pilus system can switch between at least three different energy landscapes (Fig. 8). We propose that binding of PilT to the base of the pilus sets the system into a retraction-competent state; in addition, binding and unbinding (or activation and deactivation) of PilT probably occurred on a timescale of 100 ms to 1 s (Fig. 8, a and b). Also, the duration of long-time retraction intervals decreased with increasing force, and the duration of long-time elongation intervals increased with increasing force; both these findings are consistent with the force-dependent binding/activation constant of PilT. In contrast, the duration of the short-time retraction and elongation did not demonstrate any strong dependence on force or PilT concentration. Therefore, we suggest that these short timescale directional changes may be attributed to backward steps within a preset state of the motor. In other words, when the motor is in the retraction state through PilT binding, eventual elongation may occur because the motor steps backward. Conversely, when the motor is in the elongation-competent state, the motor may occasionally retract several pilins.
Figure 8.
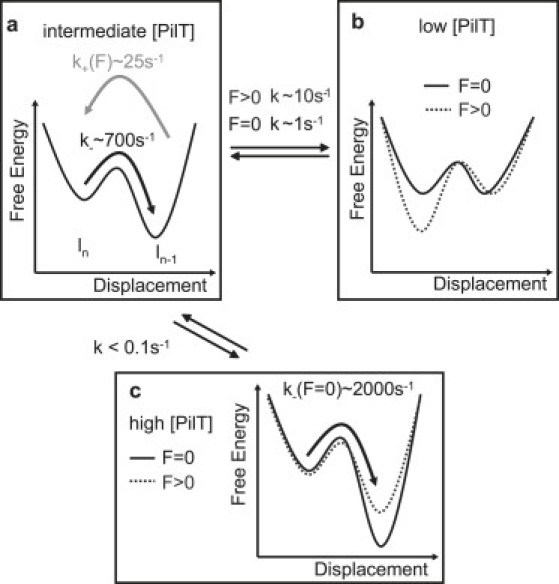
Hypothetical kinetic model describing the experimental data. Switching between different energy landscapes may explain different velocity modes and different timescales of switching. (a) Free energy of depolymerized state is energetically lower than polymerized state. Thus, the retraction rate k− is higher than the elongation rate k+. (b) With increasing external force, the free energy of the polymerized state decreases compared to the depolymerized state. The switching rate k between the landscapes depends on force. (c) The high velocity mode may be described by an energy landscape in which the free energy difference between polymerized and the depolymerized states increases compared to a.
Our results demonstrated that the velocity of type IV pili retraction shows bimodal behavior. Could multiple pili be responsible for bimodality? Previous experiments with varying concentrations of pili per cell showed that, even with wild-type bacteria retraction, events resulting from more than one pilus are negligible. The histogram of stalling force showed only one mode (15). Moreover, if multiple motors were to pull via multiple pili in parallel, the velocity would be reduced only because of the reduced load on each motor. In the case of two pili, the high velocity mode at 30 pN would equal the low velocity mode at 15 pN, and the high velocity mode at 15 pN would equal the low velocity mode at 8 pN. However, we found a threefold discrepancy, again arguing against the pilus bundling being responsible for the occurrence of two velocity modes. Thus, our results indicate that the type IV pilus system is a multistate system and that the probability for switching between, and remaining in, a certain state determines the dynamics and force generation by type IV pili (Fig. 8, a–c).
How can bimodality of pilus retraction be explained at the molecular level? The average velocity of the high velocity mode shifted to lower values with increasing force and disappeared at high force. With increasing PilT concentration, the high velocity mode persisted at higher forces. One simple explanation may be that more than one binding site exists at the base of the pilus and that the binding constant of PilT to the pilus depends on force. At low force, either one or two PilT hexamers could bind to the base of the pilus and generate different velocities. Another explanation could be that the ability of PilT to generate pilus movement is fine-tuned by other proteins and that titration of PilT changes their relative concentrations. Obvious candidates would be PilT-like proteins, such as PilU (27,28), which may even form heterohexamers with PilT.
Conclusion
We found that pilus dynamics (i.e., velocity and directional switching probability) is fine-tuned by the external force on the pilus and the concentration of PilT retraction proteins. In particular, we found that PilT concentration controls the occupation of a high velocity state and the probability of force-induced pilus elongation. What may be the biological role of the multistate system? We suggest that differential regulation of factors that determine the dynamics enables the bacterium to adjust to different adhesion surfaces and thereby regulate the tension of individual pili during twitching motility and infection of host cells.
Acknowledgments
The authors thank Finn Erik Aas, Hanne Winther-Larsen, Martin Linden, Mats Wallin, Vladimir Pelicic, and Kerstin Stingl for helpful discussions; we also thank Alexandra Friedrich for supplying the strains used in the experiments.
This work was supported by grant MA3898 from the Deutsche Forschungsgemeinschaft (DFG; the German Research Foundation).
Supporting Material
References
- 1.Merz A.J., So M., Sheetz M.P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 2.Wolfgang M., Lauer P., Park H.S., Brossay L., Hebert J. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 3.Merz A.J., So M. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 2000;16:423–457. doi: 10.1146/annurev.cellbio.16.1.423. [DOI] [PubMed] [Google Scholar]
- 4.Howie H.L., Glogauer M., So M. The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism. PLoS Biol. 2005;3:e100. doi: 10.1371/journal.pbio.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skerker J.M., Berg H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen I., Christie P.J., Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maier B., Chen I., Dubnau D., Sheetz M.P. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat. Struct. Mol. Biol. 2004;11:643–649. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P.K., Parsek M.R., Greenberg E.P., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 9.Bieber D., Ramer S.W., Wu C.Y., Murray W.J., Tobe T. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 10.Morand P.C., Bille E., Morelle S., Eugene E., Beretti J.L. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 2004;23:2009–2017. doi: 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig L., Li J. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herdendorf T.J., McCaslin D.R., Forest K.T. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 2002;184:6465–6471. doi: 10.1128/JB.184.23.6465-6471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satyshur K.A., Worzalla G.A., Meyer L.S., Heiniger E.K., Aukema K.G. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakovljevic V., Leonardy S., Hoppert M., Sogaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pili function in Myxococcus xanthus. J. Bacteriol. 2008;190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier B., Potter L., So M., Long C.D., Seifert H.S. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biais N., Ladoux B., Higashi D., So M., Sheetz M. Cooperative retraction of bundled Type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier B., Koomey M., Sheetz M.P. A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. USA. 2004;101:10961–10966. doi: 10.1073/pnas.0402305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linden M., Tuohimaa T., Jonsson A.B., Wallin M. Force generation in small ensembles of Brownian motors. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:021908. doi: 10.1103/PhysRevE.74.021908. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser D. Bacterial motility: how do pili pull? Curr. Biol. 2000;10:R777–R780. doi: 10.1016/s0960-9822(00)00764-8. [DOI] [PubMed] [Google Scholar]
- 20.Merz A.J., Forest K.T. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr. Biol. 2002;12:R297–R303. doi: 10.1016/s0960-9822(02)00806-0. [DOI] [PubMed] [Google Scholar]
- 21.Craig L., Volkmann N., Arvai A.S., Pique M.E., Yeager M. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Maier B. Using laser tweezers to measure twitching motility in Neisseria. Curr. Opin. Microbiol. 2005;8:344–349. doi: 10.1016/j.mib.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Svoboda K., Mitra P.P., Block S.M. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc. Natl. Acad. Sci. USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowther L.J., Anantha R.P., Donnenberg M.S. The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine. Mol. Microbiol. 2004;52:67–79. doi: 10.1111/j.1365-2958.2003.03963.x. [DOI] [PubMed] [Google Scholar]
- 25.Sablin E.P., Kull F.J., Cooke R., Vale R.D., Fletterick R.J. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 26.Clemen A.E., Vilfan M., Jaud J., Zhang J., Barmann M. Force-dependent stepping kinetics of myosin-V. Biophys. J. 2005;88:4402–4410. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H.S., Wolfgang M., Koomey M. Modification of type IV pilus–associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect. Immun. 2002;70:3891–3903. doi: 10.1128/IAI.70.7.3891-3903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhaya D., Bianco N.R., Bryant D., Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 29.Wolfgang M., Park H.S., Hayes S.F., van Putten J.P., Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


