Abstract
Previous studies showed that binding of water-soluble phosphatidylserine (C6PS) to bovine factor Xa (FXa) leads to Ca2+-dependent dimerization in solution. We report the effects of Ca2+, C6PS, and dimerization on the activity and structure of human and bovine FXa. Both human and bovine dimers are 106- to 107-fold less active toward prothrombin than the monomer, with the decrease being attributed mainly to a substantial decrease in kcat. Dimerization appears not to block the active site, since amidolytic activity toward a synthetic substrate is largely unaffected. Circular dichroism reveals a substantial change in tertiary or quaternary structure with a concomitant decrease in α-helix upon dimerization. Mass spectrometry identifies a lysine (K270) in the catalytic domain that appears to be buried at the dimer interface and is part of a synthetic peptide sequence reported to interfere with factor Va (FVa) binding. C6PS binding exposes K351 (part of a reported FVa binding region), K242 (adjacent to the catalytic triad), and K420 (part of a substrate exosite). We interpret our results to mean that C6PS-induced dimerization produces substantial conformational changes or domain rearrangements such that structural data on PS-activated FXa is required to understand the structure of the FXa dimer or the FXa-FVa complex.
Introduction
The process of blood clotting and the subsequent dissolution of the clot, are crucial events in controlling the normal flow of blood after vascular injury. The blood coagulation system consists of several consecutive reactions, in which activation of one zymogen leads to the activation of another. This sequential nature results in small initial signals being amplified significantly to produce first a platelet plug and then a hardened clot. It is essential that each step in the process be carefully regulated. Platelet activation is a major regulatory event in human blood coagulation. The platelet membrane contains phosphatidylserine (PS) mainly in its cytoplasmic leaflet (1), but this asymmetric arrangement of PS is lost upon platelet activation (2). It is now widely accepted that a PS-containing membrane is required for the activity of two complexes that are key to thrombin formation, the factor-X-activating complex (Xase) and the prothrombin-activating complex (prothrombinase). The conversion of prothrombin (II) to thrombin (IIa) is a vital step in the blood coagulation process. When thrombin production is not properly regulated and localized to membranes at the site of tissue damage, thrombotic disease such as stroke and heart attack can result. The prothrombinase complex functions on platelet membranes, is positioned metabolically at the intersection of the intrinsic and extrinsic paths of the traditional “coagulation cascade”, and consists of a serine protease, factor Xa, and its associated cofactor, factor Va (3,4). Thrombin is a central metabolite of blood coagulation (5) and has a key role in amplifying the response to injury since it activates additional platelets that trigger formation of more activated factor Xa and thrombin. This amplification mechanism leads to the hypothesis that inhibition of factor Xa (FXa) may be more efficient than inhibition of thrombin (6,7). In addition, it is anticipated that specific FXa inhibitors would prevent thrombus formation without compromising hemostasis, thus providing a better efficacy profile (7). This qualifies FXa as an attractive target for the development of new antithrombotic drugs (8). A recent report suggests that inhibiting factor Xa is effective in preventing both arterial and venous thrombosis (9).
The domain organization of factor Xa (FXa) is similar to that of its substrate, prothrombin. Both proteins have an N-terminus γ-carboxyglutamic acid-rich region GLA module essential for membrane binding, two Cys-rich highly folded domains, (epidermal-growth-factor (EGF)-like cassettes EGFN and EGFC in FXa and Kringle domains k1 and k2 in prothrombin), and a catalytic domain containing the catalytic triad (H/D/S) in the substrate binding site, and, in the case of FXa, factor Va interaction sites (10–12). Factor X consists of two peptides, with the membrane-binding portion (GLA-EGFNC) attached by an S-S bond to the catalytic domain. Activation of FX to FXa involves cleavage of a single small peptide from the N-terminus of the catalytic chain. Activation of prothrombin to thrombin by FXa involves proteolysis of two peptide bonds, Arg273-Thr274 between the EGFC module and the catalytic domain and Arg322-Ile323 within the catalytic domain (13).
Our studies show that PS acts as an allosteric regulator of both factors Xa and Va, and their substrate prothrombin. To demonstrate this, we introduced the use of a soluble lipid, 1,2-dicaproyl-sn-glycero-3-phospho-L-serine (C6PS), that does not occur in human physiology, but is a powerful tool that allows us to examine the details of PS binding to prothrombinase proteins in solution, a feat that would be nearly impossible to accomplish for proteins bound to a membrane surface. Using this, we showed that FXa has three C6PS binding sites, one in the GLA domain that binds C6PS only in the absence of Ca2+, one Ca2+-requiring binding site in the EGFNC domain pair (C6PS binding to this site regulates activity), and a Ca2+-dependent site in the catalytic domain (not a lipid-regulatory site and likely part of a protein recognition site) (14). Functional response to C6PS binding to the EGFNC site requires linkage of the GLA, EGFNC, and catalytic domains in the presence of Ca2+ (14) and is specific for PS (15). Binding to this site increases the rate of prothrombin activation by human FXa, changes the order in which human factor Xa catalyzes cleavage of the two peptide bonds in prothrombin (16), produces structural changes in bovine FXa as judged by intrinsic fluorescence and circular dichroism (CD) (14), and triggers bovine FXa dimer formation in a Ca2+-dependent fashion (17).
It has long been known that many enzymes undergo ligand-triggered oligomerization (18), and it has been estimated that as many as 15% of all enzymes fall into this category (19–22). Thus, ligand-triggered oligomer formation is often a regulatory mechanism (19). In some cases, the monomer is active and the oligomer is inactive (e.g., carbamoyl phosphate synthase forms inactive dimers in response to N-acety-glutamic acid binding), whereas in other cases, the oligomer is active (e.g., a threonine deaminase inactive dimer forms an active tetramer in response to AMP binding) (19). This and the critical role of both platelet membranes and FXa in human blood coagulation led us to ask whether our observations with bovine FXa might extend to human FXa, specifically whether this key human enzyme might also dimerize upon binding C6PS? In addition, we asked whether PS-induced bovine or human FXa dimer is more or less active than the monomer? Two modes of enzyme regulation by ligand-induced multimerization have been proposed: occlusion of substrate binding because of formation of a multimer interface or alteration of the active site by conformational changes induced by multimer formation (19). If FXa activity is altered in the PS-induced dimer, is this because dimerization alters the active site or substrate binding to the FXa enzyme? Finally, prothrombin undergoes large PS-triggered structural changes and domain reorganizations (23,24). A model of FXa based on x-ray structures (25) puts the active site 89 Å from the plane of Ca2+ ions that define the membrane-protein interface. Fluorescence measurements put the active site 61 Å above the surface of a PS-containing membrane (26), suggesting similar PS-induced structural reorganization in FXa. If such changes occur, any predictions about the structure of the FXa dimer or the FXa-FVa complex from crystallographic data obtained in the absence of PS would be unreliable. Thus, we ask whether structurally sensitive observations on FXa in a C6PS-triggered dimer are consistent with models of FXa structure obtained from x-ray diffraction in the absence of C6PS?
In this work, we report that human FXa indeed forms a dimer. As for bovine FXa, dimerization is sensitive to Ca2+ concentration. Second, we show that the proteolytic enzyme activities of both human and bovine FXa toward prothrombin are greatly reduced due to a substantial decrease in kcat and a slight increase in KM. However, activity toward a tripeptide synthetic substrate does not decrease in the dimer. These results suggest that the activity of the dimer is lowered due not to occlusion of the active site but rather to conformational changes or occlusion of substrate exosites. Finally, C6PS-induced dimer formation is also accompanied by a slight decrease in helical secondary structure but a substantial change in tertiary or quaternary structure. In addition, mass spectrometry and chemical modification of exposed lysines identified residues whose exposure to solvent was altered by Ca2+ and C6PS binding as well as dimer formation. We found that dimer models based on current crystallographic models of monomeric FXa were not consistent with these and other experimental observations. We conclude that PS-induced dimerization of human FXa significantly alters its structure and inhibits its activity, although additional work is needed to determine the extent to which this phenomenon might occur on membranes and thus have significance in the regulation of FXa during blood coagulation.
Experimental Procedures
Materials
C6PS in the form of sodium salt was purchased from Avanti Polar Lipids (Birmingham, AL). The FXa-specific substrate N-2-benzyloxycarbonyl-D-arginyl-L-arginine p-nitroanilide dihydrochloride (S-2765) and IIa-specific substrate H-D-phenylalanyl-L-pipecolyl-L-arginine p-nitroanilide dihydrochloride (S-2238) were purchased from DiaPharma (West Chester, OH). Human and bovine prothrombin and FXa were obtained from Hematologic Technologies (Essex Junction, VT), as well as FXa labeled in its active site with dansyl tripeptide choloromethyl ketone (DEGRXa). FXa was assayed before use using an active-site titrated standard (27). All other chemicals were ACS reagent grade; all solvents were high-performance liquid chromatography (HPLC) grade. An Atlantis dC18 100 μm × 100 mm column and an Acquity trapping column 5 μm C18 180 μm × 200 mm were purchased from Waters (Milford, MA).
Short-chain phospholipid sample preparation
Chloroform was removed from measured amounts of C6PS stock using a stream of nitrogen, and the lipid sample was dissolved in cyclohexane and lyophilized overnight. The lyophilized lipid was then suspended in an appropriate amount of water to reach a desired concentration. C6PS from this stock was used within 1–2 days.
Native polyacrylamide gel electrophoresis
FXa or DEGR-Xa (250 nM in the buffer A: 50 mM Tris, 150 mM NaCl, 5 mM Ca2+ and 0.6% polyethylene glycol (PEG), pH 7.4) were incubated at 23°C for 5 min in the absence and presence of 400 μM C6PS, and then were run together with known marker proteins (116-14 KDa, Sigma, St. Louis, MO) on an 8% native polyacrylamide gel in a BioRad Mini-Protean II minigel apparatus (Biorad Corp., Hercules, CA) and stained with colloidal Coomassie blue, as detailed previously (17,28).
Activity assay of factor Xa
The activity of factor Xa was checked using the synthetic chromogenic substrate S-2765 in an assay (29) adapted to an SLT 340-ATTC microplate reader (Tecan U.S., Hillsborough, NC).
Kinetics for thrombin appearance
The initial rates of prothrombin activation were obtained using the synthetic substrate S-2238 to follow the appearance of thrombin activity. Details of the method were described previously (30).
Effect of C6PS on factor Xa activity
The amidolytic activity of FXa in the presence of C6PS was determined using a synthetic substrate, S-2765, specific for FXa active site, as described previously (29).
Circular dichroism measurements
CD spectra were recorded from 260 to 200 nm on an Aviv Model 620S spectrometer (Aviv Associates, Lake Wood, NJ) in a 1-cm path length cell at 25°C with a bandwidth of 1 nm. Samples were prepared in buffer B (1.2 mM Tris, 150 mM NaCl, pH 7.4). Details of the method are described elsewhere (14).
Modeling kinetic data
Proteolytic activity of FXa with prothrombin as substrate
Kinetic constants for the proteolytic activation of prothrombin were determined by following the time course of thrombin activity for various prothrombin and FXa concentrations. Two types of analysis were carried out. First, data obtained at each FXa concentration were analyzed using a traditional Michaelis-Menten approach, with the use of the nonlinear regression algorithm incorporated into SigmaPlot (Systat Software, San Jose, CA). Because of FXa dimerization, the kinetic constants obtained at different FXa concentrations were not in agreement. For this reason, we carried out an analysis that took into account the presence of three FXa species, FXa not bound to C6PS, and both FXa monomer and dimer bound to C6PS. In this analysis, the observed rate (Robs) is expressed in terms of the separate kinetic constants (kcat and KM) of the three forms, the fractions of FXa present as unbound monomer (f0), C6PS-bound monomer (f1), and C6PS-bound dimer (f2), the prothrombin concentration ([S]), and the total FXa concentration ([Xa]tot):
| (1) |
The fraction of FXa bound to C6PS depends on the KdPS for C6PS binding to the regulatory site of human FXa (70 ± 10 μM (27); 86 ± 5 μM (15)):
| (2) |
The fraction of C6PS-bound FXa present as dimer was previously shown to be (17)
| (3) |
where and is the dimer dissociation constant. Using these expressions, we then fitted globally the data obtained at all prothrombin and FXa concentrations to obtain k0cat/K0M, kmcat and KmM, kdcat and KdM, and k. It quickly became apparent that the unbound monomer contributed almost nothing to the observed activity (∼95% of FXa exists as C6PS-bound species at 400 μM C6PS), so this species was eliminated to reduce the number of free parameters and obtain more reliable parameter estimates.
Amidolytic activity of FXa on synthetic substrate S-2765
These data were treated using the same formalism described under proteolytic activity, except that each species was assigned only a single rate characteristic of that species:
| (4) |
Acetylation and identification of lysine residues
Solvent-exposed primary amines of FXa were acetylated as described (31). Briefly, 700-nM aliquots of FXa in 100 mM phosphate buffer (pH 7.2) were treated with a 1000-fold molar excess of acetic anhydride (Sigma Aldrich)/amine residue (∼16.8 mM) for 30 min at room temperature. Test conditions included 3 mM Ca2+, 3 mM Ca2+ and 400 μM C6PS, 5 mM Ca2+, and 5 mM Ca2+ and 400 μM C6PS. The pH was adjusted every 5 min using 5 M NaOH to maintain a pH >7.0. The reaction was carried out under nondenaturing conditions so as to retain the protein's native structure. Finally, an equal volume of 1 M Tris pH 8.0 was added to quench any remaining unreacted acetic anhydride. Under these conditions, solvent-exposed lysine residues should be modified by the addition of an acetyl group. The modified FXa was then purified by HPLC using a Hewlett Packard (Wilmington, DE) 1100 HPLC system equipped with a C4 Vydac column (4.2 mm i.d., 5-μm particles). Elution was carried out using water/0.1% trifluoroacetic acid (v/v) and a linear gradient from 5% to 60% of acetonitrile/0.1% trifluoroacetic acid (v/v). Fractions containing purified FXa, as determined by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), were combined and lyophilized. After lyophilization, FXa was resuspended in 50 μL of 50 mM NH4HCO3, pH 7.8, and was then digested with trypsin (Promega Madison, WI) and Glu-C (pH 8) (Roche, Indianapolis, IN) using a 1:20 E/S ratio. Two proteases were used because trypsin activity is inhibited when a lysine residue is acetylated. MALDI-MS and nano LC-ESI-MS/MS were performed on the resulting peptides. For the mass spectrometric analysis we used MALDI-TOF to look for the intact protein fractions after the HPLC separation. Afterward, the digested protein was analyzed by MALDI-TOF to determine the efficiency of the digestion and then the same mixture was subjected to nano-LC-ESI-MS/MS to identify the specific lysine residues that were acetylated. This was accomplished by comparing the mass spectra of the FXa digest and modified FXa digest to identify acetylated peptides as those that corresponded in mass to a predicted proteolytic fragment of FXa plus the mass of one or more acetyl groups (+42 Da). Acetylated peptides were then analyzed by nano-LC-ESI-MS/MS to verify the peptide sequence and confirm the site(s) of modification, thus identifying the acetylated residues.
MALDI-MS
Samples (0.5 μL) were spotted onto a 100 sample stainless steel MALDI plate and mixed on target with 0.5 μL of a saturated solution of α-cyanohydroxycinnamic acid in 45:45:10 (v/v/v) water/ethanol/formic acid. Mass spectrometric analyses were then performed on an Applied Biosystems DE-STR MALDI-TOF mass spectrometer in the reflector positive ion mode with a resolution of >10,000. The MS spectra were calibrated externally using a mixture of standard peptides with a mass error of ≤0.01%. Data were acquired using an accelerating voltage of 23,000 volts, grid percentage of 65%, and delay time of 200 ns.
NanoLC-ESI MS/MS
NanoLC-ESI MS/MS mass spectrometry was performed on a Micromass Q-Tof Premier mass spectrometer equipped with a NanoAcquity LC system (Waters). Analyses were performed using a 3-μm Atlantis dC18 100 μm × 100 mm column with a flow rate of 300 nL/min and an Acquity trapping column 5 μm C18 180 μm × 200 mm was used to clean up the sample before injection into the analytical column for 3 min at 5 μL/min flow rate. Briefly, a 2-μL aliquot of the digest sample was loaded onto the column. Peptides were eluted using a linear gradient from 2% B (acetonitrile/0.1% formic acid (v/v)) ramped to 95% B over 60 min. Instrument settings for MS analyses included a capillary voltage of 3.5 kV, cone voltage of 30 V, and an m/z range of 200–2000. MS/MS data were acquired using a data-dependent acquisition method selecting collision energies based on mass and charge state. For calibration, an external lock mass was used with a separate reference spray (LockSpray) using a solution of Glu-fibrinogen peptide (300 fmol/μl) with a mass of 785.8496 (2+ ion) with errors of <20 ppm.
Results
Human factor Xa forms a dimer in the presence of soluble C6PS at 5 mM Ca+2
Bovine FXa dimerizes in the presence of 400 μM C6PS at 5 mM Ca2+ (17). We have previously demonstrated the ability of native gel electrophoresis to correctly measure both the presence and molecular weight of bovine FXa dimer (17) and both bovine and human FXa-FVa complex (16,32). This method is based on the observation that the thermodynamics of protein association reactions can be determined by gel analysis (32,33), presumably because the local high concentrations of proteins in the gel prevent complex dissociation at rates to be expected in dilute solution. Thus, to confirm that human FXa behaves similarly, native polyacrylamide (8%) gel electrophoresis was performed on a sample of 250 nM human FXa (Fig. 1, lane 1) in the presence of 400 μM C6PS and 5 mM Ca2+. A similar sample prepared in the absence of Ca2+ and C6PS ran at almost half the molecular mass (Fig. 1, lane 4) of this sample. When 250 nM DEGR-Xa was incubated in the presence of C6PS and 5 mM Ca2+, the band (Fig. 1, lane 3) corresponded to the molecular mass of monomer human FXa with no dimer band detected. These results demonstrate, first, that, like bovine FXa, human FXa dimerizes in the presence of 400 μM soluble C6PS and 5 mM Ca2+ and, second, that labeling the active site of FXa inhibits dimer formation.
Figure 1.
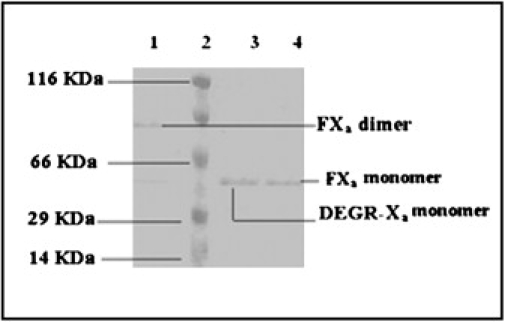
Native polyacrylamide gel electrophoresis of human FXa in the presence and absence of C6PS at 5 mM Ca2+. A 250-nM sample of FXa or DEGR-Xa was incubated with or without 400 μM C6PS at 23°C in buffer A containing 5 mM Ca2+ and then run on an 8% native polyacrylamide gel that also contains a saturating amount (400 μM) of C6PS. Lanes 1–4 contain, respectively, human FXa in the presence of 400 μM C6PS; molecular weight markers; human DEGR-Xa in the presence of 400 μM C6PS; and human FXa in the absence of C6PS.
Effect of dimerization on FXa function
Proteolytic activity of factor Xa at high (5 mM) Ca2+concentration
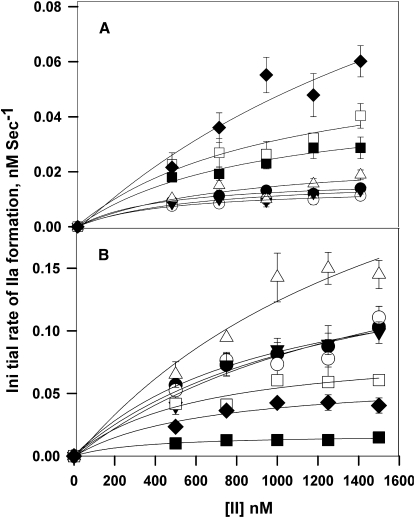
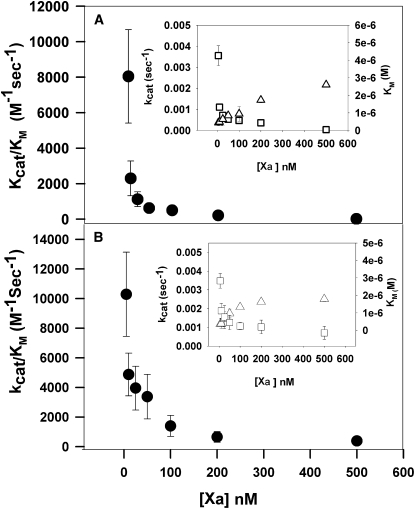
Proteolytic activation of prothrombin (II) by both human and bovine FXa was carried out in the presence of 400 μM C6PS using a range of FXa concentrations (5–500 nM) and prothrombin concentrations (0.25–1.5 μM). Initial rates of prothrombin activation were obtained by linear regression of early time courses. Fig. 2, A and B, portrays these data in the traditional Michaelis-Menten format for human and bovine FXa, respectively. These data were fitted using the Michaelis-Menten formalism to obtain kcat/KM, kcat, and KM for each FXa concentration examined. Fig. 3, A and B, shows plots versus FXa concentration of the kinetic constants (kcat/KM) for human and bovine FXa, respectively. It is evident that the apparent kcat/KM decreased with increasing FXa concentration, i.e., the enzyme's catalytic efficiency decreased dramatically and asymptotically with its concentration. The insets in Fig. 3 show that the decrease in kcat/KM is due primarily to a decrease in kcat (left vertical axis), but also involves a small increase in KM (right vertical axis). The observed variation of the apparent kinetic constants with FXa concentration under conditions that are known to trigger FXa dimerization suggests that the dimer is significantly less active than the monomer. Approximate values of kcat and KM for the monomer and dimer were taken as values obtained at 50 nM and 500 nM FXa, respectively, and are recorded in Table 1 as kcateffect and KMeffect for 5 mM Ca2+. Next, the data were fit globally to the dimer model described in Experimental Procedures (Eqs. 1–3) using five data sets and only five parameter values: KM and kcat for both monomer and dimer, and Kd2, with the fits shown as lines in Fig. 2. The five data sets could not define all five parameters uniquely, so an iterative fitting algorithm was used. We began with values for KMeffect for both the dimer and monomer and varied kcat for both species and Kd2. We repeated this procedure with kcat fixed and then again with KM fixed until the fits converged to invariant values of all five parameters. It is clear that this model provides a reasonable description of all five data sets. Fitting yielded dimer dissociation constants (Kd2) of 14 ± 1 nM for human FXa and 14 ± 1.5 nM for bovine FXa, as compared to a value of 20 nM previously estimated from intrinsic fluorescence data (17). This procedure yielded monomer kcat values of 4.0 ± 0.4 × 10−3 s−1 for human FXa and 3 ± 5 × 10−3 s−1 for bovine FXa. Values for the dimer were 2 ± 1.6 × 10−9 s−1 and 8 ± 1 × 10−9 s−1 for human and bovine FXa, respectively. KM values were 0.4 ± 0.08 μM and 0.4 ± 0.06 μM for monomer and 2.6 ± 1.5 μM and 2.5 ± 0.4 μM for dimer, for human and bovine FXa, respectively. These values are listed in Table 1 as kcatglobal and KMglobal for 5 mM Ca2+. Earlier studies performed at low concentrations of factor Xa (5 nM, i.e., monomer-forming conditions) gave KM and kcat values for prothrombin activation of 0.3 ± 0.01 μM and 3.9 ± 0.1 × 10−3 s−1, respectively (16), which agree well with the values obtained here for the FXa monomer. This analysis is therefore consistent with the C6PS-bound FXa monomer being ∼106- to 107-fold more active than the dimer in the cases of both human (kcat/KM of 104 ± 3000 vs. 0.8 ± 1 × 10−3 M−1 s−1) and bovine (7500 ± 9700 vs. 3 ± 2 × 10−3 M−1 s−1) FXa. We note that the second-order rate constants for the dimer are so small that they are not distinguishable from zero, and their exact numerical values should not be taken seriously.
Figure 2.
Prothrombin activation as a function of prothrombin concentration at different FXa concentrations and at 400 μM C6PS. The initial rates of thrombin (IIa) formation are plotted as a function of prothrombin (II) concentration. The appearance of thrombin was determined by the rate of S-2238 hydrolysis at 37°C (see Experimental Procedures). (A) Results for human FXa at concentrations of 5 nM (solid circles), 10 nM (open circles), 25 nM (solid triangles), 50 nM (open triangles), 100 nM (solid squares), 200 nM (open squares), and 500 nM (solid diamonds). (B) Results for bovine FXa at concentrations of 5 nM (solid squares), 10 nM (solid diamonds), 25 nM (open squares), 50 nM (open circles), 100 nM (solid circles), 200 nM (solid triangles), and 500 nM (open triangles). The reaction mixture contained prothrombin at concentrations from 0.25 to 1.5 μM, FXa in 50 mM Tris, 175 nM NaCl, 0.6% poly(ethylene glycol), 5 mM Ca2+, and 400 μM C6PS. The data for all FXa concentrations were fitted assuming the Michaelis-Menten model for the rate of activation by each of three species (Eqs. 1–4). The curves illustrate the success of the global fit. This yielded estimates of the kcat and KM for the monomer and dimer and dimer dissociation constants of 14 ± 1 nM for human FXa and 14.2 ± 1.5 nM for bovine FXa.
Figure 3.
Michaelis-Menten rate constants for prothrombin activation as a function of FXa concentration at 400 μM C6PS. Apparent second-order rate constants, kcat/KM, for proteolytic reaction of prothrombin activation are plotted as a function of FXa concentration of (A) human FXa and (B) bovine FXa. Values of kcat and KM at each FXa concentration were obtained by applying the Michaelis-Menten model to the individual curves in Fig. 2, with their ratios plotted here. (Insets) Decrease of kcat (open squares, left ordinate) and increase of KM (open triangles, right ordinate) with increasing FXa concentration results in the observed decrease in kcat/KM. The error bars derive from the uncertainty in the fitting parameters.
Table 1.
Apparent kcat and KM of prothrombin activation by FXa monomer and dimer at different Ca+2 concentrations
| Ca2+ concentration (mM) | Xa monomer |
Xa dimer |
||||||
|---|---|---|---|---|---|---|---|---|
| kcateffect | kcatglobal | KMeffect | KMglobal | kcateffect | kcatglobal | KMeffect | KMglobal | |
| 3 | 2.8 × 10−3 ± 4.0 × 10−4 | 2.9 × 10−3 ± 1.7 × 10−3 | 608.4 ± 212.1 | 604.99 ± 104.5 | 2.3 × 10−4 ± 4.0 × 10−4 | 1.8 × 10−9 ± 1.5 × 10−9 | 782.4 ± 363.8 | 984.98 ± 570.94 |
| 3.5 | 3.1 × 10−3 ± 4.2 × 10−4 | 3.1 × 10−3 ± 2.2 × 10−4 | 549.0 ± 191.2 | 548.9 ± 121.7 | 2.9 × 10−4 ± 3.0 × 10−5 | 1.2 × 10−9 ± 9.9 × 10−10 | 843.7 ± 209.6 | 1191.9 ± 362.5 |
| 4 | 3.0 × 10−3 ± 2.4 × 10−4 | 3.5 × 10−3 ± 3.6 × 10−4 | 526.5 ±144.4 | 509.9 ±117.2 | 3.2 × 10−4 ± 3.2 × 10−5 | 1.32 × 10−9 ± 1.1 × 10−9 | 886.8 ± 191.5 | 1499.9 ± 1214.9 |
| 5 | 3.5 × 10−3 ± 3.8 × 10−4 | 4.0 × 10−3 ± 4.0 × 10−4 | 448.0 ± 136.0 | 400.0 ± 80.0 | 7.0 × 10−4 ± 3.4 × 10−5 | 2.0 × 10−9 ± 1.57 × 10−9 | 1791.0 ± 154.0 | 2600.9 ± 1500.0 |
| 6 | 4.3 × 10−3 ± 2.8 × 10−4 | 3.8 × 10−3 ± 3.0 × 10−4 | 420.4 ± 85.2 | 449.9 ± 187.2 | 5.6 × 10−4 ± 1.2 × 10−5 | 1.7 × 10−9 ± 1.0 × 10−10 | 1942.8 ± 680.06 | 1899.9 ± 1259.2 |
Values of kcateffect and KMeffect for Xa monomer and dimer were obtained for 5 and 500 nM FXa at each Ca2+ concentration (as seen in Fig. 2 and described in Results). The kcatglobal and KMglobal values for monomer and dimer were obtained by the global fitting procedure using data from five FXa concentrations and prothrombin concentrations (see Results and Eqs. 1–3). The kcat values are in s−1 and KM values in nM.
Effect of Ca2+concentrations on factor Xa dimerization
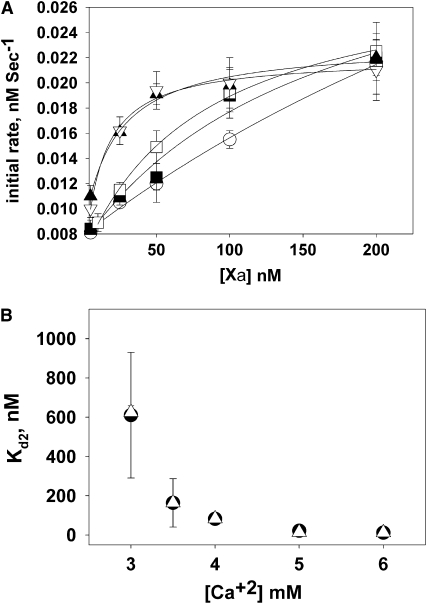
The dimerization constant of bovine FXa is very sensitive to Ca2+ concentration (17). To determine the dependence of the human FXa dimerization constant on Ca2+ concentration, we measured the initial rate of prothrombin activation as a function of FXa concentration (Fig. 4 A) at five Ca2+ concentrations (3, 3.5, 4, 5, and 6 mM) and fitted these data to a simple dimerization model (Eq. 3), which gives the value of dimer dissociation constants (Kd2) and kinetic constants (kcatglobal and KMglobal for monomer and dimer in Table 1) at each Ca2+ concentration. The Kd2 value is the inverse of the dimerization constant (k) defined in Eq. 3, and the Kd2 values obtained in this way are plotted in Fig. 4 B (open triangles) as a function of Ca2+ concentration. In addition, the apparent kcat and KM for monomer and dimer were obtained at each Ca2+ concentration from the Michaelis-Menten plots at 5 and 500 nM FXa, respectively. These are recorded in Table 1 as kcateffect and KMeffect for each Ca2+ concentration. Kd2 values obtained from the hyperbolic plots in Fig. 4 A are plotted in Fig. 4 B (solid circles). The Kd2 values obtained by both methods are quite similar. The observed hyperbolic dependence of Kd2 on [Ca2+] is similar to that reported for bovine FXa (17).
Figure 4.
Effect of calcium concentration on FXa dimerization. (A) Dependence of the initial rate of appearance of thrombin activity as a function of increasing concentration of FXa in buffer (50 mM Tris-HCl, 175 mM NaCl, and 0.6% PEG) with 400 μM C6PS at 3 (open circles), 3.5 (solid squares), 4 (open squares), 5 (solid triangles), and 6 (open triangles) mM Ca2+ concentration. The curves through the data are hyperbolas, from which we estimated dimer dissociation constants (Kd2). (B) Variation of the dimer dissociation constant (Kd2) of human FXa with Ca2+ concentration at 400 μM C6PS. Values obtained as described in A are shown as solid circles, whereas values obtained using a full global analysis of proteolytic rates (Eqs. 1–3 as applied to data shown in Fig. 2 at different Ca2+ concentrations) are shown as open triangles.
Amidolytic activity of FXa as a function of C6PS concentrations and FXa concentration
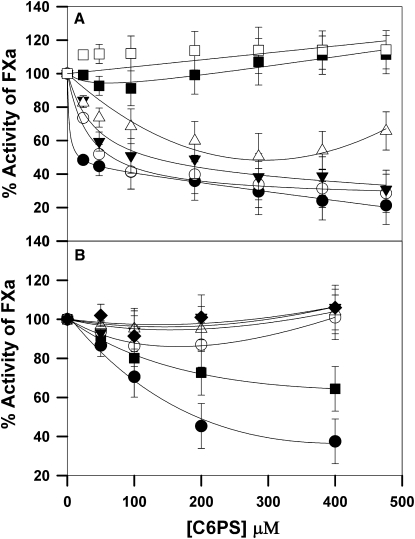
Our results show that FXa dimerization alters mostly the kcat of prothrombin proteolysis. A severe reduction in kcat can reflect significant misalignment of substrate with the active site. The active site could be occluded; the substrate might not bind to the active site or could bind inappropriately to an exosite, thereby causing misalignment at the active site; or the active site could be altered in the arrangement of key residues. To gain insight to these possibilities, we determined the amidolytic activity of FXa against the synthetic substrate S-2765, which specifically binds to the FXa active site independent of substrate binding exosites. The percent of amidolytic activity of FXa relative to the activity in the absence of C6PS is plotted in Fig. 5 as a function of C6PS concentration (as described in Experimental Procedures) for six different FXa concentrations. At low FXa concentrations (up to 50 nM), C6PS decreased FXa amidolytic activity, as previously reported for monomer human FXa (27). However, for high FXa concentrations (>50 nM), the amidolytic activity did not decrease. Thus, dimerization alters or prevents the C6PS-induced changes in the FXa active site that lower the amidolytic activity. Our most notable observation, however, is that the active site of the dimer is clearly available to the tripeptide substrate and is successful in catalyzing its hydrolysis. Thus, the active site is not occluded and its catalytic residues are appropriately arranged to affect catalysis of the synthetic substrate.
Figure 5.
Initial rate of amidolysis of synthetic substrate S-2765 as a function of C6PS concentrations and at different FXa concentrations. The rate was determined at 37°C in buffer (containing 50 mM Tris, 175 mM NaCl, 0.6% PEG, and 5 mM Ca2+), as described in Experimental Procedures. The data are expressed as a percentage of the rate of S-2765 hydrolysis by FXa in the absence of C6PS and were analyzed according to a previously described model for C6PS binding resulting in dimer formation (17), as summarized in Eq. 4 (Experimental Procedures). Rates are plotted for increasing concentrations of C6PS and several FXa concentrations: (A) human FXa at 5 nM (solid circles), 10 nM (open circles), 25 nM (solid inverted triangles), 50 nM (open triangles), 100 nM (solid squares) and 200 nM (open squares); (B) bovine FXa at 5 nM (solid circles), 10 nM (solid squares), 25 nM (open circles), 50 nM (open triangles), 75 nM (solid inverted triangles), and 100 nM (solid diamond). The data showed that amidolytic activity of monomeric FXa decreased substantially (∼70%) in response to C6PS binding, whereas such a decrease was not observed for the dimer.
Effect of dimerization on FXa structure
Previous studies have shown that C6PS binding triggers structural changes in monomeric FXa (14,17). Does dimer formation trigger further structural changes in FXa? If so, what kind?
Secondary structure by CD spectroscopy
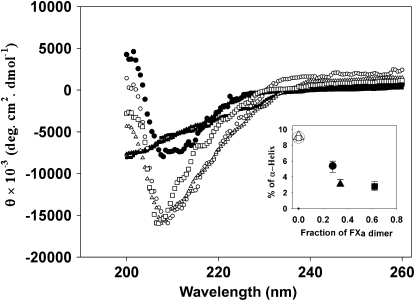
Circular dichroism (CD) spectra were recorded in the range 260–190 nm for FXa in the absence and presence of 400 μM C6PS and in the presence of 5 mM Ca2+ (Fig. 6). Analysis of these spectra using the software package CDPro (34) (analysis routines CDSSTR, CONTINLL, and SELCON3) enabled the estimation of the secondary structure composition of FXa under these different conditions. The results obtained in the absence of C6PS (open symbols) agree with previous reports (mainly β-structure and a small amount of α-helix) (14) and were the same at all three FXa concentration examined (150, 200, and 500 nM). Upon addition of C6PS to 150 nM FXa (solid circles), the percent of α-helix decreased as reported previously (14). For 200 nM (solid triangles) and 500 nM (solid squares) FXa, the shape of the spectrum changed dramatically. Although the data do not extend to wavelengths low enough to accurately estimate changes in β-structure or “other” structures, data obtained at all but the highest factor Xa concentration (500 nM) do suffice to obtain consistent estimates of α-helix from all three algorithms in the CDPro package. The averages of these estimates obtained in the presence of C6PS are plotted in the inset of Fig. 6 (solid symbols) versus the fraction of FXa as dimer (obtained using Eq. 3 and the dimer dissociation constant from Fig. 2). There was a dramatic decrease in α-helix with dimer formation in the presence of C6PS, as shown. Note that the point at 60% dimer was difficult to estimate, both because there was so little helix and because the OD of the sample was significant at 500 nM factor Xa. By contrast, all three concentrations of FXa showed the same helix content in the absence of C6PS (open symbols at 0% dimer). The decrease in helix content corresponded to a dramatic change in the shape of the CD spectra. Such dramatic changes in CD spectra are usually attributed to changes in tertiary (domain organization) or quaternary structure (35,36). These data confirm a FXa structural change (a decrease in α-helix) upon C6PS-induced dimerization and suggest a significant rearrangement of either interdomain contacts within the protein or secondary structure at the dimer interface.
Figure 6.
Change in secondary structure of FXa in the absence and presence of C6PS at monomeric and dimeric FXa concentrations. FXa was examined at 24°C at varying concentrations (150 nM (circles), 200 nM (triangles), and 500 nM (squares)) in buffer (1.2 mM Tris, 150 mM NaCl, and 5 mM CaCl2, pH 7.4) in the presence of 400 μM C6PS (solid symbols) and absence of C6PS (open symbols). The optical density of each sample was ≤0.1 at 200 nm (except for the 500 nM FXa, which had an OD of 0.16), and all spectra were reproducible for different FXa samples prepared on different days. (Inset) Variation of the percentage of α-helix with the fraction of FXa present as dimer (from Eq. 3 and dimer dissociation constant from Fig. 2). Points are presented for FXa at three concentrations in the presence (solid symbols) and absence (open symbols show 0% dimer) of C6PS.
Tertiary and/or quaternary structure changes detected by Lys acetylation and MALDI- and nanoLC ESI-MS
Acetylation of lysine residues of FXa was performed under four conditions (described in Experimental Procedures) designed to distinguish changes due to Ca2+ binding, C6PS binding, and dimer formation. Different lysine residues were found to be modified under the four different conditions, which reveals clearly that conformational changes take place in the protein upon Ca2+ binding, upon C6PS binding, with an increase in Ca2+ concentration, and upon dimer formation. Based on this analysis, lysine residues were categorized into different types: 1), residues accessible to acetylation; 2), residues not accessible to acetylation and observed unmodified in proteolytic peptides; and 3), residues not observed at all. Category 3 residues do not appear in digests because of the size of the proteolytic fragment or ionization efficiency of the fragment. Proteolysis takes place under the same conditions no matter what conditions were used for the initial acetylation. Thus, the fact that a residue can be observed for one set of conditions and not for another results from the fact that the proteolysis pattern is affected by the acetylation of lysine side chains, since trypsin does not recognize acetylated lysines. Thus, category 3 lysines provide no structural insight. Category 1 residues are exposed to the aqueous surrounding under the conditions of acetylation. Category 2 residues are not exposed to the aqueous environment under the conditions of acetylation, but they still appear in identifiable peptides. These two categories of lysines provide us with structural insights.
There are three aspects of these results that we wish to mention. First, and most relevant to this study, are changes that accompany dimer formation. Based on our experiments, we can take two paths to form a dimer. First, we add C6PS to FXa in the presence of Ca2+. C6PS binds to FXa in the presence of 3 mM Ca2+ but does not form significant dimer at this low Ca2+ concentration. However, adding C6PS to FXa in the presence of 5 mM Ca2+ does trigger dimerization. Thus, changes in acetylation that are the same in these two processes do not reflect dimer formation, but changes that differ between these two processes likely do. The results for these two processes (Path A) are summarized in Table S1 in the Supporting Material, where we use the Fung numbering system (37). The second path (Path B) involves increasing the Ca2+ concentration from 3 to 5 mM in the absence and presence of 400 μM C6PS (Table S2). The combination of observations from paths A and B demonstrate that lysine 270 becomes inaccessible to acetylation due to dimer formation. Lysine 414 went from being inaccessible to the aqueous environment in the monomer state (independent of Ca2+ concentration) to being accessible to acetylation in the dimer state. These results are summarized in terms of an atomic model for FXa structure in Fig. 7 C.
Figure 7.
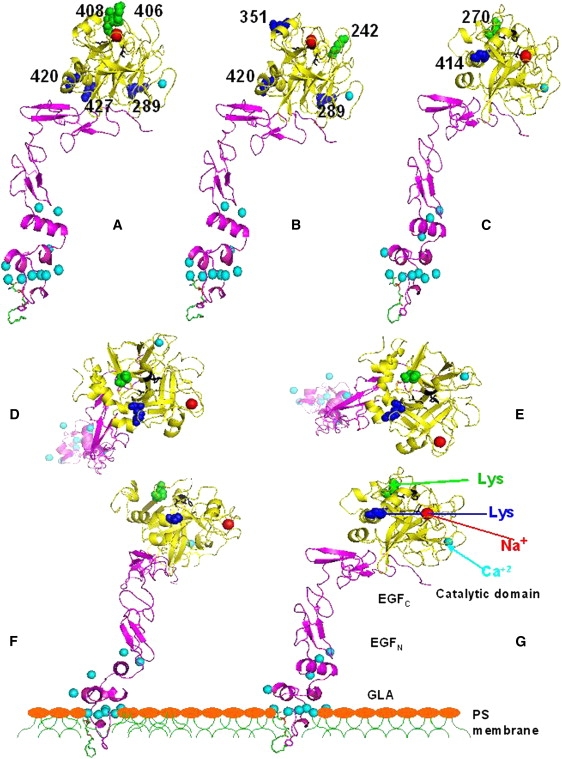
An atomic-resolution FXa model for human FXa in the presence of 3 mM Ca2+ (25) is shown. There are no crystallographic models for whole factors Xa, and certainly none that include information about how these might interact with a membrane. We felt that the model developed by the Pedersen lab had the best theoretical underpinnings, and Dr. Pedersen kindly provided us with coordinates for this model. This model is based on a crystallographic model of human GLA-domainless FXa (59) and of the EGFN GLA region from the factor VIIa-FXa-tissue factor complex (60). This model is shown with a lysophosphatidylserine molecule (green, carbon; blue, nitrogen; red, oxygen; and yellow, phosphorus) inserted (Insight II and Pymol) using the coordinates from the PT1 (bovine prothrombin fragment 1)-Ca2+-lysophosphatidylserine complex (47). The active site residues are colored black, and light and heavy chains are colored purple and yellow, respectively. Ca2+ ions are aqua (aqua arrow) and the Na+ ion is red (red arrow). (A–C) These three views highlight lysine residues whose exposure to acetylation, as determined by mass spectrometry, changes (blue, becomes exposed; green, becomes buried) upon (A) increasing Ca2+ concentration from 3 to 5 mM, (B) adding C6PS in the presence of 3 mM Ca2+ (nondimerization conditions), and (C) adding C6PS in the presence of 5 mM Ca2+ (dimerization conditions). (D–G) These views show the two lysine residues whose exposure to acetylation is changed during dimer formation (K414 (blue arrow in G) and K270 (green arrow in G)). (F) View obtained by rotating the molecule until the seven Ca2+ ions at the base of the GLA domain were roughly in a plane, as proposed previously (47). The Ca2+ planarity was tested by rotating F about the y axis by 90° to obtain G, which was adjusted and rotated to obtain F, etc., until it was clear that these Ca2+ ions were in a common plane. A slice through a hypothetical PS-containing membrane normal to the plane of the picture is shown to illustrate the arrangement of the Ca2+ plane and thus the molecule relative to a membrane. D and E were obtained by rotation of F and G, respectively, about the x axis by 90 degrees clockwise.
We wish to focus next on changes associated with increasing the Ca2+ concentration from 3 to 5 mM. These changes are summarized in the first column of Table S2. Increased Ca2+ binding leads to exposure of lysines 289, 420, and 427, all of which are on the bottom face of the catalytic domain (see Fig. 7 A). On the opposite face of the catalytic domain, we find that increased Ca2+ concentration leads to burial of lysines 406 and 408. A single Ca2+ (Fig. 7, light blue sphere) is located in the catalytic domain and has interactions with residues in the sequence D250–E260 (38). The Ca2+ binding site is linked to a Na+ (Fig. 7, red sphere) binding region defined by residues W399–I411 (39) that includes lysines 406 and 408, which become buried upon increasing the Ca2+ concentration.
Third, we consider the consequences of C6PS binding under conditions that favor the monomer state (i.e., 3 mM Ca2+). These results are summarized in column 1 of Table S1 and in Fig. 7 B. Lysine 242 becomes less accessible and lysines 289, 351, and 420 become accessible to acetylation as a result of C6PS binding.
Discussion
We showed previously that bovine factor Xa dimerizes upon C6PS binding, but we did not know whether human factor Xa forms a dimer, whether the dimer is active, or how the dimer structure differs from that of the monomer (17). We began this study to answer three questions: 1), Does human FXa form a dimer in solution? 2), If so, is the dimer active or does dimer formation block activity? and 3), Are structural changes associated with dimer formation small enough that we can think of the dimer structure in terms of the structure of two monomers? Our results clearly demonstrate that human FXa forms a dimer in solution in response to C6PS binding and that the dimer is essentially inactive. It appears that dimer formation does not occlude the active site, since the dimer still had amidolytic activity against the synthetic substrate S-2765. On the other hand, proteolytic activity against prothrombin was enormously reduced, primarily through a 107-fold decrease in kcat. Because the active site is not blocked from accessing small substrates, it is likely that dimerization interferes with binding of the substrate to exosites that are crucial for aligning the proper substrate with the active site. This is reasonable, as a dimer interface that included K270 would almost surely interfere with substrate binding to the exosite identified to include residues V262–H271 and D282–M296 (40) (see Fig. 7). Dimer formation also causes rearrangements within the active site (Fig. 5) that could also contribute to the decrease in kcat.
The results presented here provide insights into structural changes associated with Ca2+ and C6PS binding to FXa and with C6PS-induced dimerization. Because whole FXa has yet to be crystallized, even in a C6PS-free form, atomic-resolution information on these changes is not available. To illustrate our results in terms of an atomic-resolution FXa model, we have adopted a model of human FXa in the presence of Ca2+ (3 mM) (25) (Fig. 7). We begin with the effects of C6PS binding under conditions that do not favor dimer formation (3 mM Ca2+). Two molecules of C6PS bind to FXa, one to a site involving the EGFN linked to the GLA domain that regulates activity and one to a Ca2+-and-Na+-requiring site in the catalytic domain (14). These two sites are linked, with the site in the regulatory EGF region likely occupied first and then triggering binding to the second site (16). This second site is probably related to a quaternary amine site that likely defines a protein-binding region and is linked to Na+ binding (14,41). The four lysines affected by addition of 400 μM C6PS are shown in Fig. 7 B. Lysine 351 is part of a Va binding site identified by mutational study (42). Lysine 242 is adjacent to H236, one of the residues that form the FXa catalytic triad (H236, D282, and S379) shown in black in Fig. 7. Lysine 420 is located close to a proposed factor Va binding region, A404–F418 (44), and part of a substrate exosite A417–K427 (45). Also, K420 belongs to a peptide V415–R429 proposed to interfere with II binding (44). Both K420 and K427, residing in the A417–K427 region, also become exposed upon increasing Ca2+ (Fig. 7 A). Since 400 μM C6PS saturates the lipid regulatory site in the EGF region (14), it appears that binding of C6PS to the regulatory site exposes a lysine in one substrate exosite (K420), buries a lysine (K242) directly adjacent to the active site (H236), and exposes a lysine in an FVa binding region (K351). This is sensible, since binding of C6PS to FXa monomer increases the kcat and slightly decreases the KM of FXa during prothrombin activation (16) and enhances binding of FVa to FXa ∼1000-fold (46). This demonstrates remarkable cooperativity between Na+, Ca2+, C6PS, substrate, and FVa binding, painting the catalytic domain as a precisely tuned, multifaceted structure sensitive to events far from the active site (C6PS binding to the EGF-GLA).
We turn now to changes in lysines associated with C6PS-induced dimerization, which occurs at 5 mM Ca2+. Fig. 7 C shows that under this treatment K270 becomes inaccessible and K414 accessible. We note that, although increased Ca2+ concentration and C6PS addition at 3 mM Ca2+ change the exposure of five and four lysines, respectively, only two lysines are changed upon dimer formation. The exposed residue, K414, is in a region previously identified as involved in Va binding (44) and is linked to a substrate binding site (44). Since neither increasing Ca2+ from 3 to 5 mM nor adding C6PS in the presence of 3 mM Ca2+ exposed K414, it must be that dimerization triggers an FXa conformation in which K414 is exposed to the aqueous medium. It is interesting that K414 appears exposed in the particular conformation shown in Fig. 7 (based on x-ray data), but our mass spectrometry results show it as buried except in the dimer.
Interpretation of the change in K270 is less straightforward. This residue also is not changed upon increasing Ca2+ concentration and adding C6PS at 3 mM Ca2+ (Table S1 and Table S2). Since it becomes inaccessible upon dimer formation, one possibility is that it is in the dimer interface. However, it could also be that dimer formation triggers a conformational change that removes K270 from water exposure. Fig. 7, D–G, shows several views of FXa arranged so that seven Ca2+ ions believed to reside at the membrane surface (47) are in a plane. Constraining these Ca2+ ions to the membrane surface requires that FXa molecules are side-to-side in the dimer. If K270 is buried in the dimer interface, it is difficult to imagine how the conformation of FXa shown in Fig. 7 (25) could form a side-to-side dimer. It could be that the solution dimer does not have the same arrangement of FXa molecules that might exist in a membrane-bound dimer, i.e., the solution dimer may be head-to-head, whereas the membrane dimer may be side-to-side. It is not possible, on the basis of our data alone, to distinguish between these possibilities. However, several observations, discussed next, favor the dimer being side-to-side (as it would be on a membrane) and, at the same time, K270 being in the dimer interface.
First, preliminary data indicate that an inactive dimer also forms on a membrane (T. Koklic and R. Majumder, unpublished observations). Unless this dimer is fundamentally different from the inactive solution dimer, both would need to be side-to-side. Second, FXa dimer formation also interferes with FVa binding to FXa (R. Majumder and B. R. Lentz, unpublished observations). K270 is part of a synthetic peptide motif (H263–K276) previously shown to interfere with FVa binding (48). Since the FVa binding region of FXa is now widely believed to be in the Y367–D373 loop (49) and/or the Y344–K351 helix (10,43), it is likely that peptide (H263–K276)-based inhibition of FVa binding is a secondary effect caused by H263–K276 (including K270) being in another protein recognition region that communicates with the FVa binding region. Since we know that dimer formation lowers kcat substantially, it is reasonable to conjecture that K270 might be in a substrate recognition region. Third, if K270 is in the dimer interface, the conformation of FXa on the membrane or in the dimer must be very different from that shown in Fig. 7. Binding data obtained by elipsometry suggest that the conformation of FXa in the dimer may be very different from the monomer structure shown in Fig. 7. These data show twice as many FXa molecules bound to a bilayer surface as factor X, suggesting that FXa occupies half the surface area of factor X (50). The binding stoichiometries obtained from this study were 26 and 53 lipids/protein in the outer bilayer leaflet for FXa and FX, respectively. The former agrees quite well with the value of ∼26 obtained for human FXa binding to the outer monolayer of 23 nm 20% PS vesicles (51). Given that the catalytic “head” of FXa has a rotationally averaged diameter (about the FXa long axis) of 40–45 Å and that the area/lipid molecule in a bilayer is ∼60–70 Å2/lipid, depending on the lipid and the method of estimation (52), we would estimate that the stoichiometry of a cylindrical FXa perpendicular to a membrane would be 23–33 lipids/FXa. This is in reasonably good agreement with the experimentally determined values of 26. However, in Fig. 7, F and G, we show that the FXa conformation used must be at an angle to the plane of the bilayer if its membrane-binding GLA-domain Ca2+ ions are constrained to be coplanar with the membrane. From this model, we estimate that the rotationally averaged (about an axis perpendicular to the membrane) diameter of FXa would be close to 60 Å. Based on this estimation, the stoichiometry of FXa binding should be ∼50 lipids/FXa, approximately what has been reported for FX (50). This suggests that FX and FXa have different membrane-associated conformations and that the membrane conformation of FX is similar to that shown in Fig. 7 for FXa. Consistent with this, the Pedersen group's model of FXa is only slightly different from their model of FX (25). At any rate, binding stoichiometries support the FXa conformation on a membrane being quite different from that shown in Fig. 7. Finally, the Pedersen model of FXa based on x-ray structures (25) puts the active site 89 Å from the plane of Ca2+ ions that define the membrane-protein interface, whereas fluorescence resonance energy transfer puts this distance at 61 Å (26), again suggesting that the membrane- or PS-bound conformation is not that shown in Fig. 7. This is quite reasonable, since membrane- or C6PS-induced conformational rearrangements between domains within prothrombin have been reported ((23,54) and S. Sen, R. Chattopadhyay, M. Banerjee, R. Majumder, C. Zhow, and B. R. Lentz; unpublished observations), and prothrombin is structurally similar to FX. Such conformational changes could well change the orientation of the catalytic domain relative to the EGF pair, as well as the relationship between the EGF domains, so that K270 could be buried in a side-to-side dimer interface.
If this interpretation of our results is correct, it would not be possible to examine the structure of FXa dimer without achieving a PS-activated conformation. Since FVa also undergoes conformational changes and changes in interdomain interactions (55) that are responsible for tight binding to FXa (28,46,56), it is likely that the structure of the FXa-FVa complex can also not be modeled using other than structures of PS-activated forms of the individual proteins, which to date are not available.
What are the physiological implications of our observation that human FXa forms inactive dimers under the influence of phosphatidylserine? The strength of the dimer interaction is strongly Ca2+-concentration-dependent, such that significant dimer forms in solution only at or above 5 mM Ca2+. The Ca2+ concentration is 1–1.5 mM in plasma (57,58), well below the Ca2+ concentrations shown here that are needed to trigger dimer formation. However, FXa dimers would not form in solution in plasma in any case, since soluble PS must bind to trigger dimer formation, and short chain phospholipids do not occur naturally. FXa binds to PS in membranes released by activated platelets in the platelet plug, within which Ca2+ concentration increases to 3–5 mM (58). Of course, it takes a much smaller free energy of interaction to bring proteins into association on a membrane surface than in solution simply because there is less entropy loss in the surface-mediated process. We conclude that FXa dimerization could be a significant Ca2+-regulatory mechanism physiologically, although we will need to investigate FXa behavior on a membrane surface to better judge this possibility. These studies are well underway, and thus far, results demonstrate formation of an inactive FXa dimer on a membrane (T. Koklic, R. Majumder, and G. Weinreb, unpublished observations), as reported here for FXa in solution.
Acknowledgments
We acknowledge the generous help of Dr. Brenda Temple, director of the Molecular Imaging Facility at the University of North Carolina at Chapel Hill, for help in preparing Fig. 7. We also thank Drs. Lee Pederson and Chang Jun Lee of UNC-CH's Chemistry Department for providing the coordinates of the Pedersen group's FXa model (25), and for their attempts to model the FXa dimer based on this published model and our experimental constraints. The process gave us insights into how to approach determining the structure of the dimer.
This work was supported by a grant from the National Institutes of Health, National Heart, Lung, and Blood Institute (HL 072827 to BRL) and in part by the Intramural Research Program of the National Institutes of Health National Institute of Environmental Health Sciences.
Footnotes
Roxana Iacob and Shalmali Sen contributed equally to this work.
Supporting Material
References
- 1.Schick P.K., Kurica K.B., Chacko G.K. Location of phosphatidylethanolamine and phosphatidylserine in the human platelet plasma membrane. J. Clin. Invest. 1976;57:1221–1226. doi: 10.1172/JCI108390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevers E.M., Comfurius P., Zwaal R.F. Changes in membrane phospholipid distribution during platelet activation. Biochim. Biophys. Acta. 1983;736:57–66. doi: 10.1016/0005-2736(83)90169-4. [DOI] [PubMed] [Google Scholar]
- 3.Jackson C.M., Nemerson Y. Blood coagulation. Annu. Rev. Biochem. 1980;49:765–811. doi: 10.1146/annurev.bi.49.070180.004001. [DOI] [PubMed] [Google Scholar]
- 4.Mann K.G., Jenny R.J., Krishnaswamy S. Cofactor proteins in the assembly and expression of blood clotting enzyme complexes. Annu. Rev. Biochem. 1988;57:915–956. doi: 10.1146/annurev.bi.57.070188.004411. [DOI] [PubMed] [Google Scholar]
- 5.Fenton J.W., II Thrombin. Ann. N.Y. Acad. Sci. 1986;485:5–15. doi: 10.1111/j.1749-6632.1986.tb34563.x. [DOI] [PubMed] [Google Scholar]
- 6.Prager N.A., Abendschein D.R., McKenzie C.R., Eisenberg P.R. Role of thrombin compared with factor Xa in the procoagulant activity of whole blood clots. Circulation. 1995;92:962–967. doi: 10.1161/01.cir.92.4.962. [DOI] [PubMed] [Google Scholar]
- 7.Maignan S., Guilloteau J.P., Pouzieux S., Choi-Sledeski Y.M., Becker M.R. Crystal structures of human factor Xa complexed with potent inhibitors. J. Med. Chem. 2000;43:3226–3232. doi: 10.1021/jm000940u. [DOI] [PubMed] [Google Scholar]
- 8.Weitz J.I. Low-molecular-weight heparins. N. Engl. J. Med. 1997;337:688–698. doi: 10.1056/NEJM199709043371007. [DOI] [PubMed] [Google Scholar]
- 9.Wei A.Z., Alexander R.S., Duke J., Ross H., Rosenfeld S.A. Unexpected binding mode of tick anticoagulant peptide complexed to bovine factor Xa. J. Mol. Biol. 1998;283:147–154. doi: 10.1006/jmbi.1998.2069. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph A.E., Porche-Sorbet R., Miletich J.P. Definition of a factor Va binding site in factor Xa. J. Biol. Chem. 2000;276:5123–5128. doi: 10.1074/jbc.M006961200. [DOI] [PubMed] [Google Scholar]
- 11.Sabharwal A.K., Padmanabhan K., Tulinsky A., Mathur A., Gorka J. Interaction of calcium with native and decarboxylated human factor X. Effect of proteolysis in the autolysis loop on catalytic efficiency and factor Va binding. J. Biol. Chem. 1997;272:22037–22045. doi: 10.1074/jbc.272.35.22037. [DOI] [PubMed] [Google Scholar]
- 12.Padmanabhan K., Padmanabhan K.P., Tulinsky A., Park C.H., Bode W. Structure of human des(1–45) factor Xa at 2.2 Å resolution. J. Mol. Biol. 1993;232:947–966. doi: 10.1006/jmbi.1993.1441. [DOI] [PubMed] [Google Scholar]
- 13.Krishnaswamy S., Church W.R., Nesheim M.E., Mann K.G. Activation of human prothrombin by human prothrombinase. Influence of factor Va on the reaction mechanism. J. Biol. Chem. 1987;262:3291–3299. [PubMed] [Google Scholar]
- 14.Srivastava A., Wang J., Majumder R., Rezaie A.R., Stenflo J. Localization of phosphatidylserine binding sites to structural domains of factor Xa. J. Biol. Chem. 2002;277:1855–1863. doi: 10.1074/jbc.M105697200. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee M., Drummond D.C., Srivastava A., Daleke D., Lentz B.R. Specificity of soluble phospholipid binding sites on human factor X(a) Biochemistry. 2002;41:7751–7762. doi: 10.1021/bi020017p. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee M., Banerjee M., Majumder R., Weinreb G., Wang J. Role of procoagulant lipids in human prothrombin activation. 2. Soluble phosphatidylserine upregulates and directs factor X(a) to appropriate peptide bonds in prothrombin. Biochemistry. 2002;41:950–957. doi: 10.1021/bi0116902. [DOI] [PubMed] [Google Scholar]
- 17.Majumder R., Wang J., Lentz B.R. Effects of water soluble phosphotidylserine on bovine factor X(a): functional and structural changes plus dimerization. Biophys. J. 2003;84:1238–1251. doi: 10.1016/S0006-3495(03)74939-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips A.T. Ligand-induced oligomerization and regulatory mechanism. CRC Crit. Rev. Biochem. 1974;2:343–378. doi: 10.3109/10409237409105451. [DOI] [PubMed] [Google Scholar]
- 19.Traut T.W. Dissociation of enzyme oligomers: a mechanism for allosteric regulation. Crit. Rev. Biochem. Mol. Biol. 1994;29:125–163. doi: 10.3109/10409239409086799. [DOI] [PubMed] [Google Scholar]
- 20.White J.H., Wise A., Main M.J., Green A., Fraser N.J. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 21.Margeta-Mitrovic M., Jan Y.N., Jan L.Y. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 22.Owens J. Bridging the GPCR gap. Nat. Rev. Drug Discov. 2006;5:105. [Google Scholar]
- 23.Lentz B.R., Zhou C.M., Wu J.R. Phosphatidylserine-containing membranes alter the thermal stability of prothrombin's catalytic domain: a differential scanning calorimetric study. Biochemistry. 1994;33:5460–5468. doi: 10.1021/bi00184a015. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q., Lentz B.R. Fluorescence resonance energy transfer study of shape changes in membrane-bound bovine prothrombin and meizothrombin. Biochemistry. 1997;36:4701–4711. doi: 10.1021/bi961441r. [DOI] [PubMed] [Google Scholar]
- 25.Venkateswarlu D., Perera L., Darden T., Pedersen L.G. Structure and dynamics of zymogen human blood coagulation factor x. Biophys. J. 2002;82:1190–1206. doi: 10.1016/S0006-3495(02)75476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Husten E.J., Esmon C.T., Johnson A.E. The active site of blood coagulation factor Xa. Its distance from the phospholipid surface and its conformational sensitivity to components of the prothrombinase complex. J. Biol. Chem. 1987;262:12953–12961. [PubMed] [Google Scholar]
- 27.Koppaka V., Wang J., Banerjee M., Lentz B.R. Soluble phospholipids enhance factor Xa-catalyzed prothrombin activation in solution. Biochemistry. 1996;35:7482–7491. doi: 10.1021/bi952063d. [DOI] [PubMed] [Google Scholar]
- 28.Majumder R., Weinreb G., Zhai X., Lentz B.R. Soluble phosphatidylserine triggers assembly in solution of a prothrombin-activating complex in the absence of a membrane surface. J. Biol. Chem. 2002;277:29765–29773. doi: 10.1074/jbc.M200893200. [DOI] [PubMed] [Google Scholar]
- 29.Koppaka V., Lentz B.R. Binding of bovine factor Va to phosphatidylcholine membranes. Biophys. J. 1996;70:2930–2937. doi: 10.1016/S0006-3495(96)79863-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J.R., Zhou C., Majumder R., Powers D.D., Weinreb G. Role of procoagulant lipids in human prothrombin activation. 1. Prothrombin activation by factor X(a) in the absence of factor V(a) and in the absence and presence of membranes. Biochemistry. 2002;41:935–949. doi: 10.1021/bi0116893. [DOI] [PubMed] [Google Scholar]
- 31.Hochleitner E.O., Borchers C., Parker C., Bienstock R.J., Tomer K.B. Characterization of a discontinuous epitope of the human immunodeficiency virus (HIV) core protein p24 by epitope excision and differential chemical modification followed by mass spectrometric peptide mapping analysis. Protein Sci. 2000;9:487–496. doi: 10.1110/ps.9.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majumder R., Quinn-Allen M.A., Kane W.H., Lentz B.R. The phosphatidylserine binding site of the factor Va C2 domain accounts for membrane binding but does not contribute to the assembly or activity of a human factor Xa-factor Va complex. Biochemistry. 2005;44:711–718. doi: 10.1021/bi047962t. [DOI] [PubMed] [Google Scholar]
- 33.Vossen K.M., Stickle D.F., Fried M.G. The mechanism of CAP-lac repressor binding cooperativity at the E. coli lactose promoter. J. Mol. Biol. 1996;255:44–54. doi: 10.1006/jmbi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 34.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 35.Johnson W.C.J. Protein secondary structure and circular dichroism: a practical guide. Proteins Struct. Funct. Genet. 1990;78:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 36.Hennessey J.P., Jr., Johnson W.C., Jr., Bahler C., Wood H.G. Subunit interactions of transcarboxylase as studied by circular dichroism. Biochemistry. 1982;21:642–646. doi: 10.1021/bi00533a007. [DOI] [PubMed] [Google Scholar]
- 37.Fung M.R., Hay C.W., MacGillivray R.T.A. Characterization of an almost full-length cDNA coding for human blood coagulation factor X. Proc. Natl. Acad. Sci. USA. 1985;82:3591–3595. doi: 10.1073/pnas.82.11.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezaie A.R., Esmon C.T. Asp-70→Lys mutant of factor X lacks high affinity Ca2+ binding site yet retains function. J. Biol. Chem. 1994;269:21495–21499. [PubMed] [Google Scholar]
- 39.Rezaie A.R., He X. Sodium binding site of factor Xa: role of sodium in the prothrombinase complex. Biochemistry. 2000;39:1817–1825. doi: 10.1021/bi992006a. [DOI] [PubMed] [Google Scholar]
- 40.Wilkens M., Krishnaswamy S. The contribution of factor Xa to exosite-dependent substrate recognition by prothrombinase. J. Biol. Chem. 2002;277:9366–9374. doi: 10.1074/jbc.M110848200. [DOI] [PubMed] [Google Scholar]
- 41.Monnaie D., Arosio D., Griffon N., Rose T., Rezaie A.R. Identification of a binding site for quaternary amines in factor Xa. Biochemistry. 2000;39:5349–5354. doi: 10.1021/bi9926781. [DOI] [PubMed] [Google Scholar]
- 42.Reference deleted in proof.
- 43.Rezaie A.R. Identification of basic residues in the heparin-binding exosite of factor Xa critical for heparin and factor Va binding. J. Biol. Chem. 2000;275:3320–3327. doi: 10.1074/jbc.275.5.3320. [DOI] [PubMed] [Google Scholar]
- 44.Yegneswaran S., Mesters R.M., Griffin J.H. Identification of distinct sequences in human blood coagulation factor Xa and prothrombin essential for substrate and cofactor recognition in the prothrombinase complex. J. Biol. Chem. 2003;278:33312–33318. doi: 10.1074/jbc.M305906200. [DOI] [PubMed] [Google Scholar]
- 45.Murakami M.T., Rios-Steiner J., Weaver S.E., Tulinsky A., Geiger J.H., Arni R.K. Intermolecular interactions and characterization of the novel factor Xa exosite involved in macromolecular recognition and inhibition: Crystal structure of human Gla-domainless factor Xa complexed with the anticoagulant protein NAPc2 from the hematophagous nematode. Ancylostoma caninum. J. Mol. Biol. 2007;366:602–610. doi: 10.1016/j.jmb.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 46.Majumder R., Weinreb G., Lentz B.R. Efficient thrombin generation requires molecular phosphatidylserine, not a membrane surface. Biochemistry. 2005;44:16998–17006. doi: 10.1021/bi051469f. [DOI] [PubMed] [Google Scholar]
- 47.Huang M., Rigby A.C., Morelli X., Grant M.A., Huang G. Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins. Nat. Struct. Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 48.Chattopadhyay A., James H.L., Fair D.S. Molecular recognition sites on factor Xa which participate in the prothrombinase complex. J. Biol. Chem. 1992;267:12323–12329. [PubMed] [Google Scholar]
- 49.Rezaie A.R., Kittur F.S. The critical role of the 185–189 loop in the factor Xa interaction with Na+ and factor Va in the prothrombinase complex. J. Biol. Chem. 2004;279:48262–48269. doi: 10.1074/jbc.M409964200. [DOI] [PubMed] [Google Scholar]
- 50.Hathcock J.J., Rusinova E., Gentry R.D., Andree H., Nemerson Y. Phospholipid regulates the activation of factor X by tissue factor/factor VIIa (TF/VIIa) via substrate and product interactions. Biochemistry. 2005;44:8187–8197. doi: 10.1021/bi050338b. [DOI] [PubMed] [Google Scholar]
- 51.Cutsforth G.A., Whitaker R.N., Hermans J., Lentz B.R. A new model to describe extrinsic protein binding to phospholipid membranes of varying composition: application to human coagulation proteins. Biochemistry. 1989;28:7453–7461. doi: 10.1021/bi00444a045. [DOI] [PubMed] [Google Scholar]
- 52.Koenig B.W., Strey H.H., Gawrisch K. Membrane lateral compressibility determined by NMR and x-ray diffraction: effect of acyl chain polyunsaturation. Biophys. J. 1997;73:1954–1966. doi: 10.1016/S0006-3495(97)78226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reference deleted in proof.
- 54.Lentz B.R., Wu J.R., Sorrentino A.M., Carleton J.N. Membrane binding induces lipid-specific changes in the denaturation profile of bovine prothrombin. A scanning calorimetry study. Biophys. J. 1991;60:942–951. doi: 10.1016/S0006-3495(91)82128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhai X., Srivastava A., Drummond D.C., Daleke D., Lentz B.R. Phosphatidylserine binding alters the conformation and specifically enhances the cofactor activity of bovine factor V(a) Biochemistry. 2002;41:5675–5684. doi: 10.1021/bi011844d. [DOI] [PubMed] [Google Scholar]
- 56.Majumder R. A phosphatidylserine binding site in factor Va C1 domain regulates both assembly and activity of the prothrombinase complex. Blood. 2008 doi: 10.1182/blood-2008-02-138941. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lansdown A.B., Sampson B., Rowe A. Sequential changes in trace metal, metallothionein and calmodulin concentrations in healing skin wounds. J. Anat. 1999;195:375–386. doi: 10.1046/j.1469-7580.1999.19530375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grzesiak J.J., Pierschbacher M.D. Shifts in the concentrations of magnesium and calcium in early porcine and rat wound fluids activate the cell migratory response. J. Clin. Invest. 1995;95:227–233. doi: 10.1172/JCI117644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamata K., Kawamoto H., Honma T., Iwama T., Kim S.H. Structural basis for chemical inhibition of human blood coagulation factor Xa. Proc. Natl. Acad. Sci. USA. 1998;95:6630–6635. doi: 10.1073/pnas.95.12.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banner D.W., D'Arcy A., Chene C., Winkler F.K., Guha A. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.