Abstract
Objective
Growth factors, including brain-derived neurotrophic factor (BDNF), are polypeptides that are involved in the maintenance, survival, and death of central and peripheral cells. Numerous growth factors have been identified in saliva and are thought to promote wound healing and maintenance of the oral epithelium. The aim of this study was to determine if BDNF is also found in human saliva.
Methods
Whole, unstimulated saliva samples (n=30) were analyzed by SDS-PAGE and Western blot using an anti-human BDNF antibody. Proteolytic cleavage products were similarly assessed following the incubation of pooled saliva with N-glycanase F and plasmin. Subjects genotyped for the BDNF Val66Met single nucleotide polymorphism (SNP).
Results
These experiments revealed the presence of immunoreactive bands at 14, 32 and 34 kD, corresponding to mature (mBDNF) and proBDNF, as well as a truncated pro-form at 24 kD. Not every sample contained all forms of BDNF. Treatment with N-glycanase and plasmin reduced the size of the higher molecular weight bands, confirming the glycosylated pro-form of BDNF. mBDNF was detected significantly less often in subjects with the Val66Met SNP, compared to those without the polymorphism (X2 = 4.05; P<0.05).
Conclusions
While the function of salivary BDNF still requires elucidation, these findings suggest that it may be possible to use saliva in lieu of blood in future studies of BDNF and the Val66Met polymorphism.
Keywords: Brain-derived neurotrophic factor, proBDNF, saliva, Val66Met, human
Introduction
Growth factors are diffusible polypeptides essential for the development and functioning of the central and peripheral nervous systems. These proteins are categorized into families based on their homologies and shared signal transduction mechanisms.1 One of the most widely studied families is the neurotrophins, which includes brain-derived neurotrophic factor (BDNF), nerve growth factor (NGF), neurotrophin-3 (NT-3), and neurotrophin-4/5 (NT-4/5). BDNF is the most abundant neurotrophin in the central nervous system, yet has also been located in heart, spleen and skin tissue2,3, blood platelets4 and circulating plasma5, indicating that the protein affects non-neuronal populations.
BDNF is synthesized as a 32 kD N-glycosylated and glycosulfated pro-form which is proteolytically cleaved to the 14 kD mature form (mBDNF).6 ProBDNF can be cleaved intracellularly by furin and/or pro-protein convertases to yield mature dimers7, yet the majority is secreted uncleaved into the extracellular environment.8 Subsequently, a portion of the pro-protein is cleaved to the mature form by the serine protease plasmin and matrix metalloproteinases.9 ProBDNF is important for proper folding, dimerization, and targeting of mature BDNF, yet this form can also elicit distinct effects of its own which may oppose those of mBDNF.10 Accordingly, proBDNF induces cellular apoptosis9, while the mature protein promotes cell development and differentiation, survival and plasticity.11
The diverse biological functions of pro- and mBDNF are mediated through a dual-receptor system, consisting of the tyrosine kinase receptor B (trkB) and pan-neurotrophin receptor p75. mBDNF promotes cell growth and survival though activation of both the trkB and p75 receptors.12 ProBDNF promotes death in cells coexpressing p75 and another transmembrane protein, sortilin, but does not activate trkB.13 Many cells express both types of receptors, allowing interaction and fine-tuning of responses. p75 can increase affinity and specificity for trkB interactions14, yet will induce apoptosis when trkB receptors are absent.9
Recently, there has been much interest in a common single nucleotide polymorphism (SNP) which causes a valine to methionine substitution at codon 66 in the pro-region of the human BDNF protein. The amino acid substitution is believed to disrupt folding and dimerization of BDNF. Though this has no effect on the biological activity of the mature protein, it does result in defective intracellular protein trafficking and diminished activity-dependent BDNF secretion.15
Numerous growth factors have been identified in saliva, including NGF16, epidermal growth factor (EGF)17, transforming growth factor (TGF)18, and insulin-like growth factor (IGF).19 These proteins play an essential role in the protection and repair of oral and gastric soft tissue, as well as the maintenance of gustatory tissue. Numerous animal studies have reported decreased wound healing20, gastric lesions21, epithelial keratosis, and distinct changes in taste cell structure and number22,23 following sialoadenectomy (removal of the salivary glands). Furthermore, conditions of decreased saliva production in humans, such as Sjogren's syndrome, lead to increased incidence of oral infection24 and frequency of taste complaints.25
Given the extensive expression and functioning of BDNF throughout the human body, as well as the significant number of growth factors identified in saliva, the aim of the present study was to determine if BDNF is also found in human saliva. Furthermore, we sought to determine if the Val66Met polymorphism has any impact on salivary BDNF levels.
Materials and Methods
Participants
Based on early trends from a small sample, a power analysis was performed using R to test for optimal sample size. Based on the results of this test, thirty healthy volunteers (17 female and 13 male) from Cornell University in Ithaca, NY, participated in this study. Participants had a mean (± SD) age of 27 ± 6 years. All participants were nonmedicating and did not smoke. This study was approved by the Institutional Review Board for Human Participants at Cornell University. All participants gave informed consent for participation.
Sample Collection and Treatment
Saliva and blood samples were collected from each participant. Participants had not consumed any food or drink, nor brushed their teeth, for two hours before sample collection. All samples were collected between 12 and one p.m. to minimize any possible effect of diurnal variation. Approximately 3 ml of unstimulated whole resting saliva were collected from each participant by passive expectoration into a 15-ml conical tube on ice. The tubes were centrifuged at 4000 rpm for 15 minutes at 4°C to remove cellular debris. An aliquot from each sample was removed for treatment with N-glycanase F or plasmin. Protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) was added to the remaining samples, which were then aliquotted. For genotyping, 5 ml of venous blood were collected from each participant into a tube coated with EDTA to prevent coagulation. The tubes were centrifuged at 3000 rpm for 10 minutes and the buffy coat collected. All samples were stored at -80°C until use. Upon thawing, the samples were centrifuged once more to ensure complete debris removal.
SDS-PAGE and Immunoblotting
Total protein in each sample was measured using a modified version of Lowry's procedure.26 Forty ug of total protein from each sample were denatured under reducing conditions and resolved by SDS-PAGE on a 15% gel; 2.5 ng of recombinant BDNF (rBDNF, Peprotech, Rocky Hill, NJ) were run as a positive control. The movement of the proteins was monitored using Precision Plus All Blue Standards (Bio-Rad Laboratories, Hercules, CA). Proteins were transferred onto a PVDF membrane (Millipore Corp, Bedford, MA) in transfer buffer (25 mM Tris, 190 mM glycine, 20% MeOH) for 1 hour at 100 V and 4°C, then blocked for 1 hour at room temperature in blocking buffer (PBS, 0.1% tween, 1% NP-40) with 5% nonfat dry milk (NFDM; w/v). Membranes were incubated overnight at 4°C with purified polyclonal chicken anti-human BDNF (Promega, Madison, WI), diluted 1:500 in blocking buffer with 1% NFDM. After washing in PBST, membranes were incubated in rabbit anti-chicken IgY-HRP conjugate (Promega), diluted 1:1250 in blocking buffer with 5% NFDM, for 1 hour at RT. Membranes were washed again and an ECL Plus Chemiluminescence system (Amersham Biotech, Piscataway, NJ) was used for detection. The specificity of the primary antibody was tested both by peptide neutralization and by determining cross-reactivity to 2.5 ng of recombinant NGF, NT-3 and NT-4 (Peprotech). Negative controls using only the secondary antibody were also run.
Deglycosylation
The effect of N-glycanase F (PNGase F) on salivary BDNF was determined by following the manufacturer instructions (Prozyme, San Leandro, CA). Briefly, 100 ug of total protein from pooled saliva (N=8) were diluted in buffer (100 mM sodium phosphate, 0.1% sodium azide, pH 7.5). The sample was denatured using 2% SDS + 1M β-mercaptoethanol and heated at 100°C for 5 minutes. Free SDS was removed by adding 15% NP-40 in a detergent solution. The sample was incubated for 18 hours at 37°C with 12.5 mU of N-glycanase, tris buffer (50 mM Tris-HCl, pH 8). A negative control containing PBS in lieu of PNGase was incubated along with the experimental sample. Fifty ug of salivary protein from the experimental and control samples were analyzed by SDS-PAGE and Western blot, as described above.
Plasmin Treatment
Twenty ug of total salivary protein from pooled saliva were incubated with 30 mU of plasmin (Sigma-Aldrich) for 1 hr at 37°C, along with a control sample containing PBS. Thirty ul of each sample were denatured, resolved by SDS-PAGE on a 15% gel, and analyzed by Western blot.
Genotyping for the Val66Met Polymorphism
Participants were genotyped for the Val66Met SNP (rs6265) according to a previously published method27. Briefly, genomic DNA was extracted from buffy coats using a DNeasy tissue kit (Qiagen Inc., Valencia, CA). The target sequence was amplified using a HotStar Hi Fidelity Polymerase kit (Qiagen). The 50 ul reaction solution contained 150 ng DNA, 2.5 U DNA polymerase, 1 uM of each primer (Forward: 5′-GAGGCTTGACATCATTGGCT-3′; Reverse: 5′-CGTGTACAAGTCTGCGTCCT-3′) and reaction buffer, containing 200 uM dNTPs and 7.5 mM MgSO4. The DNA templates were denatured for 5 min at 95°C and then 30 PCR cycles were performed, each consisting of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. After the last cycle, samples were incubated at 4°C. Samples were then digested overnight with 2 U of Eco721 (Fermentas Life Sciences, Glen Burnie, MD). The fragments were separated on a 2.5% agarose gel at 100 V and visualized with ethidium bromide. A low molecular weight DNA ladder (New England Biolabs Inc., Ipswich, MA) was used to monitor movement down the gel. Allele A (methionine substitution) was the uncut product at 113 bp, and allele G (valine) comprised the cut bands of 78 and 35 bp.
Results
Salivary BDNF Identification
Western blot analysis confirmed the presence of BDNF in the saliva samples examined. As seen in Figure 1, four immunoreactive bands were detected with the anti-human BDNF antibody. mBDNF was detected at the predicted molecular mass of 14 kD and migrated at the same position as the recombinant positive control. The majority of proBDNF appeared as doublet bands at 32 and 34 kD; minor bands at 24 kD were also observed. Samples were highly variable in the expression and relative concentrations of each form of the protein.
Figure 1.
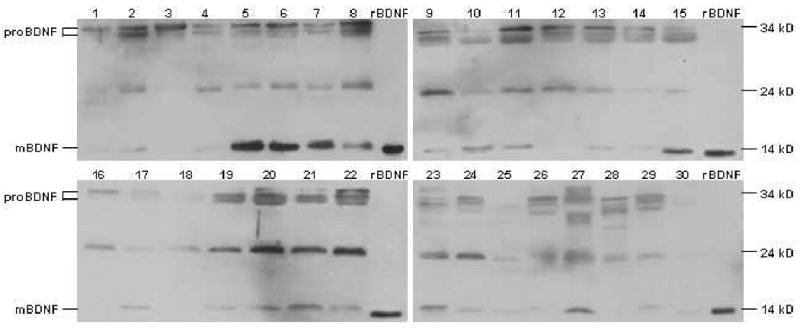
Western blot analysis of whole saliva using an anti-human BDNF antibody revealed immunoreactivity for pro- and mBDNF. Forty ug of total salivary proteins from each participant were separated by SDS-PAGE and analyzed through immunoblotting. This procedure revealed bands at 14 and 24 kD, as well as doublet bands at 32 and 34 kD; 2.5 ng of rBDNF were run as a positive control. The relative amount of each form of the protein varied considerably between individuals.
The specificity of the primary antibody for pro- and mBDNF was confirmed by control experiments. When the antibody was incubated with a 10-fold excess (by concentration) of recombinant BDNF, there were no immunoreactive bands visible for the pro- or mature forms of the protein (results not shown). The antibody also did not display cross-reactivity to 2.5 ng of NT-3, NT-4, or NGF (results not shown). There were no visible proteins when the primary antibody was omitted and the membrane was incubated with the secondary antibody only (results not shown).
PNGase Treatment of Salivary BDNF
Treatment with PNGase F reduced the intensity of the two higher molecular species to approximately 30 and 28, verifying the presence of N-glycosylated proBDNF (See Figure 2). Furthermore, the 24 kD and 18 kD immunoreactive bands observed in the control lane were reduced to approximately 17 and 15 kD, respectively. This finding suggests that the lower molecular weight species are truncated forms of proBDNF that have retained a used glycosylation site. The mature form of the protein was not affected by the enzyme. Complete deglycosylation most likely did not occur due to the complexity of the sample matrix.
Figure 2.
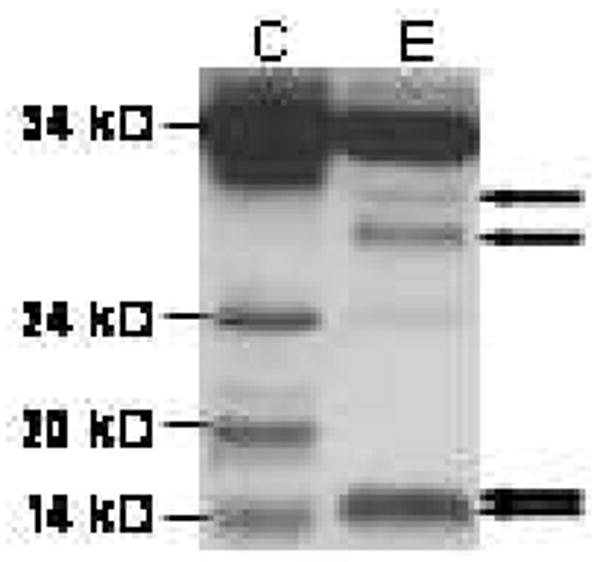
Treatment with PNGase demonstrated the presence of glycosylated proBDNF in saliva. Whole saliva was incubated for 18 hr at 37° C in the presence of PBS (“C”) or PNGase (“E”). Cleavage products were separated by SDS-PAGE and probed in Western blot analysis with anti-human BDNF. Enzyme treatment resulted in deglycosylation of the 32 and 34 kD pro-forms of BDNF, which were reduced to approximately 28 and 30 kD, respectively. The 19 and 24 kD bands were reduced to 15 and 17 kD, respectively, suggesting that these secreted products were also N-glycosylated. mBDNF was not affected by treatment with the enzyme. Positions of deglycosylated species are indicated by arrows.
Plasmin Treatment of Salivary BDNF
A number of previous experiments have demonstrated the ability of plasmin to cleave proBDNF into the mature form.9 Therefore, in order to determine whether plasmin has such an effect on salivary proBDNF, whole pooled saliva was incubated with the protease. As seen in Figure 3, the 32 and 34 kD, as well as lower molecular weight, protein forms were cleaved to mBDNF following incubation with plasmin. The size of the mBDNF present in the sample was not affected.
Figure 3.
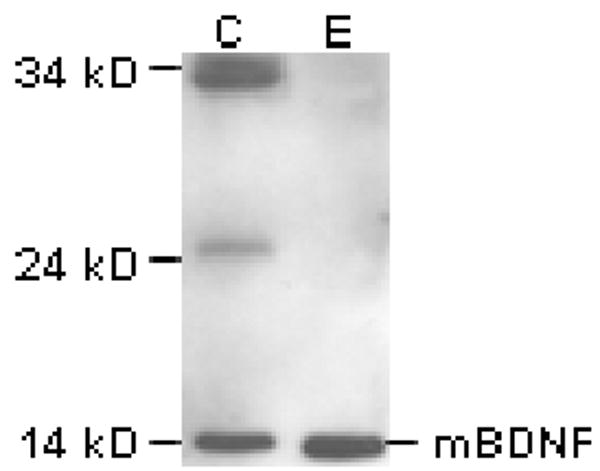
Treatment of a pooled saliva sample with plasmin demonstrated total cleavage of salivary proBDNF and lower molecular weight glycosylated forms to mBDNF. Whole saliva was incubated with either PBS (“C”) or plasmin (“E”) for 1 hr at 37°C. The products were separated by SDS-PAGE and analyzed by Western blot. Treatment with the protease resulted in the cleavage of proBDNF, as well as the lower molecular weight proteins, to mBDNF. The relative molecular weight of mBDNF was not affected by plasmin treatment.
Genotyping for the Val66Met Polymorphism
Fourteen participants (12 females and 2 males) were heterozygous for the Val66Met polymorphism. Salivary mBDNF was not detectable in five of these participants (36%), as compared to only one participant without the polymorphism (6%). See Table 1 for these results. A X2 test of independence indicated that these results were significantly different (X2= 4.05; P<0.05)
Table 1.
Val66Met SNP Analysis
| NO SNP | SNP | ||||
|---|---|---|---|---|---|
| Blot # | Sex | mBDNF present | Blot # | Sex | mBDNF present |
| 2 | M | ± | 1 | F | - |
| 5 | F | + | 3 | F | - |
| 6 | M | + | 4 | M | ± |
| 7 | M | + | 8 | F | + |
| 9 | M | ± | 12 | F | - |
| 10 | M | + | 13 | F | ± |
| 11 | F | + | 14 | M | ± |
| 15 | M | + | 16 | F | - |
| 17 | M | ± | 19 | F | ± |
| 18* | F | - | 20 | F | + |
| 22 | M | + | 21 | F | + |
| 23 | M | + | 25 | F | ± |
| 24 | F | ± | 28 | F | - |
| 26 | M | ± | 30 | F | ± |
| 27 | F | + | |||
| 29 | M | ± | |||
Subjects without mBDNF are in bold
Discussion
We demonstrate through immunoblotting and enzyme digestions that the mature and pro-forms of BDNF are present in human whole saliva. There were considerable individual differences in the expression and relative concentrations of each form of the protein; not all participants expressed every form. A significant number of participants with the Val66Met polymorphism did not express detectable levels of mature salivary BDNF.
To our knowledge, this is the first report of BDNF in salivary secretions, whether in humans or other species. BDNF mRNA expression has been observed in murine submandibular glands3,28, yet these researchers did not investigate the presence of BDNF in the animal's saliva. A recent salivary proteome analysis in humans did not identify BDNF29 and our finding highlights the importance of combining broad protein analyses with more focused approaches. In the current study, immunoreactive specificity of the anti-BDNF antibody was confirmed by peptide neutralization and by lack of cross-reactivity with the other neurotrophins, verifying that the antibody is truly binding BDNF.
The presence of multiple higher molecular weight forms of BDNF (24, 32 and 34 kD) has previously been reported in experiments using cultured neuronal and non-neuronal cells.6,13 It has been suggested that the various bands represent differentially glycosylated and glycosulfated forms of proBDNF and mBDNF. The reduction in apparent molecular weight that we observed following sample treatment with N-Glycanase and plasmin is consistent with previous reports of such proteolytic cleavage of proBDNF.6,9,30
It is particularly noteworthy that salivary proBDNF is cleaved to mBDNF by plasmin. It has been reported that plasmin is perhaps the most essential protease involved in the cleavage of proBDNF to mBDNF.30 Plasmin is usually found as the inactive zymogen, plasminogen, which is cleaved to the active form by plasminogen activators, including tissue plasminogen activator (tPA). The amount of tPA produced is tightly regulated, primarily controlled by plasminogen activator inhibitors (PAI).31 While plasmin activity, per se, is not found in saliva, plasminogen, tPA, and PAI are widely distributed in the oral cavity and salivary glands.31,32 Since proBDNF has been shown to elicit biological effects opposite to and distinct from mBDNF, regulation of protease expression and therefore, cleavage of proBDNF, is a means of controlling the direction of BDNF action.10 Whether the different forms of BDNF are released by the salivary glands already cleaved, or if the pro-form is released and then undergoes further cleavage in the oral cavity will need to be examined.
The results of this study demonstrate that there are significant individual differences in the presence and relative concentrations of each form of salivary BDNF. Since all of the samples had been treated with a protease inhibitor prior to handling, it is likely that this finding represents a real difference rather than the result of proteolytic degradation. Interestingly, more than a third of subjects with the Val66Met polymorphism did not have a detectable level of mature BDNF in their saliva. This finding raises the possibility that people with the SNP may have altered tPA-plasminogen pathway function, leading to decreased cleavage of proBDNF. Though significantly more women in our sample were heterozygous for the SNP and thus, less likely to have mBDNF, previous reports have found no effect of sex on the prevalence of the polymorphism.33 Therefore, our sample size precludes analysis of mBDNF presence according to sex.
The Val66Met gene variant is thought to affect intracellular trafficking and BDNF secretion, and has been associated with decreased neural BDNF. Brain and blood BDNF values are thought to be tightly correlated34, yet comparisons of blood BDNF concentrations between healthy participants with and without the polymorphism have found no significant differences between the two groups.35,36 Previous studies, however, have assayed for total BDNF by enzyme-linked immunosorbent assay (ELISA), a method that cannot address differences in the relative amounts of each form of BDNF. In the Western blots used in the current study, the immunoreactive antibody can simultaneously assay for both pro- and mBDNF, so we are able to observe, if not quantitate, these differences.
Cells in the oral cavity and gastrointestinal tract are subject to rapid turnover rates due to the high incidence of damage. We hypothesize that BDNF interacts with and complements other salivary growth factors in maintaining the balance among proliferation, survival and death of these cells. This theory is supported by findings that the trkB and p75 receptors are widely expressed throughout the oral cavity and gastrointestinal tract. For example, immunoreactivity for trkB is expressed in all three types of mouse taste cells37, while p75 is found in Type II and III taste cells.38 p75 has also been also detected in the intercalated ducts of human sublingual gland tissue and the ductal and acinar cells of the submandibular gland in mice.39 The presence of this receptor in the salivary glands raises the possibility that BDNF secreted into the oral cavity may exert autocrine and/or paracrine functions.
Finally, both p75 and trkB receptors have also been identified in human gastrointestinal endocrine cells.40 Subcutaneous injection in humans with rBDNF promotes gastrointestinal transit in both healthy and constipated participants, perhaps acutely effecting neurotransmission activity of the cells.41 Interestingly, Tsukinoki et al42 recently reported that increased BDNF mRNA expression in the rat submandibular gland due to immobilization stress was accompanied by concurrent increases in plasma levels. The authors suggest such increases in plasma BDNF may provide protection for the gastrointestinal organs against stress-induced ulceration. While it is likely that absorption of salivary BDNF is occurring throughout the gastrointestinal tract, it is also possible that BDNF is absorbed sublingually and undergoes systemic dissemination, as occurs with EGF.43
Since mBDNF and proBDNF appear to elicit opposing responses, it is possible that individual differences in the relative amounts of each form of BDNF may indicate different rates of cell turnover and renewal in different people. It has been suggested that increases in the duration of taste cell turnover in the elderly may be associated with reports of decreased taste sensitivity.44 Though total blood BDNF does not appear to decrease with age45, altered cell turnover in the oral cavity may result from changes in the pro- to mBDNF ratio.
In this study, the collection of whole saliva through passive expectoration did not discriminate between salivary gland excretions and the oral mucosal transudate, an ultra-filtrate of blood. It is therefore possible that the observed salivary BDNF resulted from the movement of serum macromolecules into the oral cavity. This possibility would also increase the potential for sample degradation by crevicular enzymes. The majority of studies investigating oral growth factors indicate that these proteins are synthesized in the salivary glands themselves and released through granule exocytosis.19,46 It is likely that salivary BDNF is secreted in a similar manner. Examination of glandular saliva secretions will indicate the source of salivary BDNF, as well as the extent of enzymatic protein degradation occurring in the oral cavity.
This study demonstrates that pro- and mature BDNF are present in human saliva. These findings may be of interest to advocates of using salivary parameters for diagnostics purposes. Alterations in levels of blood mature and proBDNF have been widely explored in clinical contexts and found in some studies to be associated with depression47, Alzheimer's48 and schizophrenia.49 The current results suggest that it may be possible to utilize saliva in future studies of both pro- and mBDNF in lieu of painful blood draws. Indeed, researchers have already started using saliva-derived DNA specimens when genotyping for the Val66Met polymorphism.50
Acknowledgments
Much thanks to Dr. Collynn Woeller and Cheryl Perry for their invaluable methodological advice.
This study was funded by NIH Training Grant 5 T32 DK--007158 31.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nestler EJ, Hyman SE, Malenka RC. Neurotrophic factors Molecular basis of neuropharmacology: A foundation for clinical neuroscience. New York: McGraw Hill; 2001. pp. 235–250. [Google Scholar]
- 2.Rosenthal A, Goeddel DV, Nguyen T, Martin E, Burton LE, Shih A, et al. Primary structure and biological activity of human brain-derived neurotrophic factor. Endocrinology. 1990;129:1289–94. doi: 10.1210/endo-129-3-1289. [DOI] [PubMed] [Google Scholar]
- 3.Tirassa P, Manni L, Stenfors C, Lundeberg T, Aloe L. RT-PCR ELISA method for analysis of neurotrophin mRNA expression in brain and peripheral tissues. J Biotech. 2000;84:259–272. doi: 10.1016/s0168-1656(00)00370-9. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto H, Gurney ME. Human platelets contain brain-derived neurotrophic factor. J Neurosci. 1990;10:3469–3478. doi: 10.1523/JNEUROSCI.10-11-03469.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeld RD, Zeni L, Haniu M, Talvenheimo J, Radka SF, Bennett L, et al. Purification and identification of brain-derived neurotrophic factor from human serum. Protein Expr Purif. 1995;6:465–71. doi: 10.1006/prep.1995.1062. [DOI] [PubMed] [Google Scholar]
- 6.Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, et al. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem. 2001;276:12660–6. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- 7.Seidah NG, Benjannet S, Pareek S, Chrétien M, Murphy RA. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996;379:247–50. doi: 10.1016/0014-5793(95)01520-5. [DOI] [PubMed] [Google Scholar]
- 8.Mowla SJ, Pareek S, Farhadi HF, Petrecca K, Fawcett JP, Seidah NG, et al. Differential sorting of nerve growth factor and brain-derived neurotrophic factor in hippocampal neurons. J Neurosci. 1999;19:2069–80. doi: 10.1523/JNEUROSCI.19-06-02069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 10.Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci. 2005;6:603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- 11.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF Val66Met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–69. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 12.Snider WD. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell. 1994;77:627–38. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 13.Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75 NTR and sortilin. J Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibel M, Hoppe E, Barde Y. Biochemical and functional interactions between the neurotrophin receptors trk and p75NTR. EMBO J. 1990;18:616–622. doi: 10.1093/emboj/18.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Patel PD, Sant G, Meng C, Teng KK, Hempstead BL, et al. Variant brain-derived neurotrophic fact (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF I neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipps BV. Isolation of nerve growth factor (NGF) from human body fluids; saliva, serum and urine: comparison between cobra venom and cobra serum NGF. J Nat Toxins. 2000;4:349–56. [PubMed] [Google Scholar]
- 17.Dagogo-Jack S, Atkinson S, Kendall-Taylor P. Homologous radioimmunoassay for epidermal growth factor in human saliva. J Immunoassay. 1985;6:125–36. doi: 10.1080/01971528508063025. [DOI] [PubMed] [Google Scholar]
- 18.Yeh J, Yeh YC. Transforming growth factor-alpha and human cancer. Biomed Pharmacother. 1989;43:651–9. doi: 10.1016/0753-3322(89)90083-8. [DOI] [PubMed] [Google Scholar]
- 19.Ryan J, Mantle T, McQuaid S, Costigan DC. Salivary insulin-like growth factor-I originates from local synthesis. J Endocrinol. 1992;135:85–90. doi: 10.1677/joe.0.1350085. [DOI] [PubMed] [Google Scholar]
- 20.Bodner L, Dayan D, Rothchild D, Hammel I. Extraction wound healing in desalivated rats. J Oral Path Med. 1991;20:176–8. doi: 10.1111/j.1600-0714.1991.tb00916.x. [DOI] [PubMed] [Google Scholar]
- 21.Olsen PS, Poulsen SS, Kirkegaard P, Nexø E. Role of submandibular saliva and epidermal growth factor in gastric cytoprotection. Gastroenterology. 1984;87:103–8. [PubMed] [Google Scholar]
- 22.Nanda R, Catalanotto FA. Long-term effects of surgical desalivation upon taste acuity, fluid intake, and taste buds in the rat. Dent Res. 1981;60:69–76. doi: 10.1177/00220345810600011401. [DOI] [PubMed] [Google Scholar]
- 23.Morris-Wiman J, Sego R, Brinkley L, Dolce C. The effects of sialoadenectomy and exogenous EGF on taste buds morphology and maintenance. Chem Senses. 2000;25:9–19. doi: 10.1093/chemse/25.1.9. [DOI] [PubMed] [Google Scholar]
- 24.Porter SR, Scully C, Hegarty AM. An update of the etiology and management of xerostomia. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;97:28–46. doi: 10.1016/j.tripleo.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Weiffenbach JM, Schwartz LK, Atkinson JC, Fox PC. Taste performance in Sjogren's syndrome. Physiol Behav. 1995;57:89–96. doi: 10.1016/0031-9384(94)00211-m. [DOI] [PubMed] [Google Scholar]
- 26.Bensadoun A, Weinstein D. Assay of proteins in presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 27.Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71:651–5. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukinoki K, Saruta J, Sasaguri K, Miyoshi Y, Jinbu Y, Kusama M, et al. Immobilization stress induces BDNF in rat submandibular glands. J Dent Res. 2006;85:844–8. doi: 10.1177/154405910608500913. [DOI] [PubMed] [Google Scholar]
- 29.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, et al. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- 31.Virtanen OJ, Sirén V, Multanen J, Färkkilä M, Leivo I, Vaheri A, et al. Plasminogen activators and their inhibitors in human saliva and salivary gland tissue. Eur J Oral Sci. 2006;114:22–6. doi: 10.1111/j.1600-0722.2006.00264.x. [DOI] [PubMed] [Google Scholar]
- 32.Moody GH. Plasminogen in human saliva. Int J Oral Surg. 1982;11:110–4. doi: 10.1016/s0300-9785(82)80019-7. [DOI] [PubMed] [Google Scholar]
- 33.Pivac N, Kim B, Nedić G, Joo YH, Kozarić-Kovacić D, Hong JP, et al. Ethnic differences in brain-derived neurotrophic factor Val66Met polymorphism in Croatian and Korean healthy participants. Croat Med J. 2009;50:43–8. doi: 10.3325/cmj.2009.50.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karege F, Perret G, Bondolfi G, Schwald M, Bertschy G, Aubry JM. Decreased serum brain-derived neurotrophic factor levels in major depressed patients. Psychiatry Res. 2002;109:143–8. doi: 10.1016/s0165-1781(02)00005-7. [DOI] [PubMed] [Google Scholar]
- 35.Trajkovska V, Marcussen AB, Vinberg M, Hartvig P, Aznar S, Knudsen GM. Measurements of brain-derived neurotrophic factor: methodological aspects and demographical data. Brain Res Bull. 2007;73:143–9. doi: 10.1016/j.brainresbull.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Tramontina J, Frey BN, Andreazza AC, Zandona M, Santin A, Kapczinski F. Val66met polymorphism and serum brain-derived neurotrophic factor levels in bipolar disorder. Mol Psychiatry. 2007;12:230–1. doi: 10.1038/sj.mp.4001941. [DOI] [PubMed] [Google Scholar]
- 37.Takeda M, Suzuki Y, Obara N, Tsunekawa H. Immunohistochemical detection of neurotrophin-3 and –4, and their receptors in mouse taste bud cells. Arch Cytol Hist. 2005;68:393–403. doi: 10.1679/aohc.68.393. [DOI] [PubMed] [Google Scholar]
- 38.Yee C, Bartel DL, Finger TE. Effects of glossopharyngeal nerve section on the expression of neurotrophins and their receptors in lingual taste buds of adult mice. J Comp Neurol. 2005;490:371–90. doi: 10.1002/cne.20670. [DOI] [PubMed] [Google Scholar]
- 39.De Vicente JC, Garcia-Suárez O, Esteban I, Santamaria J, Vega JA. Immunohistochemical localization of neurotrophins and neurotrophin receptors in human and mouse salivary glands. Ann Anat. 1998;180:157–163. doi: 10.1016/S0940-9602(98)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Esteban I, Hannestad J, Levanti B, Del Valle ME, Naves FJ, Vega JA. Neurotrophin receptor proteins immunoreactivity in human gastrointestinal endocrine cells. Brain Res Bull. 1995;38:539–43. doi: 10.1016/0361-9230(95)02025-9. [DOI] [PubMed] [Google Scholar]
- 41.Coulie B, Szarka LA, Camilleri M, Burton DD, McKinzie S, Stambler N, et al. Recombinant human neurotrophic factors accelerate colonic transit and relieve constipation in humans. Gastroenterology. 2000;119:41–50. doi: 10.1053/gast.2000.8553. [DOI] [PubMed] [Google Scholar]
- 42.Tsukinoki K, Saruta J, Muto N, Sasaguri K, Sato S, Tan-Ishii N, et al. Submandibular glands contribute to increases in plasma BDNF levels. J Dent Res. 2007;86:260–4. doi: 10.1177/154405910708600312. [DOI] [PubMed] [Google Scholar]
- 43.Purushotham KR, Offenmüller K, Bui AT, Zelles T, Blazsek J, Schultz GS, et al. Absorption of epidermal growth factor occurs through the gastrointestinal tract and oral cavity in adult rats. Am J Physiol. 1995;269:G867–73. doi: 10.1152/ajpgi.1995.269.6.G867. [DOI] [PubMed] [Google Scholar]
- 44.Fukunaga A, Uematsu H, Sugimoto K. Influences of aging on taste perception and oral somatic sensation. J Gerontol. 2005;60A:109–113. doi: 10.1093/gerona/60.1.109. [DOI] [PubMed] [Google Scholar]
- 45.Lommatzsch M, Zingler D, Schuhbaeck K, Schloetcke K, Zingler C, Schuff-Werner P, et al. The impact of age, weight and gender on BDNF levels in human platelets and plasma. Neurobio Aging. 2005;26:115–123. doi: 10.1016/j.neurobiolaging.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Cossu M, Perra MT, Piludu M, Lantini MS. Subcellular localization of epidermal growth factor in human submandibular gland. Histochem J. 2000;32:291–294. doi: 10.1023/a:1004036929006. [DOI] [PubMed] [Google Scholar]
- 47.Lang UE, Hellweg R, Gallinat J. BDNF serum concentrations in healthy volunteers are associated with depression-related personality traits. Neuropsychopharmacology. 2004;29:795–8. doi: 10.1038/sj.npp.1300382. [DOI] [PubMed] [Google Scholar]
- 48.Michalski B, Fahnestock M. Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer's disease. Brain Res Mol Brain Res. 2003;111:148–54. doi: 10.1016/s0169-328x(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 49.Toyooka K, Asama K, Watanabe Y, Muratake T, Takahashi M, Someya T. Decreased levels of brain-derived neurotrophic factor in serum of chronic schizophrenic patients. Psychiatry Res. 2002;110:249–57. doi: 10.1016/s0165-1781(02)00127-0. [DOI] [PubMed] [Google Scholar]
- 50.Kaufman J, Yang BZ, Douglas-Palumberi H, Grasso D, Lipschitz D, Houshyar S, et al. Brain-derived neurotrophic factor-5-HTTLPR gene interactions and environmental modifiers of depression in children. Biol Psychiatry. 2006;59:673–80. doi: 10.1016/j.biopsych.2005.10.026. [DOI] [PubMed] [Google Scholar]


