Abstract
The mammalian intestine harbors complex societies of beneficial bacteria that are maintained in the lumen with minimal penetration of mucosal surfaces. Microbial colonization of germ-free mice triggers epithelial expression of RegIIIγ, a secreted C-type lectin. RegIIIγ binds intestinal bacteria but lacks the complement recruitment domains present in other microbe-binding mammalian C-type lectins. We show that RegIIIγ and its human counterpart, HIP/PAP, are directly antimicrobial proteins that bind their bacterial targets via interactions with peptidoglycan carbohydrate. We propose that these proteins represent an evolutionarily primitive form of lectin-mediated innate immunity, and that they reveal intestinal strategies for maintaining symbiotic host-microbial relationships.
The human gut is home to a vast consortium of symbiotic bacteria. Members of this complex microflora metabolize dietary substances, such as plant polysaccharides, that are otherwise indigestible by their human hosts (1). Indigenous gut microbes thus make essential contributions to human nutrient metabolism and, in return, inhabit a protected, nutrient-rich environment. Maintaining the mutually beneficial nature of this relationship requires strict sequestration of resident bacteria in the intestinal lumen, as microbial incursions across epithelia can elicit inflammation and sepsis.
Epithelial antimicrobial proteins are evolutionarily ancient innate immune effectors. As key elements of intestinal mucosal defense, they likely play an important role in maintaining mutually beneficial host-microbial relationships by restricting contact between resident microbes and mucosal surfaces. This idea is underscored by the fact that deficiencies in antimicrobial peptide expression are associated with inflammatory bowel disease (IBD) (2, 3), a chronic inflammatory disorder thought to be triggered by resident gut microbes. However, although cationic antimicrobial peptides such as defensins are well-characterized, the full repertoire of gut antimicrobial mechanisms remains undefined. Here we show that resident gut bacteria drive intestinal epithelial expression of a C-type lectin that binds peptidoglycan and has direct antimicrobial activity, revealing a primitive mechanism of lectin-mediated innate immunity.
Paneth cells are key effectors of small intestinal antimicrobial defense. These specialized epithelial cells are located at the crypt base and harbor abundant cytoplasmic secretory granules containing antimicrobial proteins, including α-defensins. To gain new insights into how intestinal surfaces cope with microbial challenges, we used DNA microarrays to identify Paneth cell antimicrobial factors whose expression is altered by bacteria. Paneth cells were harvested by laser capture microdissection from “germ-free” (microbiologically sterile) mice and “conventionalized” mice (germ-free mice reconstituted for 10 days with an intestinal microflora from conventionally raised mice). Paneth cell mRNAs from both groups were amplified to generate complementary RNAs (cRNAs) in sufficient quantity to hybridize to Affymetrix mouse genome 430 2.0 GeneChip arrays. The results of our screen revealed 149 transcripts whose expression was changed 2- to 45-fold by microbial colonization (table S1).
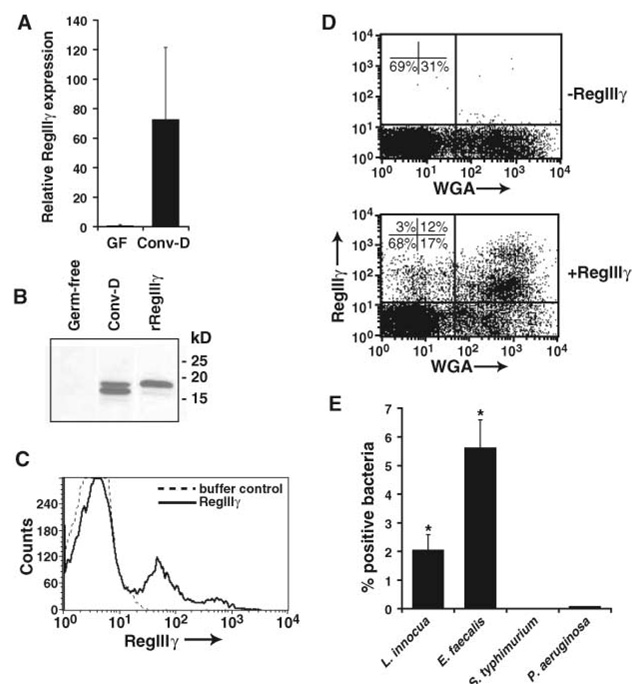
One of the most prominent responses uncovered by our analysis was a 31-fold increase in the abundance of RegIIIγ transcripts in Paneth cells from conventionalized as compared with germ-free mice (table S1). Increased expression of RegIIIγ mRNA was confirmed by quantitative real-time polymerase chain reaction (Q-PCR) (Fig. 1A) and correlated with increased protein expression (Fig. 1B).
Fig. 1.
RegIIIγ is induced by resident intestinal microbes and binds to Gram-positive bacteria. (A) RegIIIγ mRNA expression in Paneth cells. Paneth cells were harvested by laser-capture microdissection from germ-free and conventionalized small intestines. Q-PCR analysis was performed on RNAs from microdissected Paneth cells from three mice per group. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and mean ± SD is plotted (range for conventionalized samples is 42 to 309). Results are expressed relative to one of the germ-free samples. GF, germ-free; Conv-D, conventionalized. (B) RegIIIγ protein expression in small intestine. RegIIIγ was detected in mid–small intestinal protein by immunoblot with RegIIIγ-specific antiserum (13). The lower band in the protein sample from conventionalized mice likely results from proteolytic cleavage at a trypsin-like site near the N-terminus (13), similar to that described for RegIα (8). Results are representative of two independent experiments. rRegIIIγ, recombinant RegIIIγ. (C) RegIIIγ binds to intestinal bacteria. Flow cytometry reveals binding of AlexaFluor555–conjugated RegIIIγ to intestinal bacteria recovered from conventional mouse small intestine. BSA-AlexaFluor555 showed no binding (not shown). Results are representative of independent experiments with three mice. (D) RegIIIγ binds preferentially to Gram-positive intestinal bacteria. Dual-color flow cytometry analysis with WGA-AlexaFluor488 and RegIIIγ-AlexaFluor555 shows preferential binding to the WGA-positive bacterial population. Results are representative of three independent experiments. (E) RegIIIγ binds preferentially to cultured Gram-positive bacteria. Formaldehyde-fixed preparations of Gram-positive (L. innocua and E. faecalis) and Gram-negative bacteria (S. typhimurium and P. aeruginosa) were incubated with RegIIIγ, followed by detection with RegIIIγ-specific antiserum and Cy3-labeled goat secondary antibody against rabbit IgG, and analyzed by flow cytometry. Results are representative of three experiments. Asterisks indicate statistically significant differences between Gram-positive species and S. typhimurium (P < 0.05). Preimmune serum controls are shown in fig. S3.
The Reg gene family encodes a diverse group of secreted proteins that contain conserved sequence motifs found in C-type lectin carbohydrate recognition domains (CRDs). The Reg family constitutes a distinct group of mammalian C-type lectins, with each member consisting solely of a ~ 16-kD CRD and N-terminal secretion signal. The family is further classified into subgroups (I, II, III, and IV) on the basis of primary sequence. Several RegIII family members are expressed predominantly in small intestine, including mouse RegIIIγ (fig. S1) and human HIP/PAP (4, 5). Inflammatory stimuli, such as bacteria (5, 6) or mucosal damage (7), increase gastrointestinal expression of mouse RegIIIγ. Furthermore, HIP/PAP expression increases in the mucosa of patients with IBD (5, 8), a disorder characterized by increased mucosal adherence of resident bacteria and chronic intestinal inflammation (9). Although mitogenic functions have been suggested for Reg proteins in other tissues (10), the biological functions of intestinal RegIII proteins and their role in IBD have remained poorly defined. We show here that RegIIIγ and human HIP/PAP are peptidoglycan-binding proteins with direct antibacterial activity.
Immunogold electron microscopy revealed that RegIIIγ is present in Paneth cell secretory granules (fig. S2). Granule contents are released apically (11); their release indicates that RegIIIγ is targeted to the gut lumen, which harbors large resident bacterial populations. Other members of the C-type lectin family, such as the mannose-binding lectin (MBL), bind to microbial surface carbohydrates and trigger innate immune responses (12). Therefore, we hypothesized that RegIIIγ might similarly bind intestinal bacteria. Previously, we established a procedure for purification of recombinant mouse RegIIIγ and human HIP/PAP (13). We used fluorochrome-conjugated recombinant RegIIIγ to look for binding to a mixed microbial population harvested from the small intestines of conventionally raised mice. Flow cytometry revealed that RegIIIγ bound to a subpopulation of intestinal bacteria (Fig. 1C).
Given that intestinal microbial communities consist of both Gram-positive and Gram-negative species (14), we asked whether RegIIIγ bound preferentially to one of these groups. Wheat germ agglutinin (WGA) binds to surface peptidoglycan on Gram-positive bacteria, thus distinguishing between Gram-positive and Gram-negative populations (15). Dual staining with fluorochrome-conjugated RegIIIγ and WGA revealed that RegIIIγ preferentially bound to the WGA-positive bacterial population (Fig. 1D). Furthermore, we found that RegIIIγ bound to pure preparations of cultured Gram-positive bacteria, including Listeria innocua and Enterococcus faecalis, with comparatively little binding to preparations of cultured Gram-negative bacteria, including Salmonella typhimurium and Pseudomonas aeruginosa (Fig. 1E).
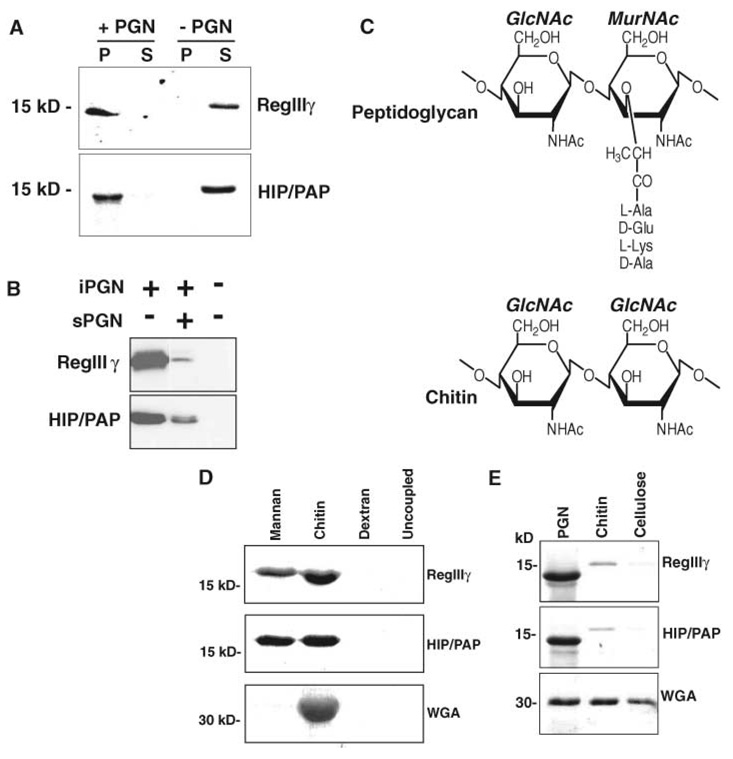
These findings suggested that RegIIIγ binds peptidoglycan, a molecule that is exposed on the Gram-positive bacterial surface, but is buried in the periplasmic space of Gram-negative bacteria. To test this idea, we performed pull-down assays using insoluble cell wall peptidoglycan (16). Purified RegIIIγ was completely removed from solution by incubation with peptidoglycan and was retained in the peptidoglycan-bound fraction after extensive washing (Fig. 2A). Human HIP/PAP is 65% identical to RegIIIγ and exhibited a similar peptidoglycan binding activity (Fig. 2A). The specificity of both interactions was confirmed by using soluble peptidoglycan (sPGN) to compete for binding to insoluble peptidoglycan (iPGN) (Fig. 2B). Furthermore, we calculated a dissociation constant (Kd) of 11 nM for RegIIIγ and 26 nM for HIP/PAP (fig. S4). These results indicate high-affinity binding to peptidoglycan and are in good agreement with the dissociation constants determined for other known peptidoglycan-binding proteins, including CD 14 (17) and members of the peptidoglycan-recognition protein (PGRP) family (18).
Fig. 2.
Mouse RegIIIγ and human HIP/PAP bind peptidoglycan. (A) Peptidoglycan pull-down assays. RegIIIγ or HIP/PAP (10 µg of either) was added to 50 µg insoluble Bacillus subtilis peptidoglycan and pelleted. Pellet (P) and supernatant (S) fractions were analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. (B) sPGN competes with iPGN for lectin binding. Pull-down assays were performed with or without 100 µM soluble B. subtilis peptidoglycan. (C) Comparison of peptidoglycan and chitin structures. The structure of a typical Gram-positive peptidoglycan is depicted. (D) Lectin binding to immobilized polysaccharides. Lectins were bound to immobilized polysaccharide for 2 hours at 4°C. After washing, bound proteins were released by boiling in SDS-PAGE sample buffer and analyzed by SDS-PAGE and Coomassie blue staining. (E) Pull-down assays comparing binding to peptidoglycan, chitin, and cellulose. Purified recombinant RegIIIγ or HIP/PAP (10 µg of either) was added to 50 µg of peptidoglycan, chitin, or cellulose and analyzed as in (A). The lower molecular weight forms of RegIIIγ and HIP/PAP in (A) and (E) result from cleavage at an N-terminal trypsinlike site by a peptidoglycan-associated proteolytic activity.
Peptidoglycan consists of extended glycan chains composed of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues cross-linked by short peptides (Fig. 2C). Because RegIIIγ and HIP/PAP contain predicted CRDs, we determined whether they recognize the carbohydrate moiety of peptidoglycan. Chitin is composed of β1,4-linked GlcNAc chains that are virtually identical to the peptidoglycan carbohydrate backbone (Fig. 2C). As shown in Fig. 2D, purified recombinant RegIIIγ and HIP/PAP bound to chitin immobilized on Sepharose beads. Both lectins also bound to mannan, a polymer of mannose residues (Fig. 2D). This is consistent with the fact that C-type lectins that bind GlcNAc-containing saccharides frequently also bind mannose-containing saccharides (19), owing to the similar arrangements of the 3- and 4-hydroxyls of these sugars. In contrast, neither lectin bound dextran-Sepharose or uncoupled Sepharose (Fig. 2D). No binding was detected to monomelic GlcNAc-Sepharose or mannose-Sepharose, which indicated that both lectins show specificity for polymeric carbohydrates (13). Although the C-type lectin family includes members that bind their ligands in a calcium-dependent manner, we found that RegIIIγ and HIP/PAP do not require calcium for binding to peptidoglycan and chitin. Taken together, these results suggest that RegIIIγ and HIP/PAP are pattern-recognition proteins that recognize the microbe-associated molecular pattern represented by the extended glycan chains of peptidoglycan.
Chitin binding activity was also detected in pull-down assays in which we assessed RegIIIγ and HIP/PAP binding to equivalent masses of peptidoglycan and chitin (Fig. 2E). Peptidoglycan bound more RegIIIγ and HIP/PAP than chitin did, which suggested that both lectins bind more avidly to peptidoglycan than to chitin. Neither lectin bound to cellulose, a β1,4-linked glucose polymer.
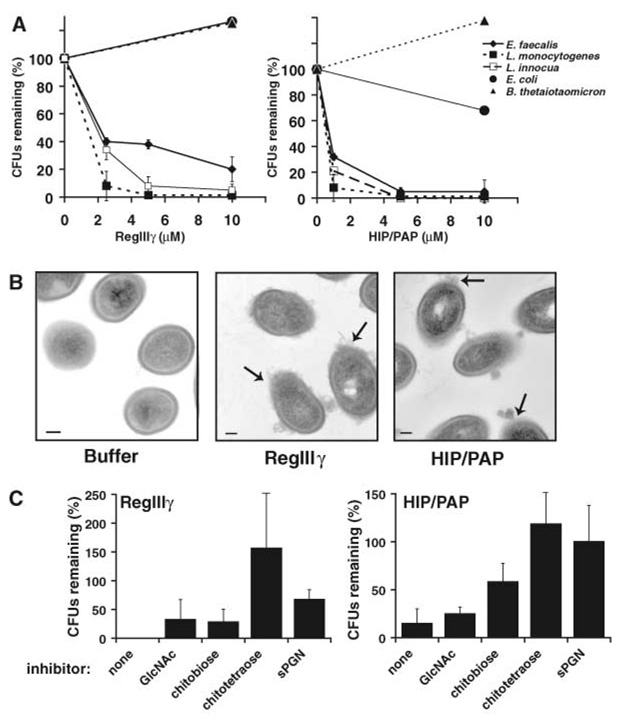
Our results reveal a carbohydrate ligand preference similar to that of mannose-binding lectin (MBL), a C-type lectin with an established role in innate immunity. As a serum protein, MBL recognizes invading microbes by binding to surface mannose residues (12) or to peptidoglycan (20). This binding triggers the lectin-activated complement pathway, which is initiated by recruitment of the serine proteases MASP1 and 2 via interactions with the MBL collagenous domain. In contrast to MBL, RegIIIγ and HIP/PAP consist of secreted CRDs that lack collagenous domains required for complement recruitment. We therefore postulated that RegIIIγ and HIP/PAP might be directly antimicrobial, without requiring additional factors to kill targeted microbes. We tested this idea by adding purified RegIIIγ and HIP/PAP to Gram-positive enteric microbes including Listeria monocytogenes, L. innocua, and E. faecalis, and observed a dose-dependent reduction in the viability of each organism (Fig. 3A). The number of colony-forming units (CFUs) of each microbe declined by 99% after a 2-hour exposure to 5 µM HIP/PAP (Fig. 3A). A similar decline in the viability of L. monocytogenes and L. innocua was observed after a 2-hour exposure to 5 µM RegIIIγ (Fig. 3A). The viability of E. faecalis declined by ~80% after a 2-hour exposure to 10 µM RegIIIγ (Fig. 3A). Thus, the effective antibacterial concentrations of both lectins are similar to those of other intestinal antimicrobial proteins (21, 22). Furthermore, we found that infusion of RegIIIγ into isolated small intestinal segments from L. innocua monocolonized mice resulted in a decline in bacterial numbers relative to a buffer infusion (fig. S5), indicating that RegIIIγ is active under native intestinal conditions.
Fig. 3.
Mouse RegIIIγ and human HIP/PAP have antibacterial activity against Gram-positive bacteria. (A) Percentage of CFUs remaining after exposure to purified RegIIIγ and HIP/PAP. L innocua, L monocytogenes, E. faecalis, Escherichia coli K12, and B. thetaiotaomicron were grown to mid–log phase and incubated with purified lectins. Initial bacterial concentrations ranged from 105 to 106 CFU/ml. After incubation for 2 hours at 37°C, viable bacteria were quantified by dilution plating. Assays were done in triplicate. Mean ± SD is plotted. (B) Transmission electron microscopy of L. monocytogenes following a 2-hour exposure to 10 µM purified recombinant RegIIIγ and HIP/PAP. Arrows indicate examples of damaged cell surfaces and cytoplasmic leakage. Scale bar, 100 nm. (C) Lectin bactericidal activity is inhibited by chitooligosaccharides and sPGN. GlcNAc, chitobiose (GlcNAc2), or chitotetraose (GlcNAc4) at 10 mM or 35 µM sPGN was added to antibacterial assays performed on L. innocua as in (A). Each percentage CFU was calculated relative to a no-lectin control assay containing an identical amount of chitooligosaccharide or sPGN.
As expected, neither RegIIIγ nor HIP/PAP was bactericidal toward the Gram-negative enteric organisms Escherichia coli or Bacteroides thetaiotaomicron (Fig. 3A). This is consistent with our observation of preferential binding to Gram-positive bacteria and the fact that pepti-doglycan is buried in the periplasmic space of Gram-negative bacteria. Additionally, neither lectin reduced the viability of fungal microorganisms, including Saccharomyces cerevisiae and Candida albicans.
We used transmission electron microscopy to visualize morphological changes in L. monocytogenes cells after exposure to RegIIIγ and HIP/PAP. Our images revealed evidence of cell wall damage and cytoplasmic leakage (Fig. 3B). These results are remarkably similar to those obtained with cationic antimicrobial peptides, such as human β-defensin-3 (21), which kill bacteria by cell wall permeabilization. Our findings indicate that lectin-mediated bacterial killing also involves cell wall damage.
RegIIIγ and HIP/PAP bactericidal activities were inhibited with sPGN and chitin fragments, linking peptidoglycan binding to antibacterial function. Addition of 35 µM sPGN to antibacterial assays inhibited the bactericidal activity of both lectins (Fig. 3C). At 10 mM, chitotetraose, a 4-sugar acid hydrolysis fragment of chitin, also fully inhibited the antibacterial activity of both RegIIIγ and HIP/PAP (Fig. 3C). Consistent with the preference of RegIIIγ and HIP/PAP for polymeric sugars, 10 mM monomelic GlcNAc or chitobiose (GlcNAc2) were less inhibitory. These results demonstrate that a soluble oligosaccharide that mimics the peptidoglycan saccharide backbone is sufficient to inhibit lectin antimicrobial activity. These findings are consistent with a model in which lectin binding to surface peptidoglycan carbohydrate precedes microbial killing.
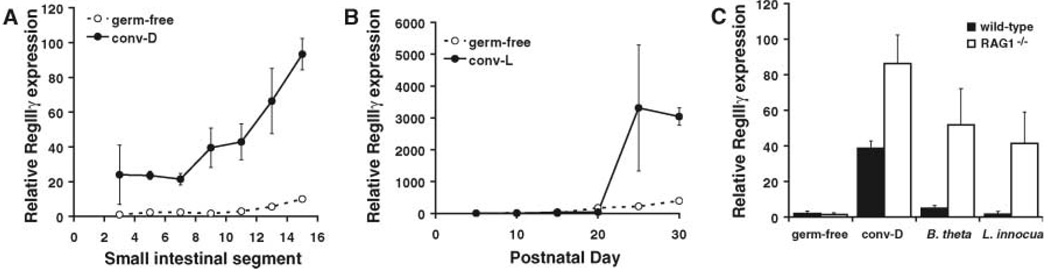
Given RegIIIγ’s bactericidal activity, we predicted that its expression patterns would reflect microbial colonization levels in the mouse small intestine. Q-PCR analysis of RegIIIγ mRNA levels along the cephalocaudal axis of conventionalized small intestines revealed increasing expression toward the distal region (ileum) (Fig. 4A), concomitant with increasing microbial densities. In contrast, germ-free mice showed minimal RegIIIγ expression throughout the small intestine (Fig. 4A). We also assayed for changes in RegIIIγ mRNA expression during postnatal intestinal development. RegIIIγ mRNA levels rose dramatically during the weaning period [postnatal days (P) 17 to 22] and remained high into adulthood (≥P28) in conventionally raised but not germ-free mice (Fig. 4B). Weaning is associated with dramatic changes in gut microflora composition, as well as withdrawal of maternal immunoglobulin A (IgA) antibodies. The antibacterial activity of RegIIIγ suggests that its expression is elicited as part of a compensatory response to maintain mucosal homeostasis in the face of changing microbial ecology and withdrawal of passive immunity.
Fig. 4.
RegIIIγ expression is triggered by intestinal bacteria. (A) RegIIIγ expression along the cephalocaudal axis of the small intestine. Small intestines from adult germ-free or conventionalized (conv-D) NMRI mice were divided into 16 equal segments (numbered proximal to distal) and RegIIIγ mRNA expression was determined in specific segments using Q-PCR. Results are representative of experiments in two sets of mice. (B) RegIIIγ mRNA increases during the weaning period (P17 to 22) in developing conventionally raised NMRI mice. Assays were performed on pooled mid–small intestinal RNAs (for three mice per time point). (C) RegIIIγ expression is triggered by single Gram-positive or Gram-negative bacterial species in immunodeficient mice. Q-PCR determinations were done on cDNAs from mid–small intestine. Each point represents the average value from three or more different mice. All Q-PCR determinations were performed in triplicate (mean ± SD plotted) and were normalized to 18S ribosomal RNA.
Because conventional microflora are composed of diverse microbial societies, we asked whether single enteric bacterial species are sufficient to drive small intestinal RegIIIγ expression. As expected, a mixed microbial community recovered from a conventional mouse elicited a ~20-fold increase in RegIIIγ expression when introduced into germ-free wild-type C57/b6 mice. In contrast, colonization with the Gram-negative symbiont Bacteroides thetaiotaomicron elicited only a 2.5-fold increase in expression, whereas the noninvasive Gram-positive L. innocua had no effect on RegIIIγ mRNA levels (Fig. 4C). These results indicate that neither organism alone was sufficient to stimulate RegIIIγ expression to conventional levels in wild-type mice. However, bacteria that are normally strictly compartmentalized in the intestinal lumen show increased mucosal adherence and invasion in mice that lack mucosal IgA (23). We therefore postulated that mucosal defenses such as secretory IgA might be sufficient to sequester B. thetaiotaomicron and L. innocua in the gut lumen and so could account for the inability of these single species to stimulate RegIIIγ expression. Indeed, we found that B. thetaiotaomicron and L. innocua trigger a 52- and 41-fold increase, respectively, in RegIIIγ mRNA expression after colonization of germ-free RAGT deficient mice, which lack mature lymphocytes and are therefore IgA-deficient (24). Wild-type and RAG1-deficient mice were colonized to virtually identical levels (~108 CFU/ml ileal contents), which indicated that differences in RegIIIγ mRNA expression did not result from differences in total microbial numbers. Our findings thus support a model in which increased bacterial-epithelial contact drives RegIIIγ expression as a mechanism to limit potential microbial penetration and maintain mucosal surface integrity.
Together, RegIIIγ and HIP/PAP represent a new family of inducible antibacterial proteins that seek out their microbial targets via interactions with bacterial peptidoglycan. Because they lack domains necessary for complement recruitment and are directly bactericidal, these proteins reveal a new function for mammalian C-type lectins. We propose that these antibacterial lectins represent an evolutionarily primitive form of lectin-mediated innate immunity, and that the lectin-mediated complement pathway may have evolved from a directly antimicrobial precursor. In support of this idea, simple model organisms such as Drosophila and Caenorhabditis elegans harbor a number of genes encoding putative C-type lectins that consist solely of a simple CRD with an N-terminal secretion signal (25, 26). The data presented here suggest that these proteins could function in innate antimicrobial defense. Furthermore, the human and mouse Reg families encompass multiple members, many of which are expressed in the gut (27). It seems likely that other members of this protein family are also antimicrobial, but may exhibit preferences for different microbial targets.
The discovery of directly antibacterial C-type lectins points to a previously uncharacterized mucosal defense mechanism that helps to sequester the gut microflora and preserve intestinal homeostasis. Our results suggest that RegIIIγ expression is triggered by increased microbial-epithelial contact at mucosal surfaces. Enhanced expression of Reg proteins such as HIP/PAP in IBD patients may therefore be a compensatory response that limits mucosal penetration by gut microbes. Because Reg proteins exhibit increased expression in IBD mucosa (5, 8), whereas α-defensin expression decreases (2, 3), these two groups of antimicrobial proteins are probably regulated by distinct mechanisms. It is not yet known whether Reg expression is triggered by direct bacterial interactions with gut epithelia, or whether other intestinal cells (e.g., macrophages) direct epithelial Reg expression. Further investigation will therefore be required to decipher the host and microbial factors that regulate antimicrobial lectin expression. These studies will contribute to a better understanding of IBD pathogenesis and will provide new insights into how symbiotic host-microbial relationships are maintained.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/313/5790/1126/DC1
Materials and Methods
Figs. S1 to S5
Table S1
References
References and Notes
- 1.Hooper LV, Midtvedt T, Gordon JI. Annu. Rev. Nutr. 2002;22:283. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 2.Wehkamp J, et al. Gut. 2004;53:1658. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehkamp J, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18129. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christa L, et al. Am. J. Physiol. 1996;271:G993. doi: 10.1152/ajpgi.1996.271.6.G993. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa H, et al. Inflamm. Bowel Dis. 2003;9:162. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Syder AJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3467. doi: 10.1073/pnas.0230380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Proc. Natl. Acad. Sci. U.S.A. 2005;102:99. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieckgraefe BK, et al. J. Investig. Med. 2002;50:421. doi: 10.1136/jim-50-06-02. [DOI] [PubMed] [Google Scholar]
- 9.Swidsinski A, et al. Gastroenterology. 2002;122:44. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 10.Livesey FJ, et al. Nature. 1997;390:614. doi: 10.1038/37615. [DOI] [PubMed] [Google Scholar]
- 11.Ayabe T, et al. Nat. Immunol. 2000;1:113. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 12.Ezekowitz RA. J. Infect. Dis. 2003;187 suppl. 2:S335. doi: 10.1086/374746. [DOI] [PubMed] [Google Scholar]
- 13.Cash HL, Whitham CV, Hooper LV. Protein Expr. Purif. 2006;48:151. doi: 10.1016/j.pep.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckburg PB, et al. Science. 2005;308:1635. [Google Scholar]
- 15.Holm C, Jespersen L. Appl. Environ. Microbiol. 2003;69:2857. doi: 10.1128/AEM.69.5.2857-2863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner T, et al. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13772. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dziarski R, Tapping RI, Tobias PS. J. Biol. Chem. 1998;273:8680. doi: 10.1074/jbc.273.15.8680. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Gelius E, Liu G, Steiner H, Dziarski R. J. Biol. Chem. 2000;275:24490. doi: 10.1074/jbc.M001239200. [DOI] [PubMed] [Google Scholar]
- 19.Drickamer K. Nature. 1992;360:183. doi: 10.1038/360183a0. [DOI] [PubMed] [Google Scholar]
- 20.Nadesalingam J, Dodds AW, Reid KB, Palaniyar N. J. Immunol. 2005;175:1785. doi: 10.4049/jimmunol.175.3.1785. [DOI] [PubMed] [Google Scholar]
- 21.Harder J, Bartels J, Christophers E, Schroder JM. J. Biol. Chem. 2001;276:5707. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 22.Porter EM, van Dam E, Valore EV, Ganz T. Infect. Immun. 1997;65:2396. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macpherson AJ, et al. Science. 2000;288:2222. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 24.Mombaerts P, et al. Cell. 1992;68:869. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 25.Drickamer K, Dodd RB. Glycobiology. 1999;9:1357. doi: 10.1093/glycob/9.12.1357. [DOI] [PubMed] [Google Scholar]
- 26.Dodd RB, Drickamer K. Glycobiology. 2001;11:71R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita H, et al. FEBS Lett. 1995;377:429. doi: 10.1016/0014-5793(95)01381-4. [DOI] [PubMed] [Google Scholar]
- 28.The authors wish to thank T. Januszewski for help with electron microscopy and D. Farrar for help with flow cytometry. This work was supported by grants from the NIH (R01 DK070855), the Crohn’s and Colitis Foundation of America, and a Burroughs Wellcome Career Award in the Biomedical Sciences (to L.V.H.). H.L.C. was supported by NIH training grant T32-AI007520. L.V.H. is on the scientific advisory board of The Dannon Company. The microarray data are held in U.S. National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and the accession number is GSE5156.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.