Abstract
Septic or endotoxic shock is a common cause of death in hospital intensive care units. In the last decade numerous antimicrobial peptides and proteins have been tested in the search for an efficient drug to treat this lethal disease. Now in phase III clinical trials, rBPI21, a recombinant N-terminal fragment of the bactericidal/permeability-increasing protein (BPI), is a promising drug to reduce lesions caused by meningococcal sepsis. We correlated structural and stability data with functional information of rBPI21 bound to both model systems of eukaryotic and bacterial membranes. On interaction with membranes, rBPI21 loses its conformational stability, as studied by circular dichroism. This interaction of rBPI21 at membrane level was higher in the presence of negatively charged phospholipid relatively to neutral ones, with higher partition coefficients (Kp), suggesting a preference for bacterial membranes over mammalian membranes. rBPI21 binding to membranes is reinforced when its disulfide bond is broken due to conformational changes of the protein. This interaction is followed by liposome aggregation due to unfolding, which ensures protein aggregation, and interfacial localization of rBPI21 in membranes, as studied by extensive quenching by acrylamide and 5-deoxylstearic acid and not by 16-deoxylstearic acid. An uncommon model of the selectivity and mechanism of action is proposed, where membrane induces unfolding of the antimicrobial protein, rBPI21. The unfolding ensures protein aggregation, established by protein-protein interaction at membrane surface or between adjacent membranes covered by the unfolded protein. This protein aggregation step may lead to membrane perturbation.
Introduction
Gram-negative bacteria have two cell envelope membranes. The outer membrane is asymmetric, with the glycolipid lipopolysaccharide (LPS) covering >90% of its outer leaflet surface and the inner leaflet with a phospholipid composition similar to the cytoplasmic membrane (1,2). The highly packed LPS moieties confer bacteria resistance to most antimicrobial peptides. Nevertheless, many antimicrobial cationic peptides have been shown to interact with LPS of Gram-negative bacteria and sometimes pass across this membrane by self-promoted uptake, followed by interaction and insertion into the negatively charged cytoplasmic membrane (3,4). When present in the mammalian bloodstream, LPS elicits a common but often fatal condition named septic or endotoxic shock, characterized by hypotension and circulatory failure, which can culminate in multiple system organ failure. When in circulation, LPS, complexed with several different proteins (mainly lipopolysaccharide binding protein [LBP]), is recognized by CD14 receptors in macrophages, monocytes or neutrophils, which triggers a cascade of events that leads to sepsis (5–7). Sepsis is a combination of both local and systemic inflammatory responses, which requires a multipronged interventional strategy to contain the various phases of disease progression. LPS sequestration by small peptides or proteins is an attractive interventional modality and many naturally occurring peptides (magainins, mellitins, cecropins, etc.) have intrinsic LPS binding activity (8–12). Most of these peptides may form amphipathic helices. Their cationic nature facilitates anchorage to the negatively charged bacterial surface, whereas their amphiphilicity and high helical propensity promote the formation of α-helices in membranes. Moreover, the insertion of such amphipathic α-helices in membranes underpins their membrane disruption activity. Two related proteins, bactericidal/permeability-increasing protein (BPI) and LBP are known to mediate the biological effects of LPS and participate in the innate immune response to bacterial infection in different ways (13–15). LBP promotes LPS transfer to innate immune cells, activating them, whereas BPI, present in azurophil granules of polymorphonuclear neutrophils, acts as a cytotoxic agent against Gram-negative bacteria and neutralizing the toxicity of LPS (16,17). The N-terminal fragment of BPI (rBPI21) not only maintains the biological activity, neutralizing LPS (18–21), but also represents a promising clinical tool to act against meningitis and septic shock (22). rBPI21 is a recombinant 21 kDa truncated protein corresponding to amino acids residues 1–193 of the N-terminal of human BPI (with the exception that a cysteine is replaced by an alanine at position 132). Although various studies with BPI and its fragments have been carried out, namely concerning antimicrobial activity, there is still a great lack of information concerning its mode of action, specially at membrane level. The interaction of rBPI21 with negatively charged lipids was reported (23,24) but detailed structural information on the interaction with LPS is needed for the development of more effective endotoxin-neutralizing agents.
In this study, fluorescence spectroscopy, circular dichroism (CD) and light scattering were combined to systematically study the interaction of rBPI21 with model systems of both eukaryotic and bacterial membranes, mainly: 1), the role of the disulfide bond and the selectivity to saccharide moiety of LPS regarding bounding/recognition of LPS by rBPI21; 2), temperature effects on the binding to negatively charged groups of phospholipid bilayers; 3), the interaction with membranes of different compositions, as a key to understand the selectivity toward bacteria and; 4), the rBPI21 conformational changes in its interaction with phospholipid membranes and/or LPS.
Materials and Methods
rBPI21, 99% pure, was a gift from XOMA Ltd. (Berkeley, CA). All lipids were obtained from Avanti Polar Lipids (Alabaster, AL), except LPS (Escherichia coli O55:B5) that was purchased from Fluka (Buchs, Switzerland). Dodecyl 2,6-dideoxy-β-L-arabino-hexopyranoside (DAHP) was a kind gift from Prof. Pilar Rauter (University of Lisbon, Portugal) and synthesized as described elsewhere (25). Acetate buffer (5 mM sodium acetate, 150 mM NaCl) was used in all the fluorescence studies. Sodium phosphate buffer was used instead of acetate buffer in the CD studies to prevent significant absorption interferences. Tris(2-cyanoethyl)phosphine (TCP) was from Molecular Probes (Eugene, OR). 5-deoxylstearic acid (5NS), 16-deoxylstearic acid (16NS), guanidinium chloride (GdnHCl), sodium phosphate, and all other chemicals were of the highest commercially available purity and were purchased from Sigma (St. Louis, MO). UV-Vis absorption measurements were carried out in a Shimadzu spectrophotometer (model UV-3101 PC), unless otherwise stated. Fluorescence spectroscopy measurements were conducted on a spectrofluorimeter SLM-Aminco 8100, equipped with a 450 W Xe Lamp and double monochromators in both excitation and emission, as well as a quantum counter.
Large unilamellar vesicles preparation
Large unilamellar vesicles (LUV), with ∼100 nm of diameter, were obtained by extrusion of multilamellar vesicles as described elsewhere (26).
Fluorescence decays
Time-resolved fluorescence decay measurements were carried out as described previously (27) with a time-correlated single-photon counting system using a Hamamatsu R-2809 MCP photomultiplier (Hamamatsu, Japan) for the detection. Excitation and emission wavelengths of 285 nm and 308 nm, respectively, were chosen for rBPI21 studies. rBPI21, stock solution of 95.2 μM, was added to previously prepared vesicle suspensions to a final concentration of 0.7 μM. Fluorescence decays were complex and described by a sum of three exponentials. The mean lifetime, , was obtained from Eq. 1 (28):
| (1) |
where ai and τi are the pre-exponential factors and the fluorescence life-time components, respectively.
The extent of incorporation of rBPI21 in model system of eukaryotic and bacterial membranes
Partition studies were carried out in LUV, prepared using different lipids (phospholipid + glycolipid or phospholipid + lipopolysaccharide). rBPI21 concentration was kept constant (0.7 μM). Fluorescence intensity was measured with excitation and emission wavelengths of 295 nm and 338 nm, respectively. The measured fluorescence intensity, If, is a balance between the fluorescence intensities of the peptide in aqueous phase (IW) and inserted in the lipid matrix (IL) as follows Eq. 2 (29). The weight factors in this balance depend on the partition coefficient, Kp, which was calculated as a fit parameter in a nonlinear regression methodology.
| (2) |
| (3) |
where [L] the lipid molar concentration and γL denotes the molar volume of the lipid; nS,i are the moles of the solute present in each phase (i = L and i = W refer to the lipid and aqueous media, respectively).
In this work we studied the lipid phase influence on rBPI21 partition to different model systems of membranes. We wanted to study not only the influence of the lipid phase (liquid-crystalline versus gel) but also the electrostatic effect (negative charged lipids versus zwitterionic lipids) and the influence of different kinds of negatively charged lipid headgroups (phosphatidylglycerol versus phosphatidylserine). Moreover we wanted to compare LPS with an unrelated membrane bound saccharide (dihydroxyacetone phosphate [DHAP] versus LPS). palmitoyloleoylphosphatidylcholine (POPC) and palmitoyloleoylphosphatidylglycerol (POPG) LUV are in liquid-crystalline phase whereas dipalmitoylphosphatidylcholine (DPPC), dipalmitoylphosphatidylglycerol (DPPG), and dipalmitoylphosphatidylserine (DPPS) LUV are in gel phase at 25°C and 37°C.
Fluorescence quenching studies of rBPI21 in aqueous phase and model system of bacterial membranes
Acrylamide 8 M was used to quench rBPI21 fluorescence in the aqueous phase. rBPI21 was dissolved in acetate buffer, pH 6.5 or 7.4. Aliquots of acrylamide were added to the mixture up to 0.5 M and the fluorescence intensity was measured at 325 nm. Quenching experiments with acrylamide were carried out to study Trp residues exposure to the aqueous phase. Acrylamide was added to samples containing rBPI21 and LUV of phospholipids with or without DAHP or LPS up to 0.5 M. Data treatment was as described elsewhere (30). Fluorescence intensity was measured with excitation and emission wavelengths of 295 nm and 330 nm, respectively. Differential quenching studies were carried out to study the in-depth location of rBPI21 in LUV of phospholipids with or without DHAP or LPS (both were in final 5%, molar, of the total lipid concentration). These quenching studies were carried out by adding small aliquots of 5NS or 16NS (70 mM solutions in ethanol) to the suspension of LUV with rBPI21 added previously. Ethanol was kept below 2% (v/v) to not affect lipid vesicles structure. When inserted in lipid bilayers, 5NS and 16NS have doxyl moieties close to the headgroup region and in the acyl chain region of the lipid palisade, respectively.
The simplest model that describes dynamic or static fluorescence quenching leads to linear Stern-Volmer plots:
| (4) |
where I0 and I denote the fluorescence intensities without and with quencher, respectively, KSV is the Stern-Volmer constant and [Q] is the quencher concentration. However, nonlinear Stern-Volmer plots are observed when there are multiple classes of fluorophores (31), such as a fluorophore population protected from the contact of the quencher and a population that is accessible to it (32).
The role of the disulfide bond of rBPI21 both in its structure in solution and in the extent of incorporation in model systems of bacterial membranes
TCP solution was prepared by dilution in acetate buffer. For modification with TCP in solution, rBPI21 was dissolved in acetate buffer, pH 6.5 or 7.4. Aliquots of TCP were added to the mixture at fixed intervals and fluorescence spectra were recorded. The final concentration of TCP ranged from 0.2 to 1.4 mM. To study the influence of a reduction in rBPI21 extent of incorporation in model systems of bacterial membranes, rBPI21 was pre-incubated with TCP (final concentration 1.4 mM) and added to LUV composed of POPC/POPG/DAHP or POPC/POPG/LPS. Fluorescence intensity was measured afterward and partition coefficients were calculated.
Conformational studies of rBPI21 in aqueous phase and in model systems of bacterial membranes by CD
Secondary structure analysis was carried out by far-UV (185–260 nm) CD in a Jasco J810 Spectropolarimeter (Jasco, Japan) equipped with a temperature control unit Julabo F25. Far UV CD spectra were recorded with 0.1 cm (linear) path length quartz cuvette. For each spectrum, three scans were averaged. rBPI21 samples in 20 mM sodium phosphate buffer with 150 mM sodium fluoride, pH 6.5 or 7.4, in the presence or absence of 0.1 mM lipid concentration were equilibrated either at 25°C or 37°C, before measurements. Minor contributions of circular differential scattering were eliminated by subtracting the CD spectrum of buffer and/or lipids from the peptide spectrum on buffer and lipids, respectively. For rBPI21 secondary structure estimation, CD spectra were deconvoluted using the CDSSTR algorithm (33) on Dichroweb (34).
Conformational stability measurements
rBPI21 conformational stability studies were carried out by CD in GdnHCl-induced protein unfolding experiments either at 25°C or 37°C, at pH 6.5 and pH 7.4, after 48 h of incubation. CD denaturation curves were constructed using the ellipticity at 222 nm. A nonlinear least squares fitting procedure yielded values for ΔG0(H2O), the conformational stability, and m, the dependence of ΔG0 on denaturant concentration. Cm, the denaturant concentration at the midpoint of the unfolding transition was calculated as and KU, the unfolding equilibrium constant, was calculated as (35). Denaturation curves for monomeric species were analyzed considering the equation developed by Santoro and Bolen (36). Parameters were obtained by fitting the derived equations to experimental data by nonlinear regression using the solver add-on for Microsoft Excel 2003 (Microsoft, Redmond, WA).
Aggregation of negatively charged vesicles in the presence of rBPI21 by dynamic light scattering
Dynamic light scattering experiments were carried out on a Malvern Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK) with a backscattering detection at 173° and equipped with He-Ne laser (λ = 632.8 nm), at 37°C, using disposable polystyrene cells. The phosphate buffer and lipid were mixed and then filtered in a syringe filter with pore size of 0.45 μm (Whatman, Florham Park, NJ). Lipid concentration was fixed at 50 μM and the rBPI21 concentration was varied from 24 nM to 238 nM. The samples were left equilibrating for 30 min at 37°C. Normalized intensity autocorrelation functions were analyzed using the CONTIN method (37,38) yielding a distribution of diffusion coefficients (D) in the whole solution through multiexponential fits. The measured D is used for the calculation of the hydrodynamic radius (RH) through the Stokes-Einstein relationship:
| (5) |
where κ is the Boltzmann constant, T the absolute temperature, and η the medium viscosity. The hydrodynamic diameter (DH) of the sample was obtained from the peak with the highest scattered light intensity in light scattering intensity distributions.
Results and Discussion
Studies in aqueous solution
On excitation at λ ≥ 295 nm, the fluorescence of rBPI21 is dominated by tryptophan fluorescence, either at pH 6.5 or 7.4. In buffer solution the Trp emission of rBPI21 occurs at shorter wavelengths, relative to the emission of Trp in aqueous environment (Table 1). This blue shift results from the shielding of the Trp residues from water by the hydrophobic core of the folded peptide. BPI structure shows a Trp residue in hydrophobic environment (39), which may explain both this large blue shift and the increased fluorescence lifetime (Fig. 1). In buffer solution, intrinsic fluorescence emission was decreased for pH 6.5, although no shift in the fluorescence spectra is observed. This result suggests that a slightly acid pH induces conformational changes that induce self-quenching but do not involve exposure of the Trp residues to the aqueous medium.
Table 1.
Photophysical parameters for Trp and rBPI21, in acetate buffer and in large unilamellar vesicles (5 mM), at pH 7.4
λAbs, λEm are maximum absorption and emission wavelengths, respectively, and is the mean fluorescence lifetime. [Trp] = 1.0 mM; [rBPI21] = 7.7 μM. W and L subscripts denote aqueous and lipid environments, respectively.
At 23°C in aqueous solution at neutral pH (28).
Figure 1.
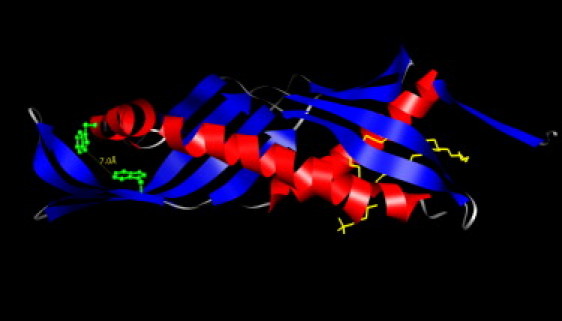
3D ribbon structure of the first 193 residues of BPI, corresponding to the sequence of the rBPI21. Secondary structure of the α-helices is represented in red online and black in print, β-sheets are in blue online and dark gray in print. Phosphatidylcholine (yellow online and gray in print), is localized in the hydrophobic pocket of the protein, and tryptophan residues are represented in green online and as a ball and stick residue in print. Structure based on the crystallographic structure of BPI (44).
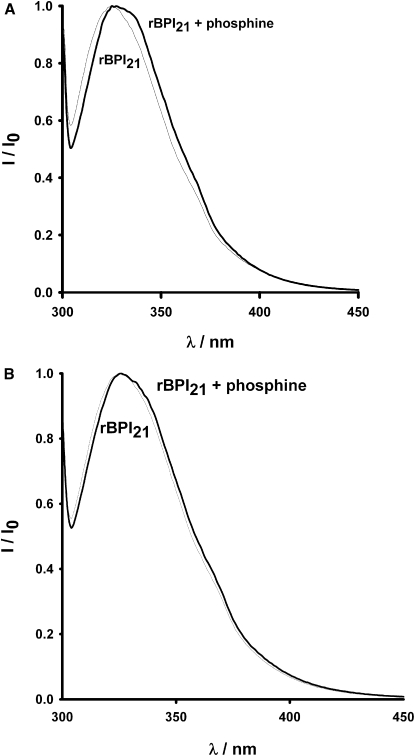
TCP was added to rBPI21 solution to study eventual changes in the Trp environment after the breaking of the disulfide bond. At pH 6.5 the emission maximum occurs at a higher wavelength (Fig. 2 A), on addition of TCP. This red shift suggests a change in the Trp residue environment to a more hydrophilic one. This is a combined effect of lowering the pH, which is close to histidines pKa, and disulfide bond breaking. No significant difference was observed in the emission spectra at pH 7.4 (Fig. 2 B), on addition of TCP, which points out to the fact that although disulfide bond reduction may induce conformational changes, these do not globally affect the Trp environment at this pH. These results in aqueous solution show that rBPI21 undergoes conformational changes when both acidification and disulfide bond reduction occurs, where rBPI21 exposes the Trp to a more hydrophilic environment.
Figure 2.
rBPI21 (7.7 μM) fluorescence emission spectra in acetate buffer before and after incubation with TCP (1.4 mM). (A) pH 6.5. (B) pH 7.4. λexc= 295 nm, at 37°C. Similar spectra were obtained at 25°C.
rBPI21 conformation and stability in aqueous buffer solution
To know if rBPI21 is folded at 37°C, thermal stability runs followed by CD were carried out (results not shown). The results show that rBPI21 is in its native conformation in aqueous solution at 25°C and 37°C.
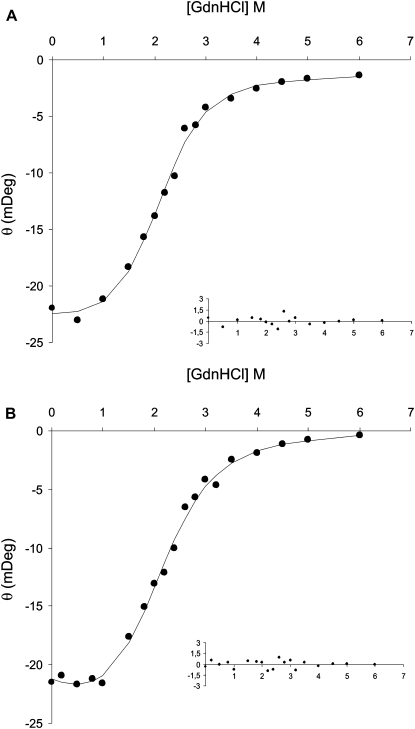
To address the importance of histidine protonation on the rBPI21 structure, we have characterized the chemical conformational stability of rBPI21 at pH 6.5 and 7.4. Fig. 3 shows the equilibrium denaturation curves at pH 7.4 and 6.5 at 25°C, using guanidinium chloride (GdnHCl) as denaturant, monitored by CD. The free energy of unfolding (ΔG0(H2O)) was directly obtained by a nonlinear least-squares fit of the experimental fluorescence data to a two-state model for monomer unfolding (35,36). GdnHCl-induced denaturation of rBPI21 was found to be reversible for both pH values studied, as judged by CD and fluorescence experiments. Analysis of GdnHCl-unfolded rBPI21 samples after dialysis, by CD and fluorescence, produced typical rBPI21 spectra parameters. Table 2 shows the values obtained from the curves in Fig. 3 for ΔG0(H2O), the conformational stability, m, the dependence of ΔG0 on denaturant concentration, and Cm, the denaturant concentration at the midpoint of the unfolding transition. The low values of ΔG0(H2O) for rBPI21 are well within typical values of conformational stability for peptides and truncated proteins.
Figure 3.
GdnHCl equilibrium denaturation curves of rBPI21 at pH 7.4 (A) and 6.5 (B), at 25°C, monitored by CD at 222 nm. The curves are nonlinear least squares fits of an equation representing the entire denaturation curves, based on a two-state model and using the linear extrapolation method to the experimental CD data (36). Insets represent the residues of the fit to the experimental data.
Table 2.
Stability parameters for rBPI21 in aqueous solution, at pH 6.5 and 7.4, obtained from a direct fit of the experimental data in Fig. 3 to a two-state unfolding model
| pH | ΔG0(H2O)/kcal.mol−1 | Cm/M | m/kcal.mol−2 × L |
|---|---|---|---|
| 6.5 | 2.05 | 1.90 | 1.07 |
| 7.4 | 2.68 | 2.04 | 1.31 |
ΔG0(H2O) is the free energy of unfolding, Cm, is the denaturant concentration at the midpoint of the unfolding transition, calculated as Cm = ΔG0(H2O)/m, and m is the ΔG0 dependence on the denaturant concentration.
The lower ΔG0(H2O) values obtained at pH 6.5 indicates a decrease in the conformational stability of rBPI21 when compared with the ΔG0(H2O) at pH 7.4 (Table 2). The smaller value of the conformational stability of rBPI21 at pH 6.5 agrees completely with the higher tendency for denaturation of rBPI21 at pH 6.5. Moreover, the denaturation midpoint, Cm, also shows this tendency, confirming the reduction of conformational stability on acidification.
It is worth noting that the slightly higher conformational stability shown by rBPI21 at pH 7.4 is also due to a higher m (Table 2). The m value is believed to reflect the extent of newly exposed surface area on unfolding (40). Thus, the higher m value observed for the unfolding of rBPI21 at pH 7.4 may indicate a more compact structure when compared with rBPI21 conformation at pH 6.5, which is in agreement with the results obtained from fluorescence techniques.
Interaction of rBPI21 with model systems of eukaryotic and bacterial membranes
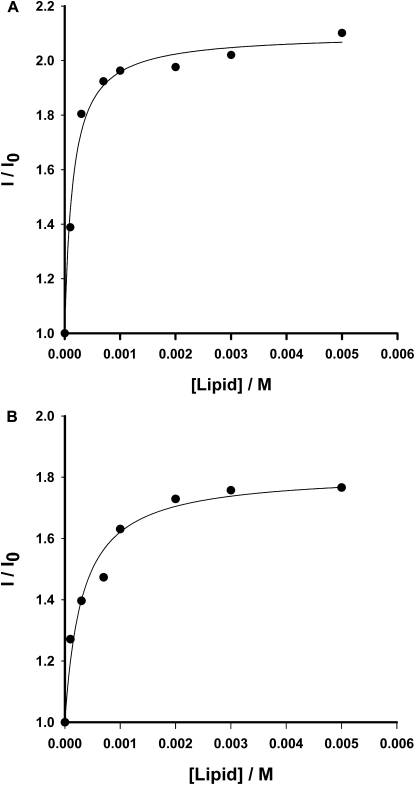
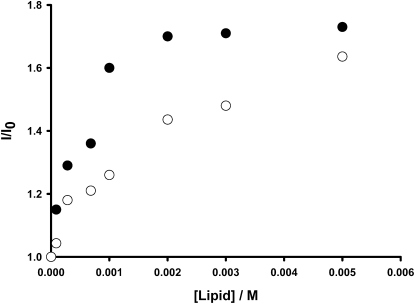
Partition of rBPI21 into model systems of eukaryotic and bacterial membranes was investigated from changes in the rBPI21 intrinsic fluorescence, as described in Materials and Methods. Table 3 shows that rBPI21 interacts more effectively with negatively charged membranes in the gel crystalline phase, the highest partition coefficients being obtained for DPPG, at 37°C, which may help to explain the preference of rBPI21 for Gram-negative bacteria. These bacteria are known for having an asymmetric outer membrane in which >90% of its outer leaflet surface is covered by LPS, which compared to typical phospholipid membranes are known to have tighter hydrocarbon chain packing and form rigid domains (41). The preference of rBPI21 for gel phase membranes with high density of negative charges is remarkable and parallels rBPI21 selectivity for the external surface of the outer membrane of gram-negative bacteria. The effect of charge alone does not suffice to explain this selectivity, as the partition coefficient for fluid POPG bilayers is considerably lower than the obtained for the rigid DPPG. Moreover, rBPI21 is sensitive to the chemical nature of the headgroups. Kp differs in mixtures containing PG and PS, where Kp(PG) > Kp(PS) (Fig. 4). This is of particular importance because PG is the major phospholipid component present in the cytoplasmic membrane of bacteria.
Table 3.
Partition coefficients (Kp) for rBPI21 in different lipid systems at pH 7.4 and 37°C
| System | Kp × 103 |
|---|---|
| DPPG | 13.6 ± 5.00 |
| DPPC/DPPG | 9.75 ± 1.82 |
| DPPC/DPPS | 3.98 ± 1.18 |
| DPPC | 0.123 ± 0.054 |
| POPG | 0.851 ± 0.213 |
| POPC | ≈0 |
| POPC/POPG/DAHP | 0.52 ± 0.17 |
| POPC/POPG/LPS | 1.12 ± 0.073 |
| POPC/POPG/DAHP (pre-incubated with TCP) | 2.14 ± 0.71 |
| POPC/POPG/LPS (pre-incubated with TCP) | 4.64 ± 1.50 |
Anionic lipid contents in lipid mixtures: 50% molar; LPS or DAHP content in mixtures: 5%.
Figure 4.
Determination of the partition constant (Kp) of the protein between the aqueous phase and phospholipid vesicles. Peptide concentration, of 0.7 μM, was constant during each set of experiments. Increase in fluorescence intensity obtained for different concentrations of (A) DPPC/DPPG and (B) DPPC/DPPS LUV. Fitting lines calculated using Eq. 2.
However, similar partition coefficients are obtained when LPS is replaced by an unrelated saccharide, which shows that rBPI21 is rather insensitive to the glycidic group. POPC is the major lipid component of eukaryotic membrane cells. If we calculate the ratio Kp(glycolipid or LPS systems)/Kp(POPC) for rBPI21 at physiological conditions (pH 7.4 and 37°C), values between 52 and 100 are obtained, which show clearly the preference of rBPI21 for bacterial membranes. Curiously, for some lipid model systems higher partition coefficients were obtained at pH 6.5, relative to pH 7.4 (data not shown). Similar fluorescence emission and CD spectra were obtained for rBPI21 at both pH values. However, a slight decrease in fluorescence intensity without a spectral shift was observed with pH decrease, which suggests that pH changes can induce an approximation between Trp and intramolecular quencher groups, rather than exposing Trp to the aqueous medium. Nevertheless, the exposure of hydrophobic segments is probably involved in the interaction of rBPI21 with the target membrane. This hydrophobic segments exposure hypothesis provides what we believe is a new insight, if we remember that conformational stability studies (see section above) showed that there is a slight conformational stability reduction on acidification, and that at pH 6.5 rBPI21 has a slightly less compact native state. Moreover, rBPI21 has several His residues near a tryptophan residue. As pH 6.5 is near the pKa value for His residues, protonation of His residues could generate positive charges on rBPI21, which may contribute both to the reduction in the compactness of the protein, due to intercharge repulsion, and to the increase electrostatic interaction required for protein membrane binding. Consequently, Kp increases and potential quencher groups are formed.
Localization of rBPI21 in model systems of bacterial membranes
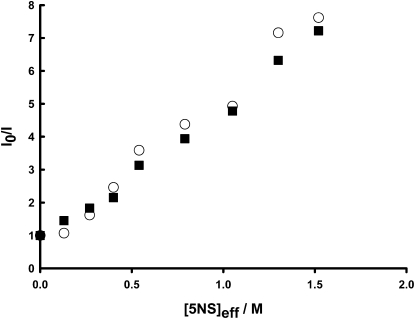
The in-depth location of rBPI21 in the model systems of bacterial membranes was studied by monitoring the relative quenching of the fluorescence of this peptide by the lipophilic spin probes 5NS and 16NS when inserted in fluid phase membranes of POPC/POPG/DAHP or POPC/POPG/LPS mixtures. The addition of any of the two nitroxide quenchers resulted in a decreased of rBPI21 fluorescence intensity. The Stern-Volmer plots of the fluorescence intensity changes were linear (Fig. 5), indicating that nearly all rBPI21 is inserted in the membrane at the used lipid concentration. The Stern-Volmer constants were obtained from their slopes. The ratio of these slopes, KSV(5NS)/KSV(16NS) was found to be always higher than 1, pointing toward a shallow location of the peptide, except for the POPC/POPG system at pH 7.4 (Table 4).
Figure 5.
rBPI21 location. Stern-Volmer plots for the quenching of rBPI21 fluorescence by 5NS in POPC/POPG/LPS (○) and POPC/POPG/DAHP (■) membranes (1 mM), pH 7.4 and 37°C. Effective 5NS concentrations in the membrane were calculated as described previously (52).
Table 4.
Location of rBPI21 in model systems of bacterial membranes
| System |
KSV (pH 6.5) |
KSV (pH 7.4) |
||
|---|---|---|---|---|
| 5NS | 16NS | 5NS | 16NS | |
| POPC/POPG | 2.07 | 0.36 | 0.82 | 1.29 |
| POPC/POPG/DAHP | 2.25 | 1.83 | 4.11 | 1.83 |
| POPC/POPG/LPS | 2.86 | 0.36 | 4.60 | 1.68 |
Stern-Volmer constants (KSV) of rBPI21 were obtained as described in Materials and Methods using 5NS and 16NS quenchers at 37°C, pH 6.5 and pH 7.4 Anionic lipid contents in lipid mixtures: 40% molar; LPS or DHAP content in mixtures: 5%.
In the case of the two bacterial model membranes composed of POPC/POPG/DAHP (or LPS) at physiological conditions (pH 7.4 and 37°C) the ratios obtained are quite similar (2.25 and 2.74 for the glycolipid and the LPS containing systems, respectively). This is indicative that rBPI21 location is not LPS-specific.
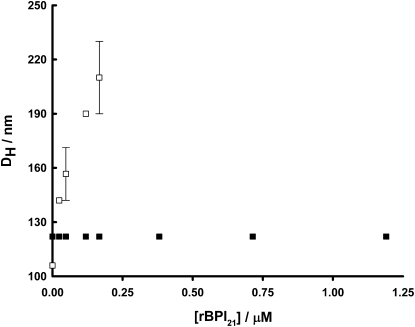
To confirm the membrane location of rBPI21, we used the hydrophilic quencher acrylamide, which quenches molecules present in the aqueous phase or in the lipid/water interface. rBPI21 is more extensively quenched by acrylamide in lipid vesicles than in buffer solution, for all the lipid systems tested (data not shown). This result is in agreement with a shallow location of rBPI21 in lipid vesicles, with an extended conformation where the Trp residues are relatively unshielded and more exposed to the aqueous environment than in the unbounded folded conformation. Tobias et al. (42) showed that BPI can increase the apparent size of LPS aggregates at 1:20 BPI/LPS ratio. Our preliminary dynamic light scattering experiments indicate a similar effect for rBPI21 on membranes composed of anionic phospholipids (Fig. 6). This means that not only rBPI21 has the capacity of forming aggregates at a low molar ratio but also that rBPI21 is capable of cross-linking LPS aggregates into larger ones. This LPS aggregation effect induced by rBPI21 may be helpful for its in vivo function and an explanation for its high biological activity. LPS interaction with leukocytes can initiate an inflammatory responses, but the LPS that binds to BPI is essentially inactive (43). This capacity for inactivating LPS due to surface binding may help to prevent septic shock. Partially unfolded proteins are prone to aggregate. Likewise, vesicle aggregation may be a direct consequence of partial unfolding.
Figure 6.
Aggregation of membrane model systems by rBPI21. Hydrodynamic diameter variation followed by dynamic light scattering of POPC (■) and POPC/POPG (□) membranes at fixed 50 μM lipid concentration, with 45% molar of negative lipid content, in the presence of increasing rBPI21 amount. Bars represents the size range from at least two independent experiments.
Role of disulfide bond in rBPI21 interaction with model systems of bacterial membranes
As the reduction of the disulfide bond between cysteines 135 and 175 with TCP is associated with a possible conformational change of the protein in buffer, we studied this effect of the chemical change on the interaction with membranes. rBPI21 partition to model system of bacterial membranes under physiological conditions was increased (pH 7.4, 37°C) (Table 3, Fig. 7). A fourfold increase of Kp is observed for both membranes on disulfide bond breaking. Cysteines 135 and 175 are both necessary for an efficient production of a biologically active N-terminal BPI fragment, putatively trough a disulfide bond (44), but at a certain cost in its LPS-binding activity, because disulfide bond reduction can lead to a more effective interaction with model systems of bacterial membranes.
Figure 7.
Implications of chemical modification on rBPI21 partition to model system of bacterial membranes. rBPI21 was dissolved in acetate buffer at pH 7.4 to a final protein concentration of 7.7 μM, and incubated with TCP 1.4 mM before partition coefficients determination, at 37°C. Partition curves of rBPI21 to POPC/POPG/DAHP LUV with (•) and without (○) pre-incubation with TCP. Kp values are obtained by fitting the experimental data with Eq. 2.
The results are in agreement with a model of action that involves unfolding/denaturation on interaction with in LPS-containing membranes. A decrease in stability renders the membrane disturbing effect more effective.
Conformational changes of rBPI21 after binding to lipid membranes
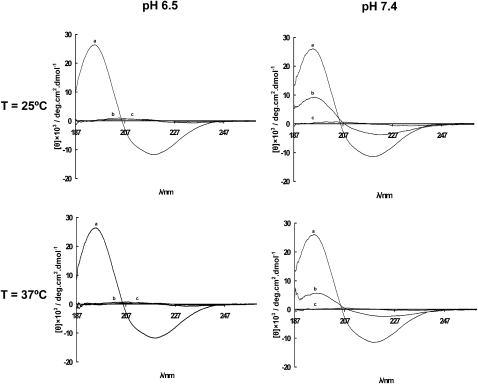
According to the CD measurements, rBPI21 adopts a tridimensional conformation with regular secondary structure in aqueous buffer solution but forming a partial or total unfolded structure in the presence of liposomes composed of POPC/POPG/LPS, either at pH 6.5 or 7.4, at 25°C and 37°C (Fig. 8). The CD spectra of rBPI21 in aqueous solution, both at pH 6.5 and pH 7.4, at 25°C and 37°C, are similar and display a positive ellipticity maximum near 194 nm and a negative ellipticity minimum around 217 nm, evidence for the presence of β-sheet conformation. In fact, secondary structure estimation from CD spectra analysis showed 33% of β-sheet, 14% of α-helix, and 30% random coil. Using POPC/POPG/LPS 0.1 mM as a bacterial membrane mimicking environment, rBPI21 completely loses its ordered native conformation independently of the pH and/or temperature (Fig. 8). CD spectra analysis showed a loss of secondary structure, with a major decrease in the α-helix content. For rBPI21 in interaction with POPC/POPG model systems of membranes, a less intense positive and negative ellipticity and wavelengths slightly different from the ones observed in aqueous solution were observed in CD spectra.
Figure 8.
Effect of pH, temperature and lipid system on the secondary structure of rBPI21. rBPI21 was dissolved in phosphate buffer (pH 6.5 or 7.4), at 25°C or 37°C, to a final concentration of 7.7 μM with or without added lipid (0.1 mM). CD spectra of rBPI21 in (a) buffer solution, and in interaction with (b) POPC/POPG/DAHP or (c) POPC/POPG/LPS membranes both at pH 6.5 and 7.4, and T = 25°C and 37°C.
Typically, peptides such as antimicrobial peptides, for instance, adopt a helical structure when interacting with lipid vesicles and not the other way around. Nevertheless, this high structure flexibility and low chemical conformational stability probably imply the loss of the hydrophobic pocket, because it is composed mainly by α-helix (44), which decreases in proportion with its increasing contact with lipid membranes. This represents further evidence that the exposure of this segment (hidden in the three-dimensional structure of rBPI21 in the aqueous phase) is a key-step in an innovative model mechanism for peptide-membrane interaction. The exposure of this segment to membranes decreases the mean fluorescence lifetime of rBPI21 (Table 1), which implies that Trp environment changes to a more hydrophilic one. Moreover, this flexibility may be the reason why rBPI21 can coat LPS containing vesicles and, consequently, be effective in inactivating the inflammatory cascade in vivo, in a first stage.
Conclusions
We have shown that rBPI21 in solution is stable at 37°C and has a higher chemical conformational stability at pH 7.4, when compared with a more acidic pH. It undergoes conformational changes when its disulfide bond is broken or pH decreases, exposing its Trp to a more hydrophilic environment when both changes occur. When in interaction with model systems of membranes, rBPI21 interacts preferentially with rigid negatively charged membranes, this being one of the reasons that may be invoked for its preference for gram-negative bacteria. Also, rBPI21 has a shallow location and completely loses its secondary structure, unfolding, while coating and aggregating LPS-containing vesicles (Fig. 8). rBPI21 is at variance with most of antimicrobial peptides that adopt a helical structure when inserted in the lipid bilayer. This may be related to a peculiar mechanism of action, where coating and aggregation of LPS-containing entities is of first importance for their neutralization. We have also shown that decreasing the pH, near the pKa of His, is important for rBPI21 interaction with membranes and that the breaking of the disulfide bond may enhance its LPS-binding activity in vivo, as a consequence of a significant increase of partition coefficients. Although there are some studies of rBPI21 in interaction with lipid systems (23,24), to our knowledge no one has identified specific amino acid residues implication in this process.
The CD spectra results have shown that rBPI21 has a predominant β-sheet secondary structure content. The results on chemical conformational stability at pH 7.4 and 6.5 contribute to explain the partial and total loss of structure of rBPI21 after binding to POPC/POPG/LPS liposomes at pH 7.4 and 6.5, respectively. The lower rBPI21 conformational stability value at pH 6.5 completely agrees with a higher loss of structure. Moreover, partial or total unfolded proteins are prone to form aggregates, which could explain liposome aggregation in the presence of rBPI21.
In summary, the results of this study highlight structural reasons that may be responsible for the selectivity of rBPI21 for Gram-negative bacteria and show that unfolding at the surface of the membrane, on interaction with LPS is an uncommon key-event of its mechanism (Fig. 9). Antimicrobial peptides are portrayed usually as molecules unstructured in solution, which gain structure when in contact with the membrane. Peptide-peptide interactions are then triggered at certain critical concentrations (45–51), which lead to supramolecular entities that cause lysis, such as a variety of pore-forming structures. rBPI21 seems to be quite peculiar as its mode of action is not at all related to this sequence of events from the start. It loses structure and stability on contact with LPS membranes and is able to cause membrane aggregation, which is quite perturbing for membranes and ultimately responsible for the bactericidal activity of this drug.
Figure 9.
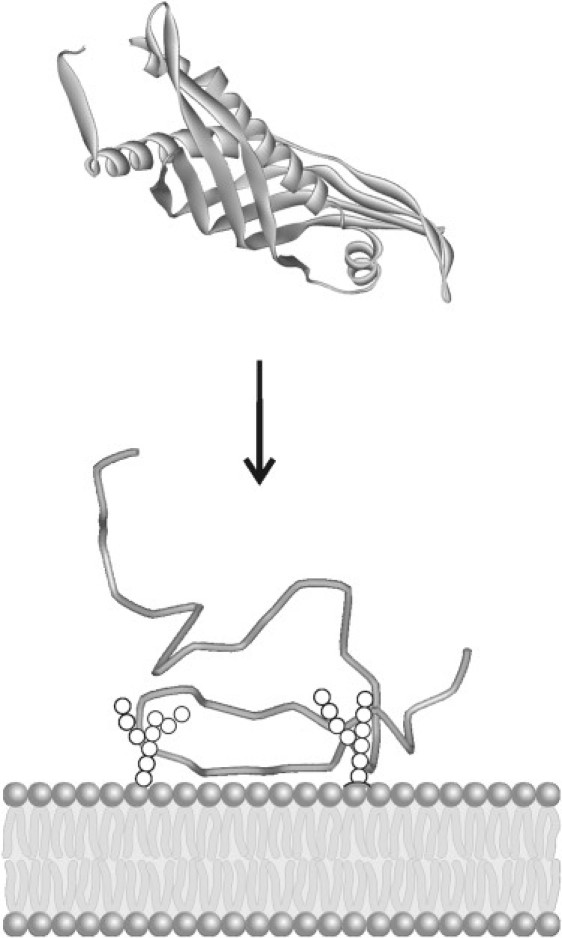
Schematic representation of the proposed mechanism of action of rBPI21 protein. The protein interacts superficially with LPS-rigid regions of the membrane of gram-negative bacteria. This interaction is followed by the unfolding of the protein covering the surface of the cell, leading to severe membrane perturbation.
Acknowledgments
We thank XOMA for a kind gift of rBPI21 and Prof. Amélia Pilar Rauters (Faculty of Science, University of Lisbon) for DAHP synthesis.
M.M.D. acknowledges Fundação para a Ciência e a Tecnologia do Ministério da Ciência, Tecnologia e Ensino Superior (Portugal) for fellowship SFRH/BD/41750/2007.
References
- 1.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rietschel E.T., Kirikae T., Schade F.U., Mamat U., Schmidt G. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 3.Hancock R.E., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu M., Maier E., Benz R., Hancock R.E. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 5.Chaby R. Lipopolysaccharide-binding molecules: transporters, blockers and sensors. Cell. Mol. Life Sci. 2004;61:1697–1713. doi: 10.1007/s00018-004-4020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumann R.R. Function of lipopolysaccharide (LPS)-binding protein (LBP) and CD14, the receptor for LPS/LBP complexes: a short review. Res. Immunol. 1992;143:11–15. doi: 10.1016/0923-2494(92)80074-u. [DOI] [PubMed] [Google Scholar]
- 7.Wright S.D., Ramos R.A., Tobias P.S., Ulevitch R.J., Mathison J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 8.Hancock R.E. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 9.Hancock R.E., Diamond G. The role of cationic antimicrobial peptides in innate host defenses. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 10.Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 11.Levy O. Antimicrobial proteins and peptides of blood: templates for novel antimicrobial agents. Blood. 2000;96:2664–2672. [PubMed] [Google Scholar]
- 12.Levy O. Antimicrobial proteins and peptides: anti-infective molecules of mammalian leukocytes. J. Leukoc. Biol. 2004;76:909–925. doi: 10.1189/jlb.0604320. [DOI] [PubMed] [Google Scholar]
- 13.Beamer L.J., Carroll S.F., Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobias P.S., Mathison J.C., Ulevitch R.J. A family of lipopolysaccharide binding proteins involved in responses to gram-negative sepsis. J. Biol. Chem. 1988;263:13479–13481. [PubMed] [Google Scholar]
- 15.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defense against Gram-negative bacteria. Biochem. Soc. Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 16.Elsbach P., Weiss J. Role of the bactericidal/permeability-increasing protein in host defense. Curr. Opin. Immunol. 1998;10:45–49. doi: 10.1016/s0952-7915(98)80030-7. [DOI] [PubMed] [Google Scholar]
- 17.Weiss J., Elsbach P., Olsson I., Odeberg H. Purification and characterization of a potent bactericidal and membrane active protein from the granules of human polymorphonuclear leukocytes. J. Biol. Chem. 1978;253:2664–2672. [PubMed] [Google Scholar]
- 18.Appelmelk B.J., An Y.Q., Thijs B.G., MacLaren D.M., de Graaff J. Recombinant human bactericidal/permeability-increasing protein (rBPI23) is a universal lipopolysaccharide-binding ligand. Infect. Immun. 1994;62:3564–3567. doi: 10.1128/iai.62.8.3564-3567.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capodici C., Chen S., Sidorczyk Z., Elsbach P., Weiss J. Effect of lipopolysaccharide (LPS) chain length on interactions of bactericidal/permeability-increasing protein and its bioactive 23-kilodalton NH2-terminal fragment with isolated LPS and intact Proteus mirabilis and Escherichia coli. Infect. Immun. 1994;62:259–265. doi: 10.1128/iai.62.1.259-265.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazzano-Santoro H., Parent J.B., Grinna L., Horwitz A., Parsons T. High-affinity binding of the bactericidal/permeability-increasing protein and a recombinant amino-terminal fragment to the lipid A region of lipopolysaccharide. Infect. Immun. 1992;60:4754–4761. doi: 10.1128/iai.60.11.4754-4761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsbach P. The bactericidal/permeability-increasing protein (BPI) in antibacterial host defense. J. Leukoc. Biol. 1998;64:14–18. doi: 10.1002/jlb.64.1.14. [DOI] [PubMed] [Google Scholar]
- 22.Levin M., Quint P.A., Goldstein B., Barton P., Bradley J.S. Recombinant bactericidal/permeability-increasing protein (rBPI21) as adjunctive treatment for children with severe meningococcal sepsis: a randomized trial. Lancet. 2000;356:961–967. doi: 10.1016/s0140-6736(00)02712-4. [DOI] [PubMed] [Google Scholar]
- 23.Wiese A., Brandenburg K., Carroll S.F., Rietschel E.T., Seydel U. Mechanisms of action of bactericidal/permeability-increasing protein BPI on reconstituted outer membranes of gram-negative bacteria. Biochemistry. 1997;36:10311–10319. doi: 10.1021/bi970177e. [DOI] [PubMed] [Google Scholar]
- 24.Wiese A., Brandenburg K., Lindner B., Schromm A.B., Carroll S.F. Mechanisms of action of the bactericidal/permeability-increasing protein BPI on endotoxin and phospholipid monolayers and aggregates. Biochemistry. 1997;36:10301–10310. doi: 10.1021/bi970176m. [DOI] [PubMed] [Google Scholar]
- 25.Rauter A.P., Lucas S., Almeida T., Sacoto D., Ribeiro V. Synthesis, surface active and antimicrobial properties of new alkyl 2,6-dideoxy-L-arabino-hexopyranosides. Carbohydr. Res. 2005;340:191–201. doi: 10.1016/j.carres.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 27.Loura L.M., Fedorov A., Prieto M. Resonance energy transfer in a model system of membranes: application to gel and liquid crystalline phases. Biophys. J. 1996;71:1823–1836. doi: 10.1016/S0006-3495(96)79383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakowicz J. 2nd ed. Academic/Plenum Publishers; New York: 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 29.Santos N.C., Prieto M., Castanho M.A.R.B. Quantifying molecular partition into model systems of biomembranes: an emphasis on optical spectroscopic methods. Biochim. Biophys. Acta. 2003;1612:123–135. doi: 10.1016/s0005-2736(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 30.Castanho M.A., Prieto M.J. Fluorescence quenching data interpretation in biological systems. The use of microscopic models for data analysis and interpretation of complex systems. Biochim. Biophys. Acta. 1998;1373:1–16. doi: 10.1016/s0005-2736(98)00081-9. [DOI] [PubMed] [Google Scholar]
- 31.Castanho M., Prieto M. Filipin fluorescence quenching by spin-labeled probes: studies in aqueous solution and in a membrane model system. Biophys. J. 1995;69:155–168. doi: 10.1016/S0006-3495(95)79886-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehrer S.S. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 1971;10:3254–3263. doi: 10.1021/bi00793a015. [DOI] [PubMed] [Google Scholar]
- 33.Johnson W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 34.Lobley A., Whitmore L., Wallace B.A. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 35.Pace C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 36.Santoro M.M., Bolen D.W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 37.Provencher S.W. A constrained regularization method for inverting data represented by linear algebraic or integral-equations. Comput. Phys. Commun. 1982;27:213–227. [Google Scholar]
- 38.Provencher S.W. Contin—a general-purpose constrained regularization program for inverting noisy linear algebraic and integral-equations. Comput. Phys. Commun. 1982;27:229–242. [Google Scholar]
- 39.Beamer L.J., Carroll S.F., Eisenberg D. The three-dimensional structure of human bactericidal/permeability-increasing protein: implications for understanding protein-lipopolysaccharide interactions. Biochem. Pharmacol. 1999;57:225–229. doi: 10.1016/s0006-2952(98)00279-2. [DOI] [PubMed] [Google Scholar]
- 40.Myers J.K., Pace C.N., Scholtz J.M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder S., Kim D., McIntosh T.J. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 42.Tobias P.S., Soldau K., Iovine N.M., Elsbach P., Weiss J. Lipopolysaccharide (LPS)-binding proteins BPI and LBP form different types of complexes with LPS. J. Biol. Chem. 1997;272:18682–18685. doi: 10.1074/jbc.272.30.18682. [DOI] [PubMed] [Google Scholar]
- 43.Leeuwenberg J.F., Dentener M.A., Buurman W.A. Lipopolysaccharide LPS-mediated soluble TNF receptor release and TNF receptor expression by monocytes. Role of CD14, LPS binding protein, and bactericidal/permeability-increasing protein. J. Immunol. 1994;152:5070–5076. [PubMed] [Google Scholar]
- 44.Beamer L.J., Carroll S.F., Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 45.Almeida P.F., Wiegel F.W. A simple theory of peptide interactions on a membrane surface: excluded volume and entropic order. J. Theor. Biol. 2006;238:269–278. doi: 10.1016/j.jtbi.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 46.Huang H.W. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Huang H.W., Chen F.Y., Lee M.T. Molecular mechanism of peptide-induced pores in membranes. Phys. Rev. Lett. 2004;92:198304. doi: 10.1103/PhysRevLett.92.198304. [DOI] [PubMed] [Google Scholar]
- 48.Melo M.N., Castanho M.A. Omiganan interaction with bacterial membranes and cell wall models. Assigning a biological role to saturation. Biochim. Biophys. Acta. 2007;1768:1277–1290. doi: 10.1016/j.bbamem.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Pan Y.L., Cheng J.T., Hale J., Pan J., Hancock R.E. Characterization of the structure and membrane interaction of the antimicrobial peptides aurein 2.2 and 2.3 from Australian southern bell frogs. Biophys. J. 2007;92:2854–2864. doi: 10.1529/biophysj.106.097238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pott T., Paternostre M., Dufourc E.J. A comparative study of the action of melittin on sphingomyelin and phosphatidylcholine bilayers. Eur. Biophys. J. 1998;27:237–245. doi: 10.1007/s002490050130. [DOI] [PubMed] [Google Scholar]
- 51.Weiss T.M., Yang L., Ding L., Waring A.J., Lehrer R.I. Two states of cyclic antimicrobial peptide RTD-1 in lipid bilayers. Biochemistry. 2002;41:10070–10076. doi: 10.1021/bi025853d. [DOI] [PubMed] [Google Scholar]
- 52.Santos N.C., Prieto M., Castanho M.A. Interaction of the major epitope region of HIV protein gp41 with membrane model systems. A fluorescence spectroscopy study. Biochemistry. 1998;37:8674–8682. doi: 10.1021/bi9803933. [DOI] [PubMed] [Google Scholar]