Abstract
Proteins containing membrane targeting domains play essential roles in many cellular signaling pathways. However, important features of the membrane-bound state are invisible to bulk methods, thereby hindering mechanistic analysis of membrane targeting reactions. Here we use total internal reflection fluorescence microscopy (TIRFM), combined with single particle tracking, to probe the membrane docking mechanism of a representative pleckstrin homology (PH) domain isolated from the general receptor for phosphoinositides, isoform 1 (GRP1). The findings show three previously undescribed features of GRP1 PH domain docking to membranes containing its rare target lipid, phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3]. First, analysis of surface diffusion kinetics on supported lipid bilayers shows that in the absence of other anionic lipids, the PI(3,4,5)P3-bound protein exhibits the same diffusion constant as a single lipid molecule. Second, the binding of the anionic lipid phosphatidylserine to a previously unidentified secondary binding site slows both diffusion and dissociation kinetics. Third, TIRFM enables direct observation of rare events in which dissociation from the membrane surface is followed by transient diffusion through solution and rapid rebinding to a nearby, membrane-associated target lipid. Overall, this study shows that in vitro single-molecule TIRFM provides a new window into the molecular mechanisms of membrane docking reactions.
Introduction
Many critical cell signaling events occur at membrane surfaces, and organisms have evolved efficient regulatory mechanisms for these processes. In particular, a broad array of intracellular signaling proteins are recruited to the plasma membrane during second messenger signals by conserved membrane-targeting domains (1–3). Such membrane recruitment events are often coordinated by second messenger appearance at a precisely controlled time in a highly localized region of the cell. The most common membrane targeting motif is the pleckstrin homology (PH) domain, which has been identified in over 600 human proteins [Pfam database (4)]. PH domain-containing proteins regulate a wide array of essential cell processes including growth, metabolism, and chemotaxis (1–3,5). Many PH domains dock to cellular membranes via specific binding to the lipid second messenger phosphatidylinositol-(3,4,5)-trisphosphate [PI(3,4,5)P3], or to other phosphatidylinositol phosphate (PIP) lipids (6). The PIP lipid specificity is dominated by headgroup recognition; however, other features of the docking reaction are less well understood. For example, even during a signaling event, the peak mole density of PI(3,4,5)P3 in the plasma membrane is typically less than 200 ppm, making this target lipid an extremely rare plasma membrane component. Yet, PI(3,4,5)P3 triggers recruitment of certain PH domains on a timescale of seconds in rapid signaling pathways such as leukocyte chemotaxis (7–10). Such PH domains must overcome an immense search problem to find this rare target lipid on the appropriate timescale. The elucidation of the molecular mechanism of rare target searching is critical for full understanding of these signaling pathways.
The PH domain from the general receptor for phosphoinositides (GRP1, also known as cytohesin-3) is a representative PI(3,4,5)P3-specific PH domain that exhibits rapid docking to this rare target lipid. GRP1 functions as a guanine nucleotide exchange factor (GEF) for Arf family GTPases, which are involved in reorganization of the cellular membrane and actin network in migrating cells (5,11,12). GRP1 is recruited to the plasma membrane in response to PI(3,4,5)P3 synthesized by phosphatidylinositol 3-kinase (11,13). Structurally, GRP1 consists of an N-terminal heptad repeat region, a catalytic GEF domain, and a C-terminal PH domain. X-ray crystal structures have been solved for the stable, isolated PH domain bound to a soluble analog of the PI(3,4,5)P3 headgroup [inositol (1,3,4,5)-tetrakisphosphate (IP4)] (14–16). Specific PI(3,4,5)P3 recognition is mediated by a binding pocket comprised primarily of lysines and arginines, as well as a titratable histidine (14,15,17–19). These residues, along with several additional basic side chains not directly involved in PI(3,4,5)P3 coordination, render the membrane-binding face highly positively charged. Correspondingly, the presence of background anionic lipids such as phosphatidylserine (PS) has been shown to enhance the PI(3,4,5)P3 association rate, and thus equilibrium affinity, by a factor of 10 (17). This enhanced on-rate arises from an efficient search mechanism involving an electrostatic attraction between the cationic membrane docking face of the PH domain and the anionic surface of the membrane, which is proposed to increase the rate of random collisions both with the membrane surface and with the rare target lipid PI(3,4,5)P3. The resulting faster target lipid on-rate is critical for rapid binding to plasma membrane PI(3,4,5)P3 on the timescale required for leukocyte chemotaxis in vivo (17).
The current molecular model for the docking of GRP1 PH domain to its target membrane leads to at least three testable predictions. First, the PI(3,4,5)P3-bound PH domain is predicted to sit on the surface of the bilayer with little penetration. This prediction is based on the low density of hydrophobic residues within its putative membrane docking face relative to other membrane targeting domains (20), and on the large size of the PIP lipid headgroup that protrudes out from the headgroup layer into solvent (21). Second, while the PH domain is bound to its PI(3,4,5)P3 target lipid on the membrane surface, multiple positively charged side chains surrounding the PI(3,4,5)P3 binding pocket are predicted to form direct contacts with background anionic lipids. Because the predominant anionic lipid in the plasma membrane inner leaflet is PS, this lipid is expected to dominate such interactions. Third, when the PH domain dissociates from the membrane, the electrostatic search mechanism is predicted to increase the probability of local membrane rebinding events before return to bulk solution. In principle, the first two predictions can be tested by measurements of PH domain diffusion on the surface of the membrane, which should be slowed by significant bilayer penetration or interactions with multiple anionic lipids. The third prediction can be tested by direct detection of membrane rebinding events immediately after dissociation. However, the predicted effects on surface diffusion and rebinding kinetics would be difficult or impossible to observe directly in standard bulk kinetic experiments. Thus, this study develops a novel application of single-molecule microscopy that enables direct mechanistic analysis of the membrane docking reaction at a molecular level in vitro.
Single-molecule techniques are rapidly emerging as tools for direct interrogation of molecular mechanisms and dynamics on a microscopic scale (22). Total internal reflection fluorescence microscopy (TIRFM), which can be readily combined with single-molecule detection, is an exquisitely sensitive technique for measuring interactions at surfaces of supported lipid bilayers or living cells (23,24). Single-molecule TIRFM methods have been used to measure diffusion of lipids and integral membrane proteins in synthetic bilayers and in cells, revealing interesting properties including compartmentalized diffusion in the plasma membrane (25–27). PH domains and other membrane binding proteins have also been observed at the single molecule level in living cells (28–30). Detection in live cells represents a major advantage of TIRFM and provides direct information about targeting to specific intracellular membranes. For the molecular analysis of membrane targeting mechanisms, however, fully controlled systems are preferable in which the membrane composition, buffer conditions, and protein concentration can be varied as needed. Such systems have been used with TIRFM at the ensemble level to measure protein-surface interactions (23,31), and are even more powerful when combined with single-molecule detection. For example, recent studies have used membrane-bound synaptotagmin proteins as platforms for single-molecule TIRFM measurements of protein complex formation (32,33). More recently, single-molecule TIRFM showed that the soluble acyl-CoA binding protein is a useful probe of lipid dynamics (34). All of these studies uncovered aspects of molecular behavior that were undetected in traditional bulk experiments.
This study shows that in vitro single-molecule TIRFM is a powerful new tool in the mechanistic analysis of membrane targeting proteins and their lipid docking reactions. By applying single-molecule TIRFM to investigate the docking of purified GRP1 PH domain to target lipid, we are able to uncover hidden features of the docking mechanism. This study is, to our knowledge, the first to use single-molecule TIRFM with purified protein and synthetic bilayers to investigate the molecular mechanisms of a membrane targeting domain docking to, diffusing on, and dissociating from a lipid membrane. Supported bilayers are prepared containing PI(3,4,5)P3 in a simple lipid background either containing or lacking anionic phospholipids. On addition, fluorophore-tagged PH domains dock to PI(3,4,5)P3 on the surface of the accessible bilayer leaflet. Single-particle tracking is used to measure statistics of both the diffusion and dissociation of the membrane-docked protein. Finally, rare microdissociation and rebinding events are detected, in which a particle appears to dissociate transiently from the membrane, diffuse briefly in solution, and rebind rapidly to PI(3,4,5)P3 a short distance away. These findings provide additional evidence supporting the electrostatic search mechanism proposed for the binding of GRP1 PH domain to its rare, membrane-associated target lipid, and show features of the membrane-bound state undetected previously.
Materials and Methods
Reagents
Synthetic phospholipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, PC); 1,2-dioleoyl-sn-glycero-3-[phospho-l-serine] (DOPS, PS); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (LRB-DOPE, LRB-PE); and 1,2-dioleoyl-sn-glycero-3-phosphoinositol-3,4,5-trisphosphate [DOPI(3,4,5)P3, PI(3,4,5)P3] were from Avanti Polar Lipids (Alabaster, AL). N-[5-(dimethylamino)naphthalene-1-sulfonyl]-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (dansyl-PE); Alexa Fluor 555 (AF555) C2-maleimide; and Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-PE) were from Invitrogen (Carlsbad, CA). 2-Mercaptoethanol was from Fluka (Buchs, Germany); dithiothreitol (DTT) was from Research Products International (Mount Prospect, IL); d-myo-inositol (1–6)-hexakisphosphate (IP6) used in bulk experiments was from Sigma (St. Louis, MO). For single-molecule experiments, a higher-purity grade of IP6 was used from Calbiochem (La Jolla, CA).
Protein mutagenesis, expression, labeling, and purification
Wild-type GRP1 PH domain (residues 255–392 of full-length human GRP1) was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein, separated from the GST domain, and purified as described previously (17). A cysteine-free version of the protein (C291S/C325A/C341S) was created using site-directed mutagenesis (QuikChange II XL, Stratagene, La Jolla, CA), and was found to retain PI(3,4,5)P3 binding properties identical to wild-type, within measurement error (J. Corbin, D. Dukellis, and J. Falke, unpublished data). For fluorescence labeling, an engineered Cys was introduced at the 303 position, and the resulting mutant (C291S/C325A/C341S/E303C) at a concentration of 15 μM was incubated with 75 μM AF555 C2-maleimide and 15 μM Tris(2-carboxyethyl) phosphine (TCEP) for 1 h at room temperature in labeling buffer (140 mM KCl, 0.5 mM MgCl2, 15 mM NaCl, 50 mM L-glutamic acid, 50 mM L-arginine, 25 mM HEPES, pH 7.1). The inclusion of 50 mM Glu/Arg increases stability of the protein during labeling (35). The reaction was quenched by addition of 1 mM DTT, and free dye was then removed by repeated exchange into labeling buffer using a 5000 MWCO centrifugal concentrator (Amicon). The labeling efficiency (>80%) and concentration of labeled protein were determined by absorbance measurements of tryptophan and Alexa Fluor 555 (AF555).
Preparation of phospholipid vesicles
Sonicated unilamellar vesicles (SUVs) were prepared as described previously (17). Briefly, phospholipids were dissolved in chloroform/methanol/water (1.0:1.2:0.4) at the desired molar ratio, dried under vacuum, and resuspended with vortexing in lipid storage buffer (140 mM KCl, 0.5 mM MgCl2, 15 mM NaCl, 0.02% NaN3, 25 mM HEPES, pH 7.5), yielding a concentration of 3.0 mM total lipid, followed by sonication with a Misonix XL 2020 probe sonicator. The resulting SUVs were stored at 4°C for no more than 1 week before use. Lipid compositions used in this study are listed in Table 1.
Table 1.
Lipid compositions used in this study
| Name∗ | Experiment | Components† | Mol % |
|---|---|---|---|
| PC/PIP3 | Bulk (wild-type protein) | PC/dansyl-PE/PI(3,4,5)P3 | 93/5/2 |
| Bulk (labeled protein) | PC/TR-PE/PI(3,4,5)P3 | 97.5/0.5/2‡ | |
| TIRFM (labeled protein) | PC/PI(3,4,5)P3 | 98/2 | |
| PC/PS/PIP3 | Bulk (wild-type protein) | PC/PS/dansyl-PE/PI(3,4,5)P3 | 69.8/23.2/5/2 |
| Bulk (labeled protein) | PC/PS/TR-PE/PI(3,4,5)P3 | 73.1/24.4/0.5/2‡ | |
| TIRFM (labeled protein) | PC/PS/PI(3,4,5)P3 | 73.5/24.5/2 | |
| PC/PS | TIRFM (labeled protein) | PC/PS | 75/25 |
Indicated names are used for lipid mixtures throughout the Results, Discussion, tables, and figures.
All lipid acyl chains used were dioleoyl, except dansyl-PE and TR-PE (dihexadecanoyl).
PI(3,4,5)P3 density used for bulk dissociation measurements with labeled protein was 3 mol %, as described in the Table 2 legend.
Bulk equilibrium FRET assay
Equilibrium fluorescence measurements were carried out on a QM-2000-6SE fluorescence spectrophotometer (Photon Technology International, Birmingham, NJ) at 21.5°C using methods developed previously (17). For measurement of inositol (1–6)-hexakisphosphate (IP6) affinity, a concentrated IP6 solution was titrated into 0.75 μM protein in assay buffer (140 mM KCl, 0.5 mM MgCl2, 15 mM NaCl, 10 mM DTT, 25 mM HEPES, pH 7.5) while monitoring the binding-induced change in tryptophan fluorescence (λex = 284 nm, λem = 332 nm). The resulting binding curve was fit to a single-site binding model to determine the equilibrium dissociation constant KD(IP6).
PI(3,4,5)P3 affinities were measured using IP6 as a competitive inhibitor in a competitive displacement binding assay (17). For wild-type PH domain, 0.75 μM protein was prebound to vesicles (SUVs) containing 3.0 μM accessible PI(3,4,5)P3 and 5% dansyl-PE. Subsequently, the displacement of membrane-bound protein was quantitated by the loss of protein-to-membrane FRET (donor tryptophan to acceptor dansyl-PE; λex = 284 nm, λem = 522 nm; slit widths 4 nm and 8 nm, respectively) as IP6 was titrated into the sample. For AF555-labeled PH domain, the analogous experiment was carried out using a different protein-to-membrane FRET pair. Thus, 0.1 μM AF555-labeled PH domain was prebound to SUVs containing 0.36 μM accessible PI(3,4,5)P3 and 0.5% TR-PE, then protein-to-membrane FRET (donor AF555 to acceptor TR-PE; λex = 515 nm, λem = 608 nm; slit widths 4 nm and 8 nm, respectively) was quantitated during IP6 titration. The excitation wavelengths used for both FRET measurements result in some direct excitation of the acceptor fluorophore, which was measured using an identical sample lacking protein and subtracted from the total FRET to yield the corrected FRET. The resulting IP6 competitive displacement curves were subjected to a nonlinear least-squares fit using Eq. 1, yielding an apparent competitive inhibition constant for IP6 [KI(IP6)app]:
| (1) |
The best-fit offset value C was subtracted from all data points, and ΔFmax was normalized to unity to simplify graphical representations. Finally, the equilibrium dissociation constant for PI(3,4,5)P3 [KD(PIP3)] was calculated using Eq. 2 (17):
| (2) |
from the measured KD(IP6) and KI(IP6)app values.
Bulk kinetic measurements
Protein-membrane association and dissociation rates were measured on an Applied Photophysics SX.17 stopped-flow fluorimeter, using protein-to-membrane FRET pairs as described above. All measurements were carried out at 21.5°C in assay buffer. For FRET from wild-type protein to dansyl-labeled lipid, excitation was set to 284 nm with a 3-nm monochromator slit width, and a 475-nm longpass filter was used for emission. For measurements with AF555-labeled protein, excitation was set to 515 nm with an 8-nm slit width, and emission wavelengths were selected with a 610-nm bandpass filter, bandwidth 60 nm.
Association measurements were initiated by rapid mixing of PH domain and vesicles (SUVs) at the concentrations indicated in the text. GRP1 PH domain membrane association has previously been shown to exhibit biphasic behavior; thus time courses were fit to a double exponential function (17):
| (3) |
In the above equation, the observed rate constants kobs are the product of the true rate constants kon and the concentration of accessible PI(3,4,5)P3. (Under the conditions used, nearly all of the PH domain binds to PI(3,4,5)P3 at equilibrium, thus the contribution of koff during the approach to equilibrium can be neglected). Because the PI(3,4,5)P3 in the interior leaflet of the SUVs is inaccessible, kon was calculated by dividing kobs by half the total PI(3,4,5)P3 concentration.
Dissociation measurements were initiated by rapid mixing of preformed PH domain-membrane complexes (SUVs) with excess IP6 at the concentrations indicated in the text. Time courses were fit to a single exponential decay function:
| (4) |
For graphical presentation of kinetic data, the best-fit offset C was subtracted from all data and the best-fit amplitude ΔFmax (or ΔFmax1 + ΔFmax2) was normalized to unity.
Sample preparation for single molecule experiments
Glass coverslips (Pella, Redding, CA) were soaked for 1 h in piranha solution (a 3:1 mixture of sulfuric acid and 30% H2O2), rinsed extensively with Milli-Q water, dried under a stream of N2, and irradiated 0.6 h in a Novascan PSD-UV ozone cleaner. A 60-μl perfusion chamber (Invitrogen, Eugene, OR) was adhered to the cleaned glass slide. To deposit supported lipid bilayers, SUVs of the desired lipid composition were mixed 1:1 with buffer containing 1M NaCl, injected into the perfusion chamber, and incubated 30 min at 22°C. After bilayer formation, samples were rinsed with 10–20 ml Milli-Q water, followed by washing with assay buffer containing 20 mM 2-mercaptoethanol in place of 10 mM DTT. All samples were imaged before and after addition of fluorescent protein, to ensure that background fluorescent contamination was negligible. Several other methods for cleaning the glass slides were also tested, and it was found in side-by-side studies that etching with piranha solution followed by UV-ozone cleaning provided the best combination of low background fluorescence and fluidity of PI(3,4,5)P3-containing bilayers.
TIRFM measurement
Measurements were taken on a home-built, objective-based TIRFM instrument (V. Fomenko, H. Lee, and A. Pardi, unpublished). The microscope is a modified Nikon TE-2000U and uses illumination from a 532-nm solid-state laser (Blue Sky Research, Milpitas, CA). Excitation intensity was varied by inserting neutral density filters in the laser beam, and was measured at the microscope objective with a power meter at various laser powers while in epifluorescence mode, and then the instrument was switched to TIR mode for measurement. For measurements involving protein samples, protein was added to the sample chamber and incubated 5 min for thermal equilibration to room temperature (20–22°C). To eliminate contributions from immobile fluorescent particles in experiments other than the direct photobleaching measurement, a high-power bleach pulse (∼30-fold higher than used for imaging) was applied for 2–5 s, and fluorescence was allowed to recover for 60 s before data acquisition. Fluorescence was detected with a Photometrics Cascade II 16-bit electron-multiplying CCD camera and recorded at 20 frames/s, with a spatial resolution of 7.0 pixels/μm unless otherwise indicated. The maximum video frame rate of our instrument at this pixel resolution is 50 frames/s. Streaming movies were acquired using MetaMorph software (AG Heinze) and subsequent analysis was carried out using ImageJ (36). Data processing and fitting were carried out using Mathematica (Wolfram Research).
Photobleaching measurement and analysis
When 100 pM AF555-GRP1PH was added to a PC/PIP3 bilayer and incubated at room temperature for 15 min, followed by equilibration with 100 μM IP6 for 30 min to displace mobile particles, a population of immobile fluorescent particles remained on the surface. By omitting the high-power bleach pulse (see above) these immobile particles were imaged and used to measure photobleaching rates at four different incident laser powers. Photobleaching time courses were generated by acquiring movies on previously unexposed areas of the slide until >90% of the particles in the area disappeared. The time at which each immobile particle disappeared in a single step was recorded. Occasionally, clusters of particles were immobilized close together; these were excluded from analysis based on high initial intensity and multi-step photobleaching. Rarely, a particle with a normal initial intensity was observed to transition to a low-intensity state before bleaching, or to “blink” on again after disappearing. Such anomalies constituted less than 10% of all particles and were excluded from analysis. The probability distribution P(t) of bleaching times for the particles with single-step bleaching (43–65 particles at each laser power) was plotted and fit to a single exponential distribution:
| (5) |
where P0 is the amplitude and kbl is the photobleaching rate.
Single particle tracking and analysis
For all single molecule analysis other than photobleaching, trajectories of individual lipid and protein molecules in acquired movies were determined using the Particle Tracker plugin for ImageJ (37). This software package determines the center position and intensity of each particle above an intensity percentage threshold in each frame, and then links particles together into trajectories. The relative intensities of mobile particles can vary from frame to frame depending on the area sampled by the randomly diffusing particle during each exposure (34). To maximize the fidelity of tracking, parameters were chosen for the Particle Tracker plugin using low intensity thresholds for detection to ensure oversampling (i.e., a significant amount of spurious detection was permitted in each frame) rather than undersampling (i.e., loss of legitimate trajectories). The resulting preliminary trajectories were imported into Mathematica, and spurious trajectories and contaminants were removed from analysis through application of a series of exclusion tests. First, the trajectories of spurious detections and dim contaminants were excluded by applying a minimum threshold for intensity averaged over the entire trajectory. Similarly, unusually bright contaminants were excluded by applying a maximum threshold for intensity averaged over the entire trajectory. The values for these thresholds were set empirically based on the distributions of average trajectory intensities. These distributions generally exhibited bimodal profiles, in which the dim population reflected spurious detections and dim contaminants, and the bright population represented fluorescently-labeled PH domains. To exclude rapidly dissociating contaminants and trajectory fragments not properly linked, trajectories shorter than five frames were eliminated. To exclude immobile contaminants that escape the high-power bleach pulse (see above), the diffusion coefficient D was calculated for each particle, and particles with D below threshold (0.5 μm2/s for lipid or protein diffusion analysis, 0.1 μm2/s for protein dissociation analysis) were eliminated. For protein dissociation measurements, fast-moving contaminants were also excluded by eliminating particles with D > 4.0 μm2/s. Finally, to exclude partial trajectories from protein dissociation measurements, those trajectories that began or ended on the first or last frame of the movie; or whose position began or ended within 6 pixels of the edge of the field of view, were eliminated. Overall, the success of these exclusion rules in removing fluorescent contaminants is clearly shown by the simple behavior of the trajectories retained for analysis, which exhibit ideal diffusion kinetics for the fluorescent lipid, and exhibit near-ideal diffusion and dissociation kinetics for the fluorescent PH domain (see Results).
Single molecule diffusion analysis
All steps from lipid or protein trajectories that passed exclusion tests (>1800 steps per movie) were combined and subjected to diffusion analysis using a method based on that of Schütz et al. (38). Briefly, the cumulative probability distribution P(r2,Δt) of square displacement r2 or greater over a given time interval Δt was plotted and fit to either a single-component or two-component model using Eqs. 6 and 7 respectively:
| (6) |
or
| (7) |
In these equations, 〈r2〉, 〈r12〉, and 〈r22〉 are mean square displacement values, and α in the two-component model is the fraction exhibiting mean square displacement 〈r12〉. An f-test was applied to determine whether two-component fitting provided statistically significant improvement, and fitting was repeated using Δt values from one to eight frames. The resulting mean square displacement values were plotted as a function of Δt. For simple, homogeneous, two-dimensional diffusion the mean square displacement is linearly related to Δt as in Eq. 8:
| (8) |
Thus, the diffusion constant D was determined by least-squares fitting to a straight line through the origin. Diffusion constants listed in the text and tables are mean ± SD from at least six movies representing at least three separate but identically prepared samples.
Single molecule dissociation analysis
For protein dissociation analysis, trajectories that passed exclusion criteria (see above) were manually validated to determine the length of time each fluorescent particle was visible on the surface. Imperfect tracking in Particle Tracker sometimes resulted in two or more shorter trajectories corresponding to the same particle; in such cases, the summed dwell time was recorded and any duplication was eliminated. The probability distribution of the validated dwell times was plotted and fit to a single-exponential distribution:
| (9) |
from which the dissociation rate constant koff was extracted. As this analysis method tends to bias against shorter trajectories (that, if split, may not have passed exclusion tests above), only trajectories whose total time was 10 frames (0.5 s) or more were included in the analysis. Bin sizes used for distributions were optimized to yield a best-fit curve with the lowest uncertainty. Rate constants listed in text and tables are mean ± SD from three separate but identically prepared samples. For the conditions yielding the slowest dissociation rate, the measurement was carried out at two different laser powers to determine the relative contribution of photobleaching, which was found to be negligible.
Single molecule jump analysis
To analyze the frequency of “jumps” arising from microdissociation, diffusion through solvent, then rebinding, the total number of frame-to-frame steps with extremely long step sizes was counted for at least three movies (2000 frames/movie, 50 ms/frame) acquired at a given condition. For two-dimensional diffusion without microdissociation and rebinding, the expected distribution of step sizes P(r,Δt) is given by the Rayleigh distribution:
| (10) |
where σ2 = 2D Δt. The threshold length rmin for the top 0.5% of this distribution was determined by numeric integration of:
| (11) |
Jumps were operationally defined as steps between adjacent frames, or within the same frame, of length between rmin and rmax = 5 μm. The latter upper limit is significantly longer than the average distance the PH domain is expected to diffuse through solution during a given microdissociation and rebinding event (because the average diffusion distance for a protein of this size in solution is 4 μm in 50 ms, the time per frame, and most microdissociation events are faster than 50 ms). Overall, apparent steps of r < rmin are predominantly due to random two-dimensional diffusion of particles that remain bound to the membrane, and apparent steps of r > rmax arise predominantly from near-simultaneous association and dissociation of two different particles. It follows that steps arising from microdissociation and rebinding will often exhibit step sizes between rmin and rmax. Exclusion of both shorter and longer steps helps maximize the fraction of jumps corresponding to true microdissociation and rebinding events.
Particle Tracker identified many jumps meeting both distance criteria, which were each further validated manually. Additional jumps not detected by Particle Tracker were identified by querying the data for sets of trajectories in which one trajectory begins in the same frame or the frame immediately after the end of another trajectory. These were also validated manually, and only those that met the distance criteria were retained.
The number of expected randomly correlated trajectories Nrc (in which one trajectory ends and another begins 0 or 1 frames later) arising from near-simultaneous association and dissociation of distinct molecules was calculated based on the ratio of the total number of trajectories NT to the total number of frames Nf:
| (12) |
where ρ is the ratio of the allowed area for observed jumps [π(rmax2 − rmin2)] to the total area of the field of view.
Results
Strategy
The overall goal of this study is to use single-molecule TIRFM measurements in vitro to elucidate protein-membrane docking mechanisms. The TIRFM system used herein provides direct detection of individual molecules bound to the membrane surface while ignoring proteins diffusing freely in solution, thereby providing a new window into the dynamics of the membrane-bound state. This surface-selective detection arises from two features of TIRFM. First, TIRFM fluorescence excitation occurs in a localized zone near a glass-water interface. This excitation zone decays to 50% intensity at a distance of ∼100 nm from the glass (24), and thus can easily accommodate a supported bilayer ∼5 nm in thickness separated from the glass by a 1–2 nm water layer (39,40). Only those fluorescent molecules bound to the surface or diffusing in solution near the surface are excited by the evanescent field. Second, small proteins freely diffusing in solution move too rapidly to be imaged by standard CCD cameras, thus only those proteins associated with the bilayer are detected. Relative to aqueous solution, the bilayer is ∼100-fold more viscous, thereby slowing diffusion rates into the range of CCD detection. The slow membrane diffusion ensures that fluorescent spots are broadened to an apparent size only ∼2-fold larger than the diffraction limit. Moreover, the selective detection of lipid-associated proteins greatly simplifies the kinetic analysis of molecular events that occur while bound to the membrane.
The PH domain from human GRP1 (GRP1-PH) was selected for analysis by single-molecule TIRFM, to test and further develop our published hypothesis that this domain uses background anionic lipids and an electrostatic search mechanism to rapidly locate its rare target lipid PI(3,4,5)P3 (17). Dissociation rate measurements, which can be carried out using both single-molecule and bulk methods, are compared to verify that the protein bound state lifetime is independent of the vastly different protein concentrations and bilayer geometries used for the two methods, thereby validating the single molecule measurements. TIRFM measurement and statistical analysis of diffusion and rare molecular events are then used to gain insight into the dynamic behavior of this protein domain on the membrane surface, both in the absence and presence of PS, the predominant, native background anionic lipid.
Lipid bilayers supported on a clean glass surface were used to create a membrane interface for GRP1-PH docking suitable for single-molecule TIRFM. For this initial study, simple mixtures of phospholipids with matched, monounsaturated, dioleoyl acyl chains were chosen to ensure that supported bilayers were as uniform and fluid as possible (Table 1). The resulting simple bilayers are ideal for a first study of the PH domain-membrane interaction, leaving important complexities such as leaflet asymmetry (40) and lipid microdomains (26,39,41) for future investigations. To maximize the homogeneity of the supported bilayer, the glass surface was etched by aggressive chemical treatment before bilayer deposition. This treatment also removes organic compounds from the glass surface, providing an extremely low level of background fluorescence ideal for single-molecule measurements.
For single-molecule fluorescence studies of GRP1-PH, the Cy3-derivative Alexa Fluor 555 (AF555) was conjugated to an engineered cysteine at position 303, well outside of the PI(3,4,5)P3 binding pocket. To directly test the effect of the probe on membrane binding, the labeled protein, hereafter referred to as AF555-GRP1PH, was compared to wild-type using established bulk fluorescence assays to determine the equilibrium affinity and kinetics of binding to PI(3,4,5)P3-containing membranes.
Characterization of AF555-labeled GRP1 PH domain docking to membranes: bulk measurements
AF555-GRP1PH exhibits a binding affinity for membranes containing PI(3,4,5)P3 similar to the wild-type domain, both in the absence of PS and in the presence of a near-native PS mole density. PI(3,4,5)P3 affinity was measured using an established assay that uses competitive displacement from membrane induced by inositol (1,2,3,4,5,6)-hexakisphosphate (IP6) to measure KD values in the nanomolar range (17). The assay is based on measurement of protein-to-membrane FRET between a donor fluorophore on the protein (tryptophan for wild-type, AF555 for labeled mutant) and an acceptor conjugated to lipid headgroups in the bilayer (dansyl for wild-type, Texas Red for labeled mutant). Titration of the competitive inhibitor (IP6) into preassembled protein-membrane complexes induces loss of FRET with a characteristic inhibition constant, KI. The KD of protein-membrane binding can then be calculated from this KI value and the independently determined affinity of the free protein for the competitive inhibitor (see Materials and Methods). The results of these measurements are shown in Supporting Material, Fig. S1, and summarized in Table 2. The fluorophore-labeled domain has an affinity for PI(3,4,5)P3 in pure PC within error of wild-type, and for PI(3,4,5)P3 in 3:1 PC/PS within 4-fold of wild-type. Notably, as observed for wild-type, the presence of background PS significantly enhances the target lipid affinity of the labeled domain, yielding 2.5-fold tighter binding (Table 2). This PS-associated affinity enhancement is modestly lower than the 10-fold enhancement previously reported for wild-type, but nevertheless shows that the labeled domain retains a functional electrostatic search mechanism that enhances target lipid association (17).
Table 2.
Thermodynamic and kinetic binding parameters from bulk experiments
| Protein domain | Lipid composition | KD (nM)∗ | kon (μM−1 s−1)† | koff (s−1)‡ | koff/kon (nM) |
|---|---|---|---|---|---|
| WT GRP1-PH | PC/PIP3 | 60 ± 10 | 2.6 ± 0.2 | 0.38 ± 0.01 | 150 ± 10 |
| WT GRP1-PH | PC/PS/PIP3 | 5 ± 1 | 19 ± 1 | 0.22 ± 0.01 | 12 ± 1 |
| AF555-GRP1PH | PC/PIP3 | 50 ± 10 | 6 ± 4 | 0.75 ± 0.03 | 130 ± 80 |
| AF555-GRP1PH | PC/PS/PIP3 | 19 ± 5 | 13 ± 7 | 0.42 ± 0.02 | 30 ± 10 |
KD values calculated from equilibrium IP6 competition titrations using Eq. 2. Measured IP6 affinities: WT 4.3 ± 0.8 μM, AF555-GRP1PH 24 ± 5 μM.
Biexponential association kinetics were observed, and the fast rate constant is reported, consistent with previous studies (17).
To improve signal/noise, membranes containing 3% PI(3,4,5)P3 were used for measurement of labeled GRP1-PH koff values, whereas 2% PI(3,4,5)P3 was used elsewhere. Corresponding measurements of koff using 2% PI(3,4,5)P3 yielded rate constants of 0.8 ± 0.2 for PC/PIP3 and 0.38 ± 0.02 for PC/PS/PIP3.
Bulk kinetic measurements further reveal that the AF555 probe has only minor effects on target membrane association and dissociation kinetics. Association rates were measured by the appearance of protein-to-membrane FRET in stopped-flow reactions where protein was rapidly mixed with lipid vesicles (Table 2, Fig. S2). Dissociation rates were similarly measured via the loss of protein-to-membrane FRET after rapid addition of excess competitive inhibitor to preformed PH domain-lipid complexes (Table 2, Fig. S3). In the latter experiment, IP6 serves as a competitive trapping agent that saturates the PH domains as they leave the membrane surface, thereby preventing rebinding to PI(3,4,5)P3. Overall, the rate constants kon and koff each differ by no more than a factor of 3 between wild-type and labeled protein. The koff/kon ratios indicate a 4-fold enhancement of PI(3,4,5)P3 binding for AF555-GRP1PH in the presence of PS and a 12-fold enhancement for wild-type, in reasonable agreement with the equilibrium measurements and again showing that both proteins possess the electrostatic search mechanism. In summary, bulk equilibrium and kinetic measurements indicate that fluorophore attachment modestly alters affinities but retains the electrostatic search mechanism of the GRP1 PH domain.
TIRFM detection of single lipid and GRP1 PH domain molecules in supported bilayers
Single-molecule diffusion measurements of a fluorescent phospholipid indicate that supported bilayers of the compositions used in this study are uniform and fluid. Glass-supported lipid bilayers were formed by vesicle fusion onto ultra-cleaned coverslips (see Materials and Methods). When the bilayer is doped with fluorescent phospholipid (lissamine rhodamine B-DOPE, or LRB-DOPE, at 150 ppb), rapidly diffusing fluorescent spots are readily observable by TIRFM (Movie S1). Using single-particle tracking, the lateral diffusion constant of the fluorescent phospholipid was measured to be 3.0 ± 0.2 μm2/s in PC/PIP3 bilayers and 2.7 ± 0.2 μm2/s in PC/PS/PIP3 bilayers (Fig. 1; Table 3). These diffusion constants are within error of each other, indicating that lipid diffusion is independent of the bilayer compositions used in this study. Moreover, the observed values are well within the range of lipid diffusion constants in supported bilayers reported previously (26,38,39,42–44)).
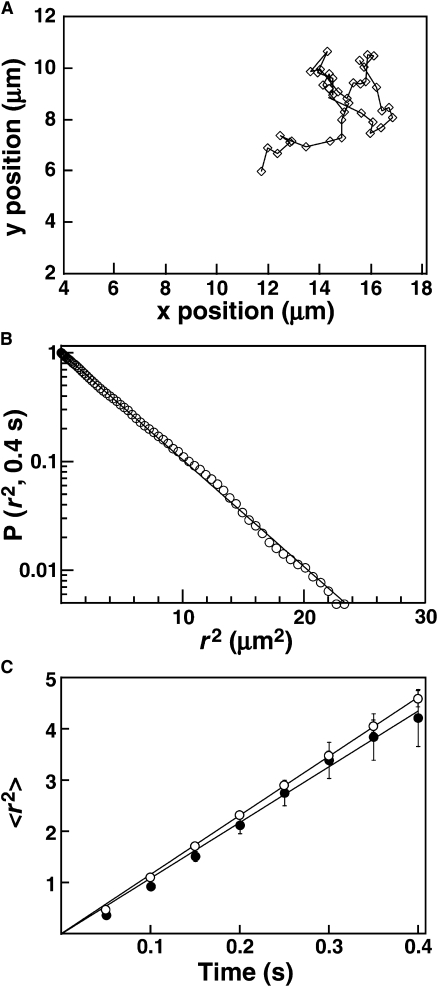
Figure 1.
Diffusion of lipids in supported bilayers. Supported lipid bilayers containing 150 ppb fluorescent lipid (LRB-DOPE) were prepared as described in Materials and Methods. (A) Representative diffusion track for a single lipid particle in PC/PIP3, acquired with an exposure of 50 ms/frame. (B) Representative cumulative distribution plot of square displacement (r2) for one movie (pooled data from 398 single particle tracks) of single lipid molecules diffusing in PC/PIP3. The vertical axis represents the probability of a particle having a square displacement of at least r2 in images acquired 8 frames apart. Solid line represents the best-fit to a single-exponential distribution, according to Eq. 6. (C) Plots of mean square displacement (〈r2〉) versus time interval for single lipid molecules in PC/PIP3 (open) or PC/PS/PIP3 (solid). The value for each point was determined by fitting a square displacement distribution from a single movie as illustrated in B. The solid line represents a linear fit according to Eq. 8. Error bars were calculated as described in Qian et al. (58).
Table 3.
Diffusion rates of lipids and PH domains
| Lipid composition | Diffusing species | D (μm2/s)∗ |
|---|---|---|
| PC/PIP3 | Lipid | 3.0 ± 0.2 |
| PC/PIP3 | GRP1 PH domain | 3.0 ± 0.3 |
| PC/PS/PIP3 | Lipid | 2.7 ± 0.2 |
| PC/PS/PIP3 | GRP1 PH domain | 2.0 ± 0.2 |
Values shown are the mean ± SD from at least six movies (20 ms or 50 ms frame rate, 2000 frames each) using at least three independent replicate samples.
In the lipid diffusion measurements, some particles are occasionally observed that i), are much brighter than the other fluorescent spots, ii), are stationary or very slowly moving, and iii), tend to appear suddenly within the field of view rather than diffusing into view from the edge of the field. Fluorescent particles exhibiting these characteristics are observed only in the LRB-DOPE samples, and are thus contaminants of the lipid probe. When these easily identified contaminants are eliminated from diffusion analysis by application of simple exclusion rules (see Materials and Methods), lipid diffusion fits well to a single population distribution, indicating that the two leaflets of the supported bilayer exhibit virtually identical lipid diffusion constants and together represent a single, homogeneous lipid phase (Fig. 1 A), as observed previously in other supported bilayer studies (43,45). Plots of mean square displacement versus time were linear, indicating no barriers to free diffusion are present on the scale of several micrometers (Fig. 1 B) (27,38). Thus, the bilayers used in this study are uniform and fluid, and provide an excellent interaction surface for peripheral membrane binding protein domains.
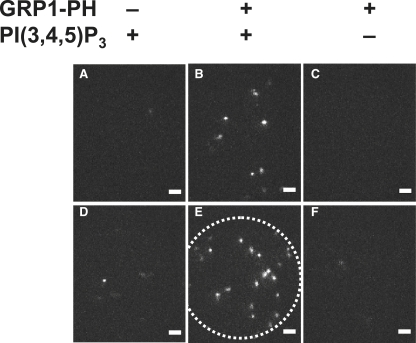
Single molecules of AF555-GRP1PH are observed to bind to supported lipid bilayers in a PI(3,4,5)P3-dependent manner (Fig. 2). Four types of supported bilayers were prepared: PC, PC/PIP3, PC/PS, and PC/PS/PIP3 (Table 1). Upon imaging in sample buffer before addition of fluorescent components, dimly fluorescent spots are observed only occasionally on all four types of bilayers (Fig. 2, A and D, and data not shown). This is indicative of a low level of fluorescent contamination, yielding a density of spots at least 10-fold smaller than observed after addition of 100 pM AF555-GRP1PH to membranes containing PI(3,4,5)P3 (Fig. 2, B and E; Movie S2 and Movie S3). By contrast, addition of AF555-GRP1PH to bilayers lacking PI(3,4,5)P3 yields no additional fluorescent particles (Fig. 2, C and F). This is consistent with our previous report using bulk FRET experiments that showed no GRP1-PH binding to PC/PS vesicles, due to a very low affinity of the PH domain for PS in the absence of the target PI(3,4,5)P3 lipid (17). Here, most of the bright fluorescent particles observed on addition of AF555-GRP1PH to PI(3,4,5)P3 seem to diffuse freely on the surface while bound to the bilayer. These mobile fluorescent protein particles appear and disappear in single steps, suggesting that single protein molecules are binding and being released from the membrane surface on the timescale of the measurement (under these conditions dissociation is ∼10-fold faster than bleaching; see below). Importantly, single-step disappearance of particles is maintained even when illumination power is increased to an intensity at which photobleaching becomes faster than dissociation. This observation confirms that the observed species are indeed single molecules. Thus, in these measurements, individual molecules of AF555-GRP1PH are observed to bind to, diffuse laterally on, and dissociate from membranes containing PI(3,4,5)P3.
Figure 2.
PI(3,4,5)P3-specific binding of AF555-GRP1PH to supported bilayers. Supported lipid bilayers were prepared containing (A and B) PC/PIP3, (C) PC, (D and E) PC/PS/PIP3, or (F) PC/PS. (A and D) Representative images of the bilayer imaged in buffer, before addition of protein. (B, C, E, and F) Representative images of the bilayer after addition of 100 pM AF555-GRP1PH. The field of illumination by the laser is outlined in E. Illumination intensity is highest near the center of the field and dim near the edge, leading to positional variation in particle intensity. Exposure time for all panels was 20 ms. Scale bars = 2 μm.
Single-molecule diffusion of membrane-bound GRP1 PH domain
Tracking of individual diffusing PH domains shows both free and constrained diffusional states, even after application of exclusion rules to eliminate fluorescent contaminants (see Materials and Methods). Although the high mobility particles are more common, low mobility particles are clearly apparent in movies of AF555-GRP1PH on either PC/PIP3 or PC/PS/PIP3, and particles are sometimes observed to transition between the two states (Movie S2 and Movie S3, circles). The diffusion and relative populations of these two states were quantified by single particle tracking, followed by a two-component analysis using established methods (38,42). Probability density plots of square displacement fit significantly better to two-component than single-component equations (Fig. 3 B). The mean square displacement of the faster, major component increases linearly with increasing time interval, indicating barrier-free diffusion for the mobile protein as well as lipid. The minor component represents 5%–15% of the membrane-associated protein and exhibits a diffusion constant <0.1 μm2/s. Similar low-mobility or immobilized states have been observed previously for lipid and protein diffusion on supported lipid bilayers, and are attributed to defects in the lipid bilayer or to glass spikes protruding through the bilayer (34,38). The chemically etched coverslips used were imaged with AFM, and glass spikes were occasionally observed, consistent with a minor contribution to protein mobility (data not shown). Because these low mobility states are not relevant to the membrane targeting mechanism of the GRP1 PH domain, all further diffusion analysis focuses on the major, mobile component of GRP1 PH domain diffusion.
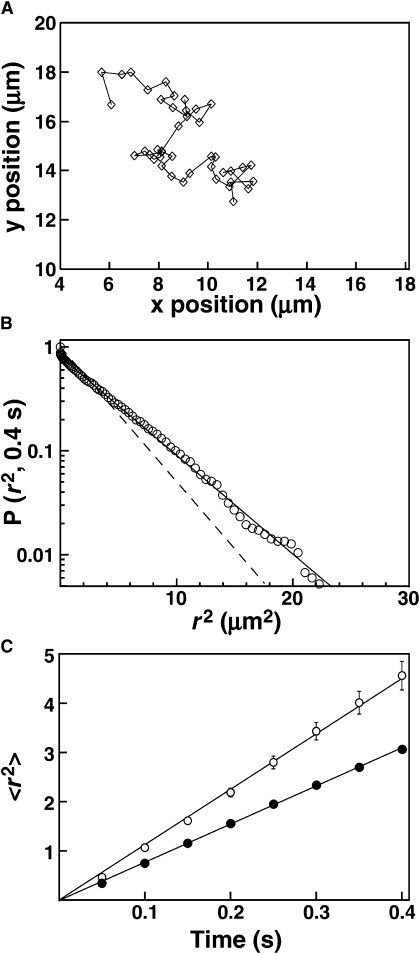
Figure 3.
Lateral diffusion of AF555-GRP1PH on supported lipid bilayers. (A) Representative diffusion track for a single fluorescent PH domain on PC/PIP3, acquired with a 50 ms exposure per frame. (B) Representative cumulative distribution plot of square displacement (r2) for PH domain on PC/PIP3, from one representative movie (94 tracks pooled). The vertical axis represents the probability of a particle having a square displacement of at least r2 in images acquired 8 frames apart (50 ms/frame). Dashed line represents the best-fit to a single-exponential distribution, according to Eq. 6. Solid line represents the best-fit to a double-exponential (two-population) distribution, according to Eq. 7. (C) Plots of mean-square displacement (〈r2〉) versus time interval for PH domain on PC/PIP3 (open) or PC/PS/PIP3 (solid). The value for each point was determined by fitting a square displacement distribution from a single movie as illustrated in B. The solid line represents a linear fit according to Eq. 8. Error bars were calculated as described in Qian et al. (58). Where not visible, error bars are smaller than the data points.
Strikingly, when bound to PI(3,4,5)P3 embedded in membranes lacking the background anionic lipid PS, AF555-GRP1PH diffuses at the same rate, within error, as the fluorescent lipid LRB-DOPE. By contrast, the PH domain diffuses significantly more slowly on bilayers also containing PS. Thus, AF555-GRP1PH exhibits diffusion constants of 3.0 ± 0.3 μm2/s on PC/PIP3 and 2.0 ± 0.2 μm2/s on PC/PS/PIP3 (Table 3, Fig. 3 C). It is highly unlikely that the reduced protein diffusion is due to lipid-lipid interactions between PI(3,4,5)P3 and PS, because i), the PI(3,4,5)P3 headgroup is nearly completely engulfed by the PH domain (14), ii), both lipids are negatively charged, and iii), the PI(3,4,5)P3 acyl chains are matched to the other lipids in the bilayer, which exhibit identical diffusion in the presence and absence of PS (Fig. 1). Rather, the change in protein diffusion suggests an interaction of the PI(3,4,5)P3-docked protein with PS. Importantly, PS in the absence of PI(3,4,5)P3 is insufficient to target the GRP1 PH domain to the membrane (Fig. 2 and (17)). Overall, the results indicate that GRP1 PH domain docked to PI(3,4,5)P3 on a bilayer surface can simultaneously associate with PS at one or more low affinity binding sites.
Single-molecule measurement of dissociation kinetics for membrane-bound GRP1 PH domain
Dissociation rates of single molecules can be measured by tracking many particles from the time each appears on the surface until the time it disappears. A probability density distribution of the resulting dwell times is an exponential distribution with a decay constant equal to the sum of the dissociation and photobleaching rate constants (29). The photobleaching rate of AF555-GRP1PH was measured explicitly by monitoring the disappearance of immobilized particles under conditions identical to those used for dissociation measurements. A high density of protein was applied to the membrane, and IP6 was added to competitively displace all freely diffusing PH domains from the membrane surface. This IP6 treatment did not displace PH domains trapped in immobile states, which remained on the surface for over 30 min. A previously unexposed region of the surface was imaged until at least 95% of the immobilized spots had disappeared, and the amount of time to disappearance was recorded for all immobile particles that bleached in a single step. Thus, for this photobleaching measurement, analysis focused on particles that, in other experiments, would have been prebleached or excluded by thresholding. The density of particles was sufficiently high that contributions from immobile contaminants were negligible. The photobleaching rate increased linearly with incident laser power, and was 0.05 ± 0.02 s−1 at the power densities used for dissociation measurements of AF555-GRP1PH (Fig. S4). All measured dissociation rates of AF555-GRP1PH are 8- to 16-fold faster than this rate, thus photobleaching is a minor contribution to the apparent dissociation kinetics.
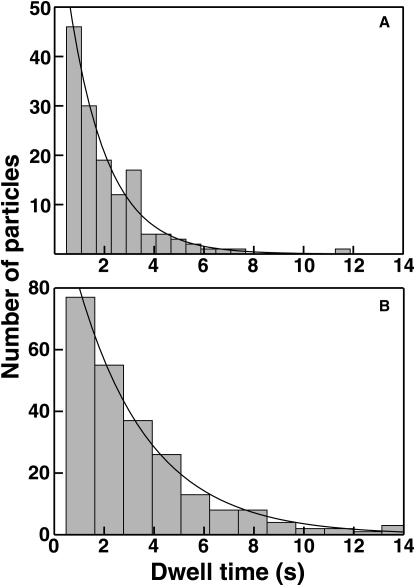
For GRP1 PH domain, the single-molecule TIRFM measurements yielded dissociation rate constants within error of those observed in bulk measurements. Mobile molecules of AF555-GRP1PH were monitored on supported bilayers at densities optimal for dissociation measurement (Movie S4 and Movie S5), and particles were tracked automatically. In the analysis of this measurement, it was found that automated particle tracking software occasionally failed to correctly link particles from frame to frame. This occurred frequently enough that the dissociation rate was overestimated by up to a factor of 2 (data not shown). Thus, all tracks were validated by hand, and individual dwell times were adjusted as necessary to ensure that such tracking artifacts did not contribute to the measured distribution. Only particles whose entire tracks fell within the field of view were included in the dwell time analysis; i.e., molecules whose tracks began or ended at the edge of the view area were discarded for this measurement. Single-exponential fits to the resulting dwell time distributions yield dissociation rate constants of 0.4 ± 0.1 s−1 from PC/PS/PIP3 and 0.7 ± 0.1 s−1 from PC/PIP3 (Fig. 4, Table 4). Relative to these rates, photobleaching is slow, and in fact is within the uncertainty of the dissociation measurement. To independently verify that photobleaching did not significantly contribute to the distribution of dwell times, analogous measurements were made at a 2-fold higher laser power for dissociation from PC/PS/PIP3 (the longest bound state lifetime). The resulting koff value was within error of that measured under standard conditions, confirming that photobleaching is negligible relative to dissociation. Thus, these measurements show that the off-rate, or, equivalently, the bound state lifetime, is indistinguishable for GRP1 PH domain docked to PI(3,4,5)P3-containing vesicles and supported bilayers. Furthermore, the measurements demonstrate the utility of single-molecule TIRFM for the measurement of protein-membrane dissociation rates without perturbing equilibrium.
Figure 4.
Dissociation rates of AF555-GRP1PH from PI(3,4,5)P3, determined from single molecule measurements. The length of time each PH domain molecule was visible on the surface (dwell time) was determined as described in Materials and Methods. Histograms of these dwell times are shown for (A) 20 pM AF555-GRP1PH on PC/PIP3, and (B) 7 pM AF555-GRP1PH on PC/PS/PIP3. Solid curves are best-fits to single-exponential decays according to Eq. 9, from which the koff values in Table 4 were obtained.
Table 4.
Dissociation rates measured from single-molecule experiments
| Lipid composition | IP6 (mM) | koff (s−1)∗ |
|---|---|---|
| PC/PIP3 | 0 | 0.7 ± 0.1 |
| PC/PIP3 | 0.1† | 0.8 ± 0.2 |
| PC/PS/PIP3 | 0 | 0.4 ± 0.1 |
| PC/PS/PIP3 | 0.5† | 0.6 ± 0.1 |
Values shown are the mean ± SD from three independent samples, from single-exponential fits to dwell time distributions of at least 33 tracks per sample.
Observation of microdissociation and rebinding events
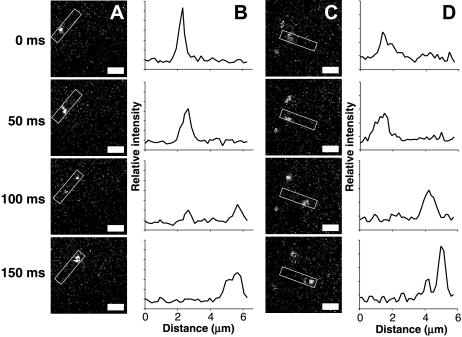
One advantage of single-molecule techniques is the ability to directly observe events that may remain hidden in bulk measurements. For a protein molecule bound to a membrane, dissociation enables diffusion through solution, sometimes followed by rapid rebinding to membrane when the local concentration of target is high near the membrane surface (46). This study detects kinetic events we term “microdissociation and rebinding” for AF555-GRP1PH using single-molecule TIRFM. [Note that we avoid the term “hop diffusion” already used in the literature to describe a different phenomenon–translocation of membrane-embedded molecules between regions of cell membrane confined by the cytoskeleton (27)]. During microdissociation and rebinding events, illustrated in Fig. 5, the particle moves in an apparent “jump” that is farther than expected for membrane-bound diffusion. If a microdissociation and rebinding event occurs near the middle of the 50-ms exposure of a given frame, the molecule appears as two spots in that frame, each with roughly half the intensity of a typical protein molecule (Fig. 5 A and B). If an event occurs near the beginning or end of an exposure, the particle may simply seem to jump a large distance between frames (Fig. 5 C). In either case, it is important to note that a given observed jump may represent one of three possibilities: i), microdissociation and rebinding, ii), an unusually large two-dimensional diffusion step from the tail of the step size distribution, or iii), near-simultaneous association and dissociation of distinct molecules. Statistical analysis was used to distinguish whether the rate of observed jumps was significantly greater than that expected for the latter two classes of random events. Here, jumps were operationally defined as possessing a length between defined rmin and rmax values, where the minimum distance threshold rmin corresponds to the longest 0.5% of the two-dimensional diffusion steps, and the maximum distance threshold rmax is set to a value, 5 μm, greater than the average distance a microdissociated PH domain is likely to diffuse in a single 50-ms frame. This jump analysis excludes steps shorter than rmin or longer than rmax, because such steps will often represent simple diffusional events or the near-simultaneous association and dissociation of two different molecules, respectively (see Materials and Methods).
Figure 5.
Direct observation of single molecule jumps. A series of screen shots is shown for a representative GRP1 PH domain jump event occurring (A) near the middle of a 50-ms exposure frame, or (C) near the end of a frame. (B and D) Line scans integrating along the path of the jump, in the regions highlighted with boxes in A and C. The two sets of images are from separate experiments, thus illumination intensity and particle brightness may vary slightly. Scale bars = 2 μm.
For AF555-GRP1PH, the number of observed jumps is 2.6-fold greater than the number of expected random jumps on PC/PS/PIP3, but only 0.5-fold greater than the number expected from random events on PC/PIP3 (Table 5). Histograms of observed jump distances clearly show that a large fraction of the jumps on PS-containing membranes are longer than expected for simple two-dimensional diffusion in the bilayer (Fig. S5). Thus, most of the observed jumps on membranes containing both PI(3,4,5)P3 and PS are true microdissociation and rebinding events, and removal of the PS causes the loss of most rebinding events as predicted by the electrostatic search hypothesis.
Table 5.
Frequency of jump events observed
| PC/PIP3 | PC/PS/PIP3 | |
|---|---|---|
| Minimum threshold distance (μm)∗ | 1.783 | 1.456 |
| Maximum threshold distance (μm) | 5.0 | 5.0 |
| Total number of tracks analyzed | 266 | 244 |
| Total number of steps analyzed | 8041 | 8850 |
| Expected random 2-D steps | 40 | 44 |
| Expected randomly correlated trajectories† | 2 | 3 |
| Total expected random jumps | 42 | 47 |
| Total observed jumps | 62 | 123 |
Minimum distance was calculated as the distance equivalent to the top 0.5% of the expected distribution of step sizes for a 50-ms time window (Eq. 10) using diffusion constants reported in Table 3.
Number of expected random correlations was calculated as described in Materials and Methods.
In principle, microdissociation and rebinding could increase the apparent bound-state lifetime, or decrease the apparent dissociation rate, if the probability of rebinding is sufficiently high. For AF555-GRP1PH on PC/PS/PIP3, roughly 76 microdissociation and rebinding events are observed for 244 tracked particles (Table 5). This corresponds to roughly 30% of the total trajectories. A simple simulation indicates that when rebinding is blocked by addition of IP6, which occupies the PIP3 binding site and thereby prevents rebinding, the single molecule off-rate should increase from 0.4 s−1 to 0.6 s−1 (Fig. S6). Indeed, measurement of the single molecule dissociation kinetics in the absence and presence of IP6 yields off-rates of 0.4 ± 0.1 s−1 and 0.6 ± 0.1 s−1, respectively (Table 4). Although any difference between these off-rates is near the limits of our current precision, the findings are consistent with the predictions of the microdissociation and rebinding model. Moreover, the observation that the off-rate measured in the absence of PS (0.7 ± 0.1 s−1) is within error of the off-rate measured in the presence of the rebinding blocker suggests that the electrostatic search mechanism, which requires background PS, is essential for efficient rebinding (Table 4). Overall, the findings indicate that GRP1 PH microdissociation and rebinding events are observable both directly, by tracking analysis of jumps, and indirectly, by effects on dissociation kinetics. Thus, in principle, microdissociation and rebinding could be detected by bulk kinetic measurements of dissociation rates. However, because the effect of jumps on dissociation kinetics is subtle and the observation is indirect, the single molecule approach, which analyzes both jumps and dissociation kinetics, is far superior to bulk analysis.
Discussion
The measurements of GRP1 PH domain docking and diffusion on supported lipid bilayers show that single-molecule measurements made with supported bilayers yield the same membrane residency times, within error, as bulk measurements made with sonicated vesicles. In addition, the single molecule approach shows three features of the membrane-docked PH domain undetected previously: i), in the absence of background anionic lipids, GRP1 PH domain bound to its membrane-embedded target lipid PI(3,4,5)P3 diffuses at the same rate as a single lipid molecule, ii), GRP1 PH domain bound to its target lipid can interact with PS via one or more secondary binding sites, which when occupied slow PH domain diffusion on the membrane surface, and iii), an electrostatic search mechanism not only speeds GRP1 PH domain binding to its rare target lipid PI(3,4,5)P3 during membrane association, but also promotes target lipid rebinding events on microdissociation. These observations provide what we believe are new mechanistic insights, inaccessible via standard bulk methods, into the target membrane docking reaction. Overall, this study shows the power of in vitro single-molecule TIRFM to analyze the molecular mechanisms of peripheral proteins docking to their target membranes.
Single-molecule measurement of peripheral membrane protein docking and diffusion
Diffusion measurements provide important information about the dynamics of the supported bilayer. Lipids in supported bilayers are known to diffuse ∼5-fold more slowly than in unsupported bilayers or vesicles (34,42). This is likely due to weak frictional coupling between the bilayer and the glass support through the intervening water layer (47). Numerous studies have shown that strong frictional coupling exists between monolayers of a supported bilayer, resulting in diffusion constants that are usually indistinguishable in the two leaflets (43,45), and differ by no more than a factor of 2 even when the proximal leaflet is connected to the substrate by a flexible linker (26). Consistent with these observations, this study measures a single diffusion constant of 3 μm2/s for lipid diffusion in supported bilayers (Table 3, Fig. 1). This value is fully consistent with the previously observed range of lipid diffusion constants (1–10 μm2/s) observed in various supported bilayer systems (26,38,39,42–44).
Supported lipid bilayers are excellent membrane substrates for single-molecule TIRFM studies of PH domains and other peripheral membrane binding proteins (48). The protein is added to the solution after the bilayer is deposited on glass, ensuring that it is excluded from the glass-exposed leaflet and docks only to the solvent-exposed leaflet. However, support-induced membrane defects can lead to small immobile fractions of protein or lipid. Care must be taken to exclude these immobile particles from analysis: this study focuses only on the mobile particles. For the mobile state, both simple diffusion and established theory for supported bilayers predict that diffusion will slow as the number of membrane contacts increases, due to increased drag from the high-viscosity bilayer (47,49). Thus, in principle, diffusion measurements of peripheral membrane proteins can provide meaningful information on protein-membrane contacts.
The dissociation kinetics of single protein molecules from the surface can provide additional information about protein-membrane contacts. However, measurement of such kinetics requires carefully optimized parameters, and is thus limited to experiments wherein certain requirements can be met. For example, the membrane-bound fluorescent protein must be bound at sufficiently high density, and possess one or more distinguishing features, to prevent fluorescent contaminants from dominating the analysis. At the same time, however, the protein density must be low enough that fluorescent particles do not overlap with each other too often, so that individual particle tracks can be distinguished. The maximum measurable off-rate is governed by the maximum video frame rate, which is ∼100 s−1 for the system used here. Limits to measurement of slow off rates are a), photobleaching, and b), the amount of time a typical particle spends in the field of view before diffusing out. The latter factor is determined by both the size of the field of view (in this case, ∼490 μm2) and the diffusion rate, with slower dissociation rates measurable for slower-diffusing particles. For this field of view and an organic fluorophore such as AF555, dissociation rates approaching 0.1 s−1 are measurable. Thus, when the other requirements are met, this method can directly measure membrane dissociation rates spanning nearly three orders of magnitude without perturbing the binding equilibrium.
An obvious advantage of any single-molecule measurement is the requirement of vanishingly small amounts of labeled macromolecule. As with any study using labeled protein, care must be taken to ensure that the extrinsic probe does not destroy the activity being investigated. The second-generation, red-shifted fluorophores with the best photophysical properties for single-molecule studies are bulky aromatics that typically possess a net charge, thus it is important to measure their effects on activity. In this study, probe effects were tested directly by producing large enough amounts of labeled protein for bulk measurements in parallel with the single-molecule measurements, and were found to be detectable but small.
Diffusion of a PI(3,4,5)P3-docked PH domain in a supported bilayer
Strikingly, when bound to PC/PIP3 bilayers, the GRP1 PH domain diffuses at a rate indistinguishable from free lipid (Figs. 1 and 3 A). This observation suggests that the contact between the PH domain and bilayer is dominated by the high-affinity interaction between the protein and the headgroup of its target lipid PI(3,4,5)P3. This headgroup, like that of other PIP lipids, is significantly larger than the non-PIP headgroups of the plasma membrane inner leaflet, projecting as much as 10 Å out of the bilayer surface into solvent (21). Structural studies of PH domain-PIP complexes suggest that the protein can almost completely engulf the headgroup in its specific binding pocket without penetrating deeply into the bilayer (14,15). Indeed, many PH domains that target PIP lipids, including GRP1 PH domain, produce only minor increases in lateral surface pressure when bound to membrane monolayers (19,20). The observation that the GRP1 PH domain-PI(3,4,5)P3 complex experiences essentially the same lateral friction in the membrane as a single lipid molecule strongly suggests that diffusion is governed almost entirely by the lipid acyl chains.
When the PI(3,4,5)P3-associated GRP1 PH domain binds simultaneously to PS in the same membrane, its diffusion is significantly slowed. The 33% decrease in diffusion constant triggered by a physiological membrane density of PS suggests that one or more secondary binding sites for PS exist on the surface of GRP1 PH domain. Furthermore, PS coordination by the PI(3,4,5)P3-docked domain decreases the membrane dissociation rate by a factor of 2, as observed for both AF555-labeled and wild-type PH domain (Table 2, Fig. S3). PS binding likely involves one or more cationic side chains exposed on the membrane-proximal surface of the PH domain. Examination of the protein surface surrounding the PI(3,4,5)P3 docking cleft shows seven candidate Arg or Lys residues. Further studies are needed to determine whether the site is saturated by native PS levels when the PH domain is bound to its target PI(3,4,5)P3 lipid, and whether the site is specific for PS or also binds other anionic lipids. Further work will also probe which of the seven cationic residues comprise the secondary PS site and generate its effects on diffusion and dissociation kinetics, and whether these same residues, or different ones, underlie the electrostatic search process that occurs during PI(3,4,5)P3 docking.
Notably, the PS affinity of the secondary binding site is too low to drive detectable equilibrium binding to membranes lacking target lipid. Indeed, our previously published FRET measurements (17) and current findings provide no evidence for GRP1-PH binding to PC/PS membranes in the absence of PI(3,4,5)P3. Furthermore, our ensemble TIRFM measurements with ∼100 nM AF555-GRP1PH indicate that the same background fluorescence intensity is observed in the evanescent field region of both PC and PC/PS bilayers (data not shown). Together, these observations indicate that if the secondary site has any direct interaction with PS in the absence of PI(3,4,5)P3, it must be both transient and weak (lifetime << 20 ms, KD > 10 mM). By contrast, for a PH domain bound to PI(3,4,5)P3 on the surface of a membrane composed of 25 mol % PS, the effective local PS concentration is extremely high (>100 mM). Thus, it is reasonable to propose that the secondary site is significantly occupied by PS only when the PH domain is bound to target lipid on the membrane surface, thereby altering the interaction of the GRP1 PH domain with the bilayer surface and slowing PH domain surface diffusion.
The presence of secondary lipid binding sites is an emerging theme for membrane-targeting protein domains. Biophysical methods of interrogating membrane interactions, along with the use of more complex, physiologically relevant lipid mixtures, have aided in the recent identification of secondary lipid binding sites in the PKCα C2 domain, the synaptotagmin C2B domain, the p47phox PX domain, and the SOS PH domain (50–54). In cells, such secondary sites can confer additional specificity to membrane targeting, and can provide regulation of binding affinity, enzymatic activity, and membrane residency time by altering the orientation of the protein relative to the bilayer surface, as well as the rate of membrane dissociation (21,55).
Implications of microdissociation and rebinding for GRP1 search mechanisms
The findings include direct observation of transient microdissociation and rebinding events for single molecules of GRP1 PH domain. Rebinding is a general property of surface-associated proteins, and evidence for rebinding of other proteins after dissociation has been provided previously by ensemble TIRFM measurements (56). The rebinding of GRP1 PH domain is likely to be dominated by the same electrostatic search mechanism that enhances the PI(3,4,5)P3 on-rate. In the simplest physical model, illustrated in Fig. 6, PI(3,4,5)P3 binding and rebinding rates are enhanced by the same local, electrostatic search mechanism, in which transient interactions with anionic background lipids, primarily PS, increase the rate of PH domain collisions with the membrane surface (Fig. 6). The increased membrane collision rate speeds binding to the rare target lipid PI(3,4,5)P3, but the transient background lipid contacts do not result in detectable levels of membrane binding in the absence of target lipid. The resulting dependence of membrane targeting on the presence of target lipid is essential to cellular function, since the PH domain drives plasma membrane docking only during a PI(3,4,5)P3 signal.
Figure 6.
Working model for the role of electrostatic searching in rare target lipid acquisition, microdissociation and rebinding. Shown is a simplified view of a PH domain (green) with its positively charged PI(3,4,5)P3 binding face (blue), together with a lipid bilayer containing high levels of anionic background lipids (red, primarily PS) and a rare target lipid (purple). As the PH domain approaches the membrane, a long-range electrostatic interaction between the PI(3,4,5)P3 binding face and the anionic membrane surface drives rotational steering. Subsequently, the positive face of the PH domain interacts weakly and transiently with anionic background lipids as it “bounces” multiple times along the membrane surface. Usually, the PH domain returns to solution, but occasionally the domain encounters and binds a PI(3,4,5)P3 target molecule. The residency time of the PI(3,4,5)P3-bound GRP1 PH domain is typically over 1 s, during which time the lipid-bound domain undergoes two-dimensional diffusion on the membrane surface (not shown). After PI(3,4,5)P3 dissociation, the PH domain may bounce along the anionic membrane surface before diffusing back into bulk solution. During a typical microdissociation and rebinding event, the domain diffuses up to several microns through solution (a large distance on the scale of this figure, as indicated by the break in the membrane) before returning to the same membrane. The membrane-proximal domain then engages in a new electrostatic search that yields binding to a second PI(3,4,5)P3 target lipid at location L2, several microns away from the original target lipid at location L1. Ultimately, the domain undergoes a macroscopic dissociation event and returns to bulk aqueous solution.
During a microdissociation and rebinding event, the PH domain dissociates from PI(3,4,5)P3 and diffuses through aqueous solution, but instead of diffusing away from the membrane as in macrodissociation, the protein returns to the membrane and finds a second PI(3,4,5)P3 molecule via another electrostatic search (Fig. 6). It is clear that the microdissociation and rebinding events detected here by single molecule tracking, which yield jumps over micrometer distances, involve aqueous diffusion rather than rapid diffusion over the membrane surface, because the PH domain would encounter many PI(3,4,5)P3 molecules during such a surface scan (on average, PI(3,4,5)P3 molecules are ∼6 nm apart in our membranes). Moreover, the observation that the PH domain disappears from TIRFM detection during the jump is consistent with rapid aqueous diffusion, rather than slow, lipid-bound diffusion. Strong evidence supporting the proposal that electrostatic searching is essential to efficient rebinding is provided by the observation that rebinding is stimulated by the anionic lipid PS, and the degree of PS stimulation is nearly identical for the on-rate and the rebinding rate (2- to 4-fold, Tables 2 and 5). This model predicts that, in the presence of PS, the rebinding rate of the wild-type PH domain will be a further 4-fold faster than that of the probe-labeled PH domain used herein, because the probe slightly weakens the electrostatic search mechanism, such that the PS stimulation of the PI(3,4,5)P3 on-rate is 4-fold higher for the native domain than for the probe-labeled domain (Table 2).
The current rebinding hypothesis makes a prediction that will be tested in a future two-color TIRFM system (57): in a mixture of PH domains labeled with two different fluors, most of the operationally identified “jumps” are predicted to retain the same color as required by true microdissociation and rebinding of the same particle, except in the absence of PS when relative proportions of same-color and different-color jumps will approach random behavior.
For GRP1 PH domain, the most important functional effect of the electrostatic search mechanism is to speed binding to PI(3,4,5)P3 by 10-fold during the membrane association reaction (17). In cells, this 10-fold rate enhancement is predicted to speed the domain response time from 20 s to 2 s, which is essential for the rapid response time of chemotaxing leukocytes (17). By contrast, the available evidence indicates that although the electrostatic search mechanism significantly speeds target lipid acquisition and generates detectable rebinding events, the effect on the macroscopic membrane dissociation rate is more subtle, even for the native PH domain that exhibits the most robust electrostatic searching.
More broadly, the association, dissociation, and diffusion rates of membrane targeting domains are precisely tuned for optimal interactions with target lipids and effector proteins during cell signaling. The GRP1 PH domain possesses an electrostatic search mechanism that significantly speeds its docking to rare target lipid during the membrane association reaction. Other targeting domains that possess similar electrostatic search mechanisms may also exhibit large effects on dissociation kinetics. In principle, efficient electrostatic stimulation of rebinding could both enable jumps into new membrane regions where substrate or downstream effector proteins are located, and significantly extend the membrane residency time. Thus, like electrostatic searching for rare target lipids, efficient microdissociation and rebinding mechanisms are likely part of a toolkit used by evolution to optimize the biophysical parameters of signaling events at membrane surfaces.
Conclusions
This study shows the power of single molecule TIRFM methods, when applied in vitro to well-defined protein and lipid systems, to uncover the hidden dynamics and protein-lipid interactions of membrane targeting proteins. The application of these methods to the GRP1 PH domain has revealed three previously undetected features of its interactions with membranes: i), exclusive interaction with a single target PI(3,4,5)P3 headgroup in the absence of other anionic lipids, yielding a membrane surface diffusion equivalent to a single lipid molecule, ii), simultaneous interaction with both its target PI(3,4,5)P3 headgroup and one or more PS molecules when both types of lipids are present, yielding a slowing of diffusion and dissociation, and iii), transient microdissociation and rebinding events when both lipids are present. Simultaneous binding of the PI(3,4,5)P3 headgroup to the primary site and PS binding to one or more weak, secondary sites are stable features of the membrane-bound state, whereas rebinding uses transient, collisional electrostatic interactions to speed redocking to rare target lipid. Identification of these and similar hidden properties of membrane docking reactions is crucial to a mechanistic understanding of specific membrane targeting in a wide array of cellular signaling pathways. Thus, in vitro TIRFM studies of membrane targeting proteins are expected to have broad applications in the field of signaling biology.
Supporting Material
Six figures and five movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(08X)00093-3.
Supporting Material
Acknowledgments
The authors thank Prof. Arthur Pardi, Dr. Vasiliy Fomenko, and Haemi Lee for training with TIRFM; Prof. Jennifer Hovis for training with supported lipid bilayers; and Profs. David Daleke, Jeff Gelles, Elizabeth Rhoades, David Nesbitt, and Thomas Perkins for helpful discussions.
Research support was provided by National Institutes of Health grant R01 GM-063235 (J.J.F.). TIRFM instrumentation was supported largely by a W. M. Keck Foundation grant to the University of Colorado, Boulder.
References
- 1.Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 2.Cho W., Stahelin R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 3.DiNitto J.P., Cronin T.C., Lambright D.G. Membrane recognition and targeting by lipid-binding domains. Sci. STKE. 2003;2003:re16. doi: 10.1126/stke.2132003re16. [DOI] [PubMed] [Google Scholar]
- 4.Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson T.R., Kearns B.G., Theibert A.B. Cytohesins and centaurins: mediators of PI 3-kinase-regulated Arf signaling. Trends Biochem. Sci. 2000;25:489–495. doi: 10.1016/s0968-0004(00)01644-3. [DOI] [PubMed] [Google Scholar]
- 6.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins P.T., Jackson T.R., Stephens L.R. Platelet-derived growth factor stimulates synthesis of PtdIns(3,4,5)P3 by activating a PtdIns(4,5)P2 3-OH kinase. Nature. 1992;358:157–159. doi: 10.1038/358157a0. [DOI] [PubMed] [Google Scholar]
- 8.Insall R.H., Weiner O.D. PIP3, PIP2, and cell movement–similar messages, different meanings? Dev. Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolsch V., Charest P.G., Firtel R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillou H., Lecureuil C., Anderson K.E., Suire S., Ferguson G.J. Use of the GRP1 PH domain as a tool to measure the relative levels of PtdIns(3,4,5)P3 through a protein-lipid overlay approach. J. Lipid Res. 2007;48:726–732. doi: 10.1194/jlr.D600038-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Cohen L.A., Honda A., Varnai P., Brown F.D., Balla T. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol. Biol. Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langille S.E., Patki V., Klarlund J.K., Buxton J.M., Holik J.J. ADP-ribosylation factor 6 as a target of guanine nucleotide exchange factor GRP1. J. Biol. Chem. 1999;274:27099–27104. doi: 10.1074/jbc.274.38.27099. [DOI] [PubMed] [Google Scholar]
- 13.Klarlund J.K., Guilherme A., Holik J.J., Virbasius J.V., Chawla A. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 14.Lietzke S.E., Bose S., Cronin T., Klarlund J., Chawla A. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson K.M., Kavran J.M., Sankaran V.G., Fournier E., Isakoff S.J. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 16.DiNitto J.P., Delprato A., Gabe Lee M.T., Cronin T.C., Huang S. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbin J.A., Dirkx R.A., Falke J.J. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronin T.C., DiNitto J.P., Czech M.P., Lambright D.G. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 2004;23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J., Haney R.M., Vora M., Verkhusha V.V., Stahelin R.V. Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manna D., Albanese A., Park W.S., Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf K.E., Malmberg N.J., Falke J.J. Effect of PIP2 binding on the membrane docking geometry of PKCalpha C2 domain: an EPR site-directed spin-labeling and relaxation study. Biochemistry. 2008;47:8301–8316. doi: 10.1021/bi800711t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joo C., Balci H., Ishitsuka Y., Buranachai C., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 23.Thompson N.L., Drake A.W., Chen L.X., VandenBroek W. Equilibrium, kinetics, diffusion and self-association of proteins at membrane surfaces: measurement by total internal reflection fluorescence microscopy. Photochem. Photobiol. 1997;65:39–46. doi: 10.1111/j.1751-1097.1997.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 24.Axelrod D. Total internal reflection fluorescence microscopy. In: Periasamy A., editor. Methods in Cellular Imaging. University Press; Oxford: 2001. pp. 362–380. [Google Scholar]
- 25.Iino R., Koyama I., Kusumi A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys. J. 2001;80:2667–2677. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiessling V., Crane J.M., Tamm L.K. Transbilayer effects of raft-like lipid domains in asymmetric planar bilayers measured by single molecule tracking. Biophys. J. 2006;91:3313–3326. doi: 10.1529/biophysj.106.091421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuoka S., Iijima M., Watanabe T.M., Kuwayama H., Yanagida T. Single-molecule analysis of chemoattractant-stimulated membrane recruitment of a PH-domain-containing protein. J. Cell Sci. 2006;119:1071–1079. doi: 10.1242/jcs.02824. [DOI] [PubMed] [Google Scholar]
- 29.Mashanov G.I., Tacon D., Peckham M., Molloy J.E. The spatial and temporal dynamics of pleckstrin homology domain binding at the plasma membrane measured by imaging single molecules in live mouse myoblasts. J. Biol. Chem. 2004;279:15274–15280. doi: 10.1074/jbc.M312140200. [DOI] [PubMed] [Google Scholar]
- 30.Vazquez F., Matsuoka S., Sellers W.R., Yanagida T., Ueda M. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheets E.D., Chen L., Thompson N.L. Decreased IgG-Fc gamma RII dissociation kinetics in the presence of a protein antigen. Mol. Immunol. 1997;34:519–526. doi: 10.1016/s0161-5890(97)00057-6. [DOI] [PubMed] [Google Scholar]
- 32.Li Y.L., Augustine G.J., Weninger K. Kinetics of complexin binding to the SNARE complex: Correcting single molecule FRET measurements for hidden events. Biophys. J. 2007;93:2178–2187. doi: 10.1529/biophysj.106.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowen M.E., Weninger K., Ernst J., Chu S., Brunger A.T. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys. J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharonov A., Bandichhor R., Burgess K., Petrescu A.D., Schroeder F. Lipid diffusion from single molecules of a labeled protein undergoing dynamic association with giant unilamellar vesicles and supported bilayers. Langmuir. 2008;24:844–850. doi: 10.1021/la702600w. [DOI] [PubMed] [Google Scholar]
- 35.Golovanov A.P., Hautbergue G.M., Wilson S.A., Lian L.Y. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 36.Image, N. I. H. J. http://rsb.info.nih.gov/ij/. Accessed January 2008.
- 37.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Schutz G.J., Schindler H., Schmidt T. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 1997;73:1073–1080. doi: 10.1016/S0006-3495(97)78139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seu K.J., Pandey A.P., Haque F., Proctor E.A., Ribbe A.E. Effect of surface treatment on diffusion and domain formation in supported lipid bilayers. Biophys. J. 2007;92:2445–2450. doi: 10.1529/biophysj.106.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crane J.M., Kiessling V., Tamm L.K. Measuring lipid asymmetry in planar supported bilayers by fluorescence interference contrast microscopy. Langmuir. 2005;21:1377–1388. doi: 10.1021/la047654w. [DOI] [PubMed] [Google Scholar]
- 41.Redfern D.A., Gericke A. pH-dependent domain formation in phosphatidylinositol polyphosphate/phosphatidylcholine mixed vesicles. J. Lipid Res. 2005;46:504–515. doi: 10.1194/jlr.M400367-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.Sonnleitner A., Schutz G.J., Schmidt T. Free Brownian motion of individual lipid molecules in biomembranes. Biophys. J. 1999;77:2638–2642. doi: 10.1016/S0006-3495(99)77097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L.F., Granick S. Lipid diffusion compared in outer and inner leaflets of planar supported bilayers. J. Chem. Phys. 2005;123:211104. doi: 10.1063/1.2138699. [DOI] [PubMed] [Google Scholar]
- 44.Tamm L.K., McConnell H.M. Supported phospholipid-bilayers. Biophys. J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merkel R., Sackmann E., Evans E. Molecular friction and epitactic coupling between monolayers in supported bilayers. J. Physiol. (Paris) 1989;50:1535–1555. [Google Scholar]
- 46.Lagerholm B.C., Thompson N.L. Theory for ligand rebinding at cell membrane surfaces. Biophys. J. 1998;74:1215–1228. doi: 10.1016/S0006-3495(98)77836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans E., Sackmann E. Translational and rotational drag coefficients for a disk moving in a liquid membrane-associated with a rigid substrate. J. Fluid Mech. 1988;194:553–561. [Google Scholar]
- 48.Wagner M.L., Tamm L.K. Reconstituted syntaxin1a/SNAP25 interacts with negatively charged lipids as measured by lateral diffusion in planar supported bilayers. Biophys. J. 2001;81:266–275. doi: 10.1016/S0006-3495(01)75697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sackmann E. Supported membranes: scientific and practical applications. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 50.Evans J.H., Murray D., Leslie C.C., Falke J.J. Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell. 2006;17:56–66. doi: 10.1091/mbc.E05-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbin J.A., Evans J.H., Landgraf K.E., Falke J.J. Mechanism of specific membrane targeting by C2 domains: localized pools of target lipids enhance Ca2+ affinity. Biochemistry. 2007;46:4322–4336. doi: 10.1021/bi062140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karathanassis D., Stahelin R.V., Bravo J., Perisic O., Pacold C.M. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao C., Du G.W., Skowronek K., Frohman M.A., Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 54.Bai J., Tucker W.C., Chapman E.R. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 55.Li L., Shin O.H., Rhee J.S., Arac D., Rah J.C. Phosphatidylinositol phosphates as co-activators of Ca2+ binding to C2 domains of synaptotagmin 1. J. Biol. Chem. 2006;281:15845–15852. doi: 10.1074/jbc.M600888200. [DOI] [PubMed] [Google Scholar]
- 56.Lagerholm B.C., Starr T.E., Volovyk Z.N., Thompson N.L. Rebinding of IgE Fabs at haptenated planar membranes: measurement by total internal reflection with fluorescence photobleaching recovery. Biochemistry. 2000;39:2042–2051. doi: 10.1021/bi9917434. [DOI] [PubMed] [Google Scholar]
- 57.Friedman L.J., Chung J., Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian H., Sheetz M.P., Elson E.L. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.