Abstract
Certain guanine-rich (G-rich) DNA and RNA molecules can associate intermolecularly or intramolecularly to form four stranded or “quadruplex” structures, which have unusual biophysical and biological properties. Several synthetic G-rich quadruplex-forming oligodeoxynucleotides have recently been investigated as therapeutic agents for various human diseases. We refer to these biologically active G-rich oligonucleotides as aptamers because their activities arise from binding to protein targets via shape-specific recognition (analogous to antibody-antigen binding). As therapeutic agents, the G-rich aptamers may have some advantages over monoclonal antibodies and other oligonucleotide-based approaches. For example, quadruplex oligonucleotides are non-immunogenic, heat stable and they have increased resistance to serum nucleases and enhanced cellular uptake compared to unstructured sequences. In this review, we describe the characteristics and activities of G-rich oligonucleotides. We also give a personal perspective on the discovery and development of AS1411, an antiproliferative G-rich phosphodiester oligonucleotide that is currently being tested as an anticancer agent in Phase II clinical trials. This molecule functions as an aptamer to nucleolin, a multifunctional protein that is highly expressed by cancer cells, both intracellularly and on the cell surface. Thus, the serendipitous discovery of the G-rich oligonucleotides also led to the identification of nucleolin as a new molecular target for cancer therapy.
Keywords: AS1411, aptamer, G-rich oligonucleotides, quadruplex, G-quartets, nucleolin, NF-kappaB, PRMT5, T-oligos, Dz13
Oligonucleotides as Therapeutic Agents
Since automated DNA synthesizers became widely available in the 1980s, there has been substantial interest in developing synthetic oligonucleotides for use as therapeutic agents [1-9]. Initial strategies aimed to prevent translation or transcription of specific viral or cellular genes via sequence-specific hybridization of oligodeoxynucleotides to target mRNA (the antisense approach) or genomic DNA (the antigene or triplex approach) [1,2]. More recently, the phenomenon of RNA interference has garnered much attention and there is considerable excitement about the therapeutic potential of modified double-stranded RNA oligonucleotides (small interfering RNAs or siRNAs) that can specifically knockdown the expression of disease-associated genes [3,4]. In addition to these approaches, in which the synthetic oligonucleotide recognizes a specific nucleic acid target via base pairing, several classes of oligonucleotides have been developed to target proteins or small molecules. These include double-stranded DNA oligonucleotides that act as “decoys” for specific transcription factors [5] and aptamer oligonucleotides, which bind to small molecule or proteins by shape-specific recognition [6-8]. Other prospective therapeutic oligonucleotides include those containing immunostimulatory or “CpG” motifs. These molecules act as agonists of toll-like receptor 9 (TLR9) and thereby induce release of various cytokines, making them potentially useful as vaccine adjuvants or in cancer immunotherapy [9]. Some of the strategies for using oligonucleotides as therapeutics are illustrated in Figure 1.
Figure 1. Strategies for the Therapeutic Use of Oligonucleotides.
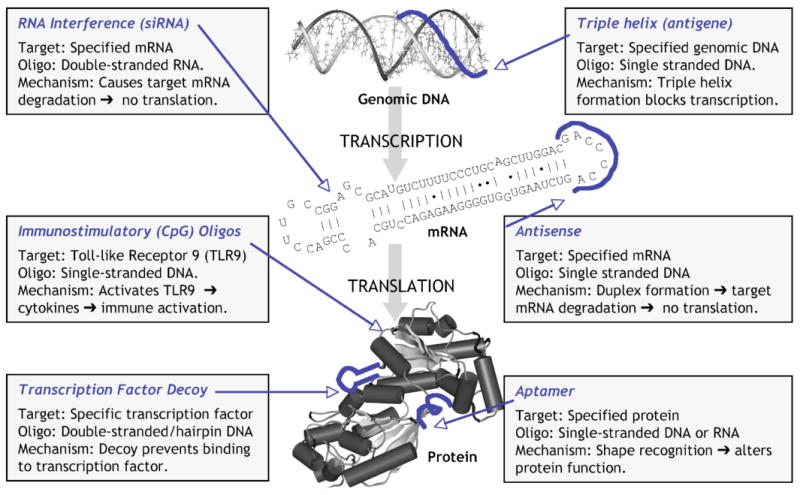
Cartoon showing different approaches using oligonucleotides (oligos) as targeted therapeutic agents. G-rich oligonucleotides appear to function as aptamers to various proteins. In the case of AS1411, the target protein is believed to be nucleolin.
While many of these strategies demonstrate considerable therapeutic promise, there are two universal problems that may limit the in vivo usefulness of oligonucleotide-based medicines. These two critical issues are the susceptibilities of oligonucleotides to degradation by serum or cellular nucleases and their ineffcient internalization by cells [10] (although this latter concern may not be applicable to aptamers that target cell surface or extracellular targets). The stability problem has been largely addressed by using oligonucleotides with chemical modifications to the nucleic acid backbone or sugars. However, the increased resistance to nuclease digestion for some of these modified oligonucleotides, especially first generation phosphorothioate analogs, is offset by their increased toxicity and reduced specificity. In cultured cells, the poor uptake of oligonucleotides has been countered by the use of various transfection methods, such as electroporation and complexation with lipids or liposomes, but none of these approaches are easily translatable to in vivo use. Several of the hybridization-dependent approaches, including antisense and siRNAs, have now progressed to clinical testing [1,4]. Early trials using phosphorothioate antisense molecules have indicated significant toxicity and off-target effects of that backbone [1], highlighting the need for alternative strategies.
Unusual Biological Properties of G-rich Oligonucleotides
Throughout the history of therapeutic oligonucleotide development, it has become apparent that sequences containing runs of contiguous guanine (G) bases or those that are generally G-rich often have rather distinct biological properties. In the early days of antisense research, several researchers noted [11-20] that the biological activities of certain oligonucleotides were not due to a true antisense effect, but rather were linked to the presence of contiguous guanines and the propensities of the oligonucleotides to form quadruplex structures containing G-quartets (Figure 2). Subsequently, a number of groups have described various quadruplex-forming and G-rich oligonucleotides that have biological activities that are not mediated by an antisense mechanism, but are most likely attributable to the protein-binding (aptameric) effects of these oligonucleotides [21-61]. While many of these observations were made by chance, aptamers generated using combinatorial methods such as SELEX (systematic evolution of ligands by exponential enrichment) frequently turned out to be capable of forming G-quadruplexes as well [62-74]. Researchers studying immunostimulatory oligonucleotides have also noted that inclusion of G-rich sequences in CpG-containing oligonucleotides can alter their biological properties, leading to enhanced uptake and activity in some cases [75-81]. Thus, although the antisense research community originally viewed the “non-specific” effects of G-rich sequences as highly undesirable [82], it has become clear that G-quadruplex-forming oligonucleotides can have distinct biological properties that may make them useful therapeutic agents. Indeed, since nuclease susceptibility and inefficient cellular uptake have proved to be universal hurdles in the development of therapeutic oligonucleotides, the enhanced biostability and cellular internalization of quadruplex oligonucleotides may prove to be major advantages.
Figure 2. Structures of Quadruplex and Duplex DNA.
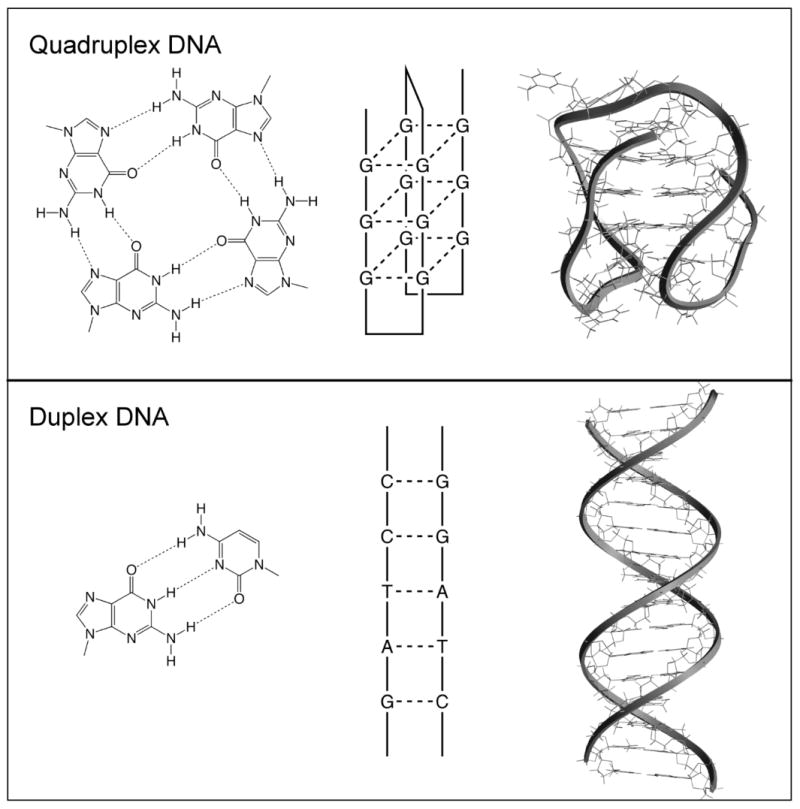
This figure shows the hydrogen-bonding arrangements (left) for a G-quartet (top) and a G•C base pair (bottom), as well as schematic illustrations (middle) and molecular models (right) for quadruplex and duplex DNA. The quadruplex shown is one possible conformation of the human telomere sequence (PDB accession code 143d). AS1411 has been shown to form a quadruplex and its detailed molecular structure is currently being investigated.
Nuclease Resistance of G-rich Oligonucleotides
Most unmodified (phosphodiester) oligonucleotides have serum half-lives in the order of minutes and it had long been the dogma in the field that phosphodiester oligonucleotides could never be clinically useful because of their digestion by serum exonucleases. This problem can be addressed in most cases by the use of more nuclease-resistant backbones, such as phosphorothioates or 2′-modified analogs. However, these approaches may not be appropriate in the case of quadruplex aptamers. For example, 2′-O-modification of the ribose in quadruplex aptamers may significantly alter their three-dimensional structure and preclude binding to their intended target [27].
Fortunately, because of the increased nuclease resistance afforded by the quadruplex structure, extensive modification may not be necessary for quadruplex-forming aptamers and it appears that phosphodiester oligodeoxynucleotides with unmodified or minimally modified (e.g. only terminal phosphorothioate linkages) backbones can be used in vivo. It is already well established that guanine nucleotides within the core of a G-quadruplex are relatively protected from chemical attack (indeed, this is the basis of several footprinting techniques that are used to investigate quadruplex structure). On the other hand, although there are numerous anecdotal references [ref] to the enhanced nuclease resistance of G-rich oligodeoxynucleotides, there have been only a few reports that specifically address this issue. Bishop et al. [ref] evaluated internally 33P-labeled anti-HIV G-rich oligodeoxynucleotides that had phosphodiester (PO) backbones, with or without terminal phosphorothioate (PS) linkages. They found that totally unmodified PO and terminal-PS modified quadruplex oligonucleotides had half-lives in bovine serum of 5 h and 4 days, respectively. Under the same conditions, unmodified and terminal-PS modified random sequence (non-quadruplex) oligodeoxynucleotides had half-lives of about 3 min and 7 min, respectively. The role of the quadruplex structure in the enhanced nuclease-resistance was confirmed by the finding that a single G→A mutation that was expected to disrupt quadruplex formation resulted in a dramatically decreased half-life in serum (to ∼3 min). Similar findings regarding the enhanced serum stability of quadruplex-forming phosphodiester oligonucleotides were also reported by our group in Dapic et al. [27]. In that paper, we reported on 5′-32P-labeled analogs of GRO29A (a nucleolin-binding antiproliferative oligonucleotide, which is described below) with backbones consisting of phosphorothioate DNA, phosphodiester DNA (with or without 3′-amine modification), or 2′-O-methyl RNA. All of these analogs were similarly resistant to degradation when placed in serum-containing medium. Interestingly, the unmodified phosphodiester DNA version of GRO29A was completely intact after 5 days in serum-containing medium, whereas for a random sequence of phosphodiester DNA, no full-length oligonucleotide remained after one hour [27]. Recently, the nuclease resistance of a quadruplex oligonucleotide inside live cells was demonstrated for the first time by fluorescence anisotropy [87].
Enhanced Cellular Uptake of Quadruplex Aptamers
As with their increased nuclease-resistance, observations concerning the efficient cellular uptake for G-rich oligonucleotides are quite common, but systematic studies are few. In the study mentioned above, Bishop et al. [86] found that both the PO and terminal-PS quadruplex-forming anti-HIV oligonucleotides were efficiently internalized by HeLa cervical carcinoma cells, reaching an intracellular concentration that was 8 to 10-fold higher than that in the medium. In contrast, the cellular uptake of non-quadruplex sequences was negligible. Several groups have noted that the inclusion of polyguanine runs at the ends of PS, PO and mixed PS/PO oligonucleotides can improve cellular uptake [34,75-81,83,88-90]. For example, addition of a G10 sequence to the 3′-end of an antisense oligodeoxynucleotide increased its uptake in J774E macrophage cells by 10-fold compared to the unmodified or 3′-C10 oligonucleotide [89]. Evidence for the differential uptake of G-rich oligonucleotides has also come from the relatively new field of immunostimulatory or “CpG” oligonucleotides. These molecules mimic bacterial DNA (since mammalian CpG dinucleotides are mostly methylated) and activate a variety of immune effector cells via their recognition by toll-like receptor 9 (TLR9). They have a number of properties that might be useful for therapeutic applications, including improving vaccine response, boosting innate immunity to pathogens, reducing allergic response and inducing anti-tumor immunity [9]. It has become clear that the activity of these oligonucleotides is directly related to their ability to bind to their target cells and become internalized [9]. Several research groups have recently discovered, either by chance or by utilizing SELEX technology to optimize cell uptake, that CpG oligonucleotides containing G-rich sequences have altered uptake [75-81]. Inclusion of G-rich sequences at the 3′-terminus was generally found to increase uptake and immunostimulatory activity, especially for phosphodiester oligonucleotides, in a variety of cells, including B lymphoma cells, dendritic cells, peripheral blood monocytes, macrophages and microglial cells [75-81]. These effects were found to be dependent on the position of the G-rich sequence [75,81] and were associated with the formation of quadruplex structures [79,81]. The increased cellular internalization of CpG oligonucleotides with 3′-G-rich tails was also related to their enhanced binding to cellular proteins [81], but was independent of TLR9 [78].
This body of evidence supports the idea that certain cell types (including cancer cells and some immune cells) preferentially internalize G-quadruplex-forming oligonucleotides, compared to unstructured sequences. However, the mechanism of their internalization remains unclear. In fact, despite more than 20 years of research, the uptake and cellular trafficking of synthetic oligonucleotides in general is rather poorly understood. Uptake appears to be dependent on oligonucleotide concentration, backbone chemistry and cell type, and most researchers believe that receptor-mediated endocytosis is the predominant mechanism [10,91,92]. Various proteins have been suggested as “oligonucleotide receptors”, including an uncharacterized 110 kDa protein, the integrin Mac-1 on leukocytes and scavenger receptors on hepatocytes [93-96], but no consensus has yet been reached. We believe that cell surface nucleolin may play an important role in the uptake of some G-rich oligonucleotides, such as AS1411 (see discussion later in this article), but the precise mechanism is not yet defined.
Biological Effects of G-Rich Oligonucleotides
Treatment of various cell types with G-rich oligonucleotides containing PO, PS, or mixed PO/PS backbones is associated with a diverse array of biological effects. In many cases, reports of these effects have come from researchers in the antisense or antigene fields who have noted that certain G-rich oligonucleotides have activity that is not consistent with sequence-specific hybridization to their target mRNA or DNA [11-61]. The biological effects reported include inhibition of cell proliferation, induction of cell death, changes in cellular adhesion, inhibition of protein aggregation, and antiviral activity [11-61]. The non-antisense antiviral effects of G-rich oligodeoxynucleotides were first noted more than a decade ago, and activity was reported against herpes simplex virus, a murine leukemia virus and HIV-1 [97-100]. The antiproliferative and pro-apoptotic effects of G-rich oligonucleotides have been observed in various cell types, including vascular smooth muscle cells and numerous cell lines derived from solid tumors, leukemias and lymphomas [11-61]. For some oligonucleotides, in vivo antiproliferative effects were also demonstrated in tumor-bearing mice [33,41,45,48,50-53]. For the most part, biologically active G-rich oligonucleotides have been shown to form G-quartet-containing structures, although the role of quadruplex formation is unclear for some [21-24,39,40]. In many cases, antiproliferative effects have been found to correlate with protein-binding properties [21-25,37,38,56], but specific targets have been identified for only a few. Proteins reported as direct targets for G-rich aptamer oligonucleotides with unexpected biological activity (i.e. those not derived by SELEX) include nucleolin [25-31], Stat3 [50-53], eEF1A [21-24] and interferon-γ [18-20]. Interestingly, several reports also point to the ability of G-rich oligonucleotides to stimulate immune responses, which are distinct from those of CpG-containing sequences. These effects include pro-proliferative effects on macrophages, co-stimulation of T cells, activation of NK cells, and induction of cytokine production in peripheral blood mononuclear cells [39,40, 101-104].
Discovery of Antiproliferative G-rich Oligonucleotides and AS1411
The science fiction writer, Isaac Asimov, once said, “The most exciting phrase to hear in science, the one that heralds new discoveries, is not ‘Eureka!’ but ‘That's funny …’”. This was certainly true in our case! In the late 1990s, we (Bates, Miller, Thomas and Trent) were evaluating the possibility of using triplex-forming oligodeoxynucleotides to modulate the expression of specific genes, with the goal of developing novel cancer-selective therapies. We decided to test the activities of our triplex-forming oligonucleotides in cultured prostate cancer cells by adding them directly to the medium (i.e. without transfection) of plated cells. However, we were not particularly optimistic that they would have any effect because of the well-known difficulties associated with using oligonucleotides in biological systems, including degradation by serum nucleases and poor cellular uptake. Our oligodeoxynucleotides were designed with phosphodiester linkages (i.e. the natural DNA backbone) and were modified at the 3′ terminus with an aminoalkyl linker to increase exonuclease resistance, based on previous reports of success with this approach in cells [105]. We were using the “purine motif” approach to triplex formation, which uses G bases to recognize G•C base pairs and T for A•T base pairs and, thus, our sequences consisted entirely of guanines and thymines. Somewhat to our surprise, we observed moderate inhibition of prostate cancer cell growth following treatment with a triplex-forming oligonucleotide specific for the promoter of our target gene (uPA). However, what was most remarkable about the experiment was that the control oligonucleotide, which was a G/T-containing sequence with no complementarity to the target gene, produced by far the most robust inhibition of cancer cell growth. After confirming our observations and testing several more G/T oligonucleotides in a variety of cell lines, we concluded that the antiproliferative effects we were seeing were clearly not due to the intended mechanism of triplex formation or any other hybridization-dependent mechanism. Nonetheless, this appeared to be an interesting phenomenon, so we resolved to further investigate the therapeutic potential and mechanism of these unusual G-rich oligonucleotides or “GROs”, as we called them.
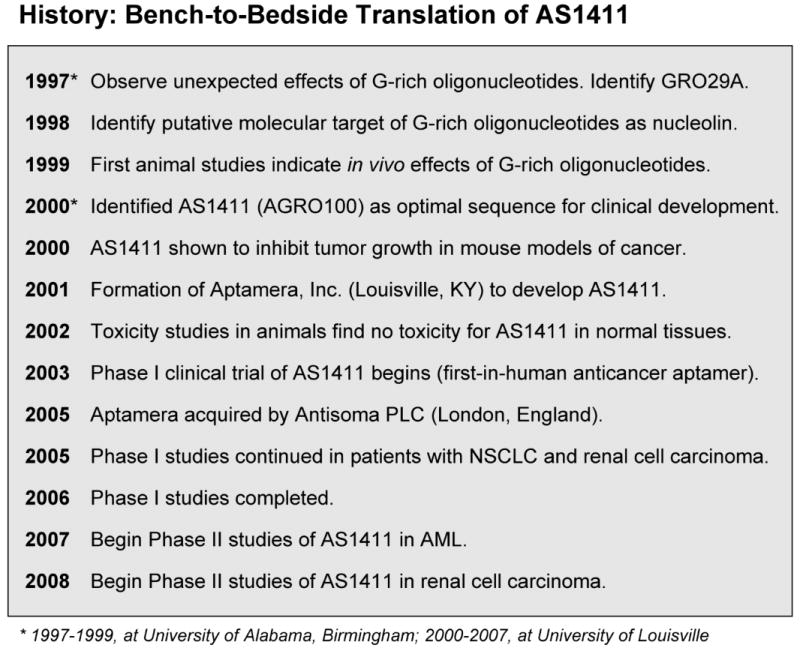
Pre-clinical Development of AS1411 (formerly AGRO100)
Initial studies of GROs were carried out using GRO29A, a 3′-modified 29-nucleotide phosphodiester oligodeoxynucleotide [25-28]. However, we found that the 3′-modification was not required for either nuclease resistance or activity [25,27] and that the 5′-terminus “tail” (5′-TTT) of GRO29A was not essential for activity [26,30,31]. Therefore, we chose to pursue pre-clinical and clinical studies using the truncated 26-mer sequence, 5′-GGTGGTGGTGGTTGTGGTGGTGGTGG, which became known as AGRO100 while being developed by Aptamera, and as AS1411 after acquisition by Antisoma in 2005 (see Figure 3). In every assay where we have tested both sequences, AS1411 has performed similarly to the original GRO29A.
Figure 3. Timeline of AS1411 Development.
One of the first things we wanted to determine was the relative activity of G-rich oligonucleotides in various cancer cell types compared to normal cells. Consequently, we evaluated AS1411 activity in numerous cell lines in our laboratories and submitted the compound for testing in the 60 tumor cell line panel at the Developmental Therapeutics Program (DTP) within the National Cancer Institute (NCI). More recently, Antisoma has examined several additional cell lines, such that AS1411 has now been tested in more than 80 human cell lines in total. Remarkably, AS1411 was found to display antiproliferative activity in almost every cancer cell type that was tested and, thus, appears to have broad therapeutic potential, as illustrated in Table 1. Typical GI50 values (the concentration required for 50% growth inhibition) for AS1411 are in the low micromolar range for cancer lines [ref] with a significant therapeutic window, reflected by very little effect on normal cells at these concentrations. In our earlier studies, we established the tumor-selectivity of G-rich oligonucleotides by showing that non-malignant Hs27 cells (human diploid skin fibroblasts) were unaffected by GRO29A at 10 μM, whereas cancer cell lines underwent cell cycle arrest and cell death in response to GRO29A [26]. A similar selectivity was also observed for AS1411 [30] and this is exemplified by the photomicrographs in Figure 4. Recently, another group led by Dr. Daniel Fernandes have reported on the tumor-selectivity of AS1411 for breast cancer cells (MCF7) compared to non-malignant breast epithelial cells (MCF10A) [106], and for acute myeloid leukemic blasts compared to normal B cells [107]. Another intriguing aspect of AS1411 activity is the time course of its effects on cancer cells [26]. Unlike most chemotherapy agents, AS1411 does not cause rapid cytotoxity when added to cells. Instead, cytostasis occurs as a result of cell division being inhibited, and induction of cell death occurs only after prolonged exposure to AS1411 (2 - 4 days, depending on the cell line). This was one reason why a continuous infusion of AS1411 for 4 or 7 days was chosen as the route of administration for subsequent clinical studies.
Table 1. Cancer Cell Lines that are Responsive to AS1411 or GRO29A.
| CANCER TYPE | CELL LINES |
|---|---|
| Lung Cancer | A549a,f, NCI-H322Ma, NCI-H460a, EKVXa, HOP-92b, NCI-H299c, CaLu1c, NCI-H1385c, NCI-H82c, CaLu6e |
| Breast Cancer | MCF7a,f, T-47Da, BT-549b, MDA-Nb, MDA-MB-231b,f, ZR7S-1e |
| Prostate Cancer | DU145a,f, PC-3b, CA-HPV-10c |
| Colon Cancer | HCC 2998a, HT-29a, KM12a, HCT-116b, SW620b, HCT-15b, LS174Te |
| Pancreatic Cancer | PANC-1c, MIA-PaCa-2c, PANC-1e |
| Renal Cell Carcinoma | 786-0a, CAKI-1a, RXF393a, TK10a, A498b, ACHNb, SN12Cb |
| Ovarian Cancer | IGROV1b, OVCAR-3b, OVCAR-4b, OVCAR-5b |
| Cervical Cancer | HeLac,f |
| Leukemia and Lymphoma | CCRF-CEMa, SRa, HL60b, K-562b, RPMI-6226b, U937c, Meg01c, MV4-11e |
| Melanoma | LOX-IMVIb, SK-MEL-2b, A375c, SK-MEL-28e, MDA-MB-435b,g |
| Glioblastoma | SF-268a, U87-MGe |
| Neuroblastoma | IMR 32c,d, Lan 5c,d |
| Sarcoma | HT-1080c |
| Gastric Cancer | KATOIIIe, HGC27e |
Data from NCI Tumor Cell Line Screen of AS1411 (≥ 50% growth inhibition at 6.3 μM).
Data from NCI Tumor Cell Line Screen of AS1411 (30-49 % growth inhibition at 6.3 μM).
Bates, Miller, Thomas, et al., unpublished data using AS1411 or GRO29A.
In collaboration with Thomas H. Inge.
Data reported by Antisoma (see reference 171).
Reported previously for AS1411 or GRO29A (see references 25-31).
This line was previously believed to be derived from breast cancer.
Figure 4. Effect of AS1411 on Cancer Cells compared to Non-malignant Cells.
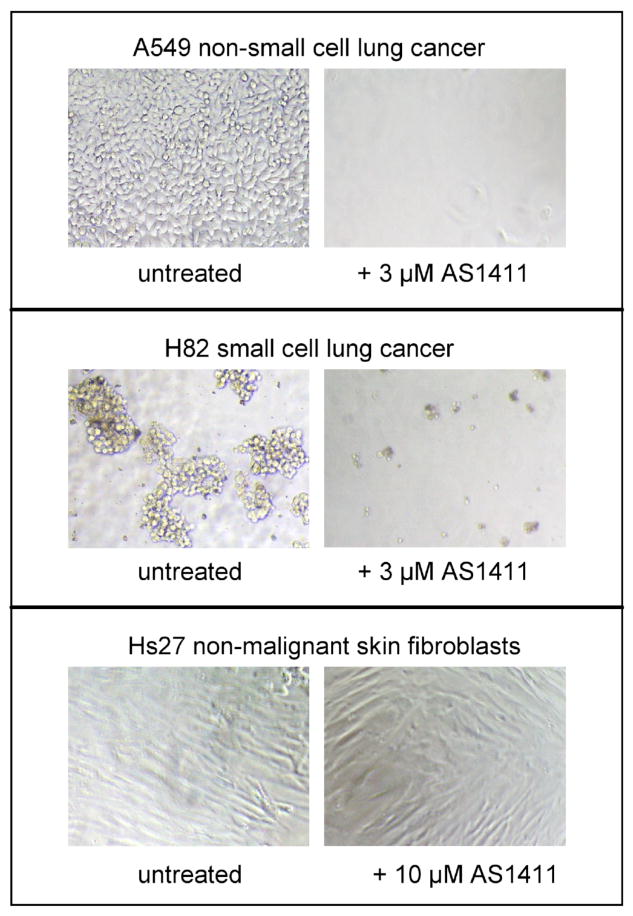
Photomicrographs showing the effect of treatment with AS1411 on various cell lines (indicated above), compared to controls (untreated cells). Cells were treated by adding AS1411 directly to the medium at the final concentration shown and photographed after 5 days.
Our next step was to assess the activity of AS1411 in animal models. Proof-of-concept data for the in vivo efficacy of AS1411 were generated in our laboratories using nude mice bearing subcutaneous xenografts derived from DU145 cells (human hormone-refractory prostate cancer). In these pilot experiments, treatment of mice with established tumors (∼ 100 mm3) by intraperitoneal injection of AS1411 at doses equivalent to approximately 5 mg/kg/day, daily for the first three days and then every other day for an additional three treatments led to a significant reduction in tumor growth, whereas a control oligonucleotide had no effect (Bates, Miller, Thomas, Xu, unpublished data).
In parallel with all of the previously mentioned studies, we were also investigating the mechanism of GRO29A and AS1411 (which will be discussed later in this review) and had identified the probable molecular target of these oligonucleotides as the cancer-associated protein, nucleolin. Thus, after several years of research into the activity and mechanism of G-rich oligonucleotides, which was funded largely by the Department of Defense (DOD) Prostate Cancer Research Program, we now had strong evidence that AS1411 had significant potential as a novel anticancer agent. At this point, with very few grants available for translational research at that time, we concluded that the most efficient way to transfer our discovery to the clinical setting would be via a biotechnology company. Hence, we (Bates, Miller, Trent) founded Aptamera, Inc., based in Louisville, Kentucky, with the goal of taking AS1411 from the bench to the bedside.
The company, in the capable hands of experienced business and scientific officers, and with support from local investors, state incentives, city development programs and the University of Louisville, was able to achieve this goal in a little over two years. In pre-clinical toxicology studies, AS1411 was administered to rats as a single intravenous (i.v.) infusion (over 30 min) at doses of up to 100 mg/kg, and was given to both rats and dogs as a 96 h continuous infusion at doses of up to 10 mg/kg. No toxic effects of AS1411 were observed in any animals at any dose tested, as assessed by gross necropsy, histopathology of all organs, clinical chemistry and hematology [8,108]. The pharmacokinetics of AS1411 were studied in nude mice bearing tumor xenografts using a radiolabeled (tritiated) oligonucleotide. After single i.v. injections at 1, 10, 25 mg/kg, the 3H-labeled AS1411 was eliminated in a biphasic manner, with 63% of the AS1411 being rapidly eliminated in urine within 5 h and a β-phase elimination half-life in blood of 2 days [8,108]. Biodistribution studies also suggested that AS1411 could accumulate selectively in the tumors of these animals [8,108]. Additional in vivo efficacy studies (using i.v. dosing at 5 – 40 mg/kg/day for 5 days) were carried out and demonstrated activity in other xenograft models, including A549 non-small cell lung cancer, A498 renal cell carcinoma, SKMES lung cancer, and MX1 breast cancer. There was also evidence of activity in pancreatic cancer xenografts (PANC-1) for the combination of AS1411 with gemcitabine [8,108].
Clinical Studies of AS1411
In September 2003, Dr. Damian A. Laber as the Principal Investigator initiated the first phase I study of AS1411 (known at the time as AGRO100) at the University of Louisville's James Graham Brown Cancer Center (Louisville, KY), sponsored by Aptamera. To the best of our knowledge, this represented a number of firsts, including the first-in-human anticancer aptamer and the first-in-class drug known to specifically target nucleolin.
Initially, this phase I clinical trial involved patients with a variety of advanced solid tumors who had documented progressive metastatic disease at the start of the trial (many had received multiple previous treatments) and who were incurable with currently available therapies. The design of the trial was a standard dose escalation scheme, enrolling three patients per dose, beginning at 1 mg/kg/day. The AS1411 was delivered by continuous intravenous infusion for 4 days and later escalated to 7 days. Patients were assessed every 28 days with laboratory and imaging studies. All patients received 1 cycle of AS1411. A second cycle was allowed, at the discretion of the investigator, in patients who achieved at least stable disease after more than 2 months. The results from these first 17 patients indicated that AS1411 was very well tolerated with no reports of any severe adverse events related to drug administration [109-111]. Also, there was evidence of clinical activity, with one patient who had metastatic renal cell carcinoma (RCC) achieving a partial response (PR) at 4 months after AS1411 treatment, which eventually became a complete response (CR) by 11 months, and an additional seven patients having disease stabilization for 2 months or more [109-111]. These results looked particularly promising for renal cell carcinoma, with all three of the RCC patients on this trial achieving some clinical benefit (the one patient having a CR and two additional patients having disease stabilization of ≥ 9 months duration [109-111]).
With this early stage clinical data supporting the safety and therapeutic potential of AS1411, Aptamera Inc. was acquired in early 2005 by the British pharmaceutical firm, Antisoma, PLC. They continued the Phase I trial at the Brown Cancer Center in order to determine the maximum tolerated dose (MTD), or to reach a pre-determined limit for dose escalation of 40 mg/kg/day, whichever came first. The inclusion criteria were restricted to patients with advanced renal cell carcinoma or non-small cell lung cancer (NSCLC) in order to clarify the early promising results in these groups of patients. The preliminary results of the completed Phase I study (a total of 30 patients) have been presented at several national and international meetings [112,113] and appear to confirm the lack of serious toxicity and encouraging signs of activity, including objective responses; detailed findings have now been submitted for publication. Several features of the clinical effects of AS1411 are especially interesting from a mechanistic perspective. For example, the timing of the responses (tumor shrinkage over a period of several months after a single dose of AS1411) and the duration of clinical benefit (several patients with long term disease stabilization or responses) are unusual [109-113].
On the basis of these findings and other supporting preclinical data, Antisoma has initiated two phase II clinical trials, one in acute myeloid luekemia and one in renal cell carcinoma. The first Phase II trial opened at several institutions in late 2007 to treat patients with relapsed or refractory acute myeloid leukemia (AML). The choice of AML as an indication was based on positive pre-clinical experiments performed by Antisoma and the Fernandes group [107,114]. The trial is designed to compare the activity of AS1411 in combination with cytarabine versus cytarabine alone in subjects with relapsed and refractory AML. Therapy will consist of two cycles of AS1411 as 7-day continuous intravenous infusions at 10 or 40 mg/kg/day combined with 1.5 g/m2 of cytarabine every 12 h during the last four days of treatment, or cytarabine alone. The preliminary results, which were recently announced in a press release from Antisoma, are encouraging. AS1411 has also recently progressed to Phase II clinical testing in renal cell carcinoma (RCC). A multi-institutional trial will involve 30 RCC patients who are intolerant to or who have relapsed after treatment with a tyrosine kinase inhibitor. Subjects will receive AS1411 monotherapy as two 4-day infusions at 40 mg/kg/day 28 days apart.
Mechanism of Action of AS1411 and Related G-rich Oligonucleotides
Clearly, elucidating a detailed molecular mechanism of action for a drug that was discovered by serendipity rather than rational design represents a rather formidable challenge! Thus, our strategy for investigating the mechanism of antiproliferative G-rich oligonucleotides was to first pursue the following fundamental questions: What are the characteristics of GROs that are important for activity? What is the molecular target of GROs? What are the effects of GROs on cells? What is the role of the presumed molecular target in mediating the cellular effects of GROs? The results of our studies attempting to address these questions are described below.
Identification of Nucleolin as the Primary Molecular Target of GROs
In our early studies, we focused on four G-rich oligodeoxynucleotides (all with a phophodiester backbone and 3′-aminoalkyl modification) with unrelated sequences and varying degrees of antiproliferative activity against cancer cells [25]. For the two most active GROs (GRO29A, GRO15A), there was evidence of stable quadruplex formation [25], which was assessed using a UV melting technique. We also observed the formation of a specific protein complex in the presence of active GROs by performing electophoretic mobility shift assays (EMSAs) using radiolabeled oligonucleotides incubated with cancer cell extracts. In contrast, there was no evidence for formation of the quadruplex structure and the specific protein complex was greatly decreased for the two GROs that were less active (GRO26A, GRO15B). Hence, we concluded that the biological activities of GROs likely depend on quadruplex formation and arise from an aptameric effect via targeting of a specific GRO-binding protein.
Clues to the identity of this putative target protein came from UV crosslinking studies, which showed that the GRO-binding protein migrated at ∼ 110 kDa on a denaturing polyacrylamide gel, and from southwestern blotting, which identified a band of the same size [25]. In accord with the EMSAs, the band corresponding to the GRO-protein complex in both the southwestern and crosslinking assay was evident when using active GROs, but was barely visible with the less active GROs. We also carried out competition studies using a radiolabeled 24-mer DNA oligonucleotide corresponding to the human single-stranded telomere region (“TEL”, 5′-T2AG3T2AG3T2AG3T2AG3), which is well known for its ability to fold into a quadruplex structure. Our experiments indicated that TEL could also bind the 110 kDa GRO-binding protein. Furthermore, when unlabeled GROs were co-incubated with radiolabeled TEL and cancer cell extracts, active GROs were able to compete with TEL for protein binding (i.e. the shifted radiolabeled band disappeared), whereas inactive GROs did not [25]. We subsequently adopted this competition EMSA as a standard method to assess GRO protein binding [27,28], since it requires labeling of only one oligonucleotide and allows the GROs to be compared in their natural (non-labeled) forms. Additional studies examining several series of G-rich oligonucleotides confirmed our original observations that antiproliferative activity requires both quadruplex structure and formation of this specific protein complex [27,28]. Notably, although necessary, quadruplex formation in itself was not sufficient for activity [27,28].
Based on observations of TEL-binding and apparent molecular weight, we hypothesized that the GRO-binding protein might be nucleolin, a protein that migrates at ∼ 110 kDa and which had been reported previously as a protein that binds telomere DNA [115,116]. To test this idea, we used biotinylated GROs and streptavidin-linked magnetic beads to capture GRO-associated proteins, and then carried out western blots using nucleolin antibodies. This showed specific precipitation by an active GRO of a protein recognized as nucleolin [25]. Our confidence in these results was further increased by two reports from the group of Dr. Nancy Maizels, which appeared shortly after we carried out these experiments, identifying nucleolin as a protein that bound selectively to quadruplex structures from IgG switch regions and ribosomal genes [117,118]. Using constructs kindly provided by Dr. Maizels [117], we were able to confirm binding of AS1411 to recombinant fragments of nucleolin consisting of the RNA binding domains and RGG region (Bates, unpublished). In later studies, we also showed by western blotting and mass spectrometry fingerprinting that biotinylated AS1411 could precipitate endogenous nucleolin from cell extracts or whole cells [30,31].
A question we are frequently asked is whether nucleolin is the only protein that is pulled down by AS1411? In fact, using mass spectrometry analysis, we were able to identify at least 15 proteins that were specifically precipitated by AS1411 after in vitro incubation with cell extracts [30,119]. However, there are a number of reasons we have focused on nucleolin as the primary target of GROs. The first is that our studies have suggested that this is the only protein that binds directly to active GROs (as evidenced by UV crosslinking and southwestern blotting [25]). Secondly, we have shown that the antiproliferative activities of several series of GROs are correlated with formation of the specific nucleolin-containing protein complex [25,27,28]. Another reason is that nucleolin is present on the cell surface and therefore easily accessible. We also know that nucleolin can be precipitated by AS1411 after incubation with cancer cells, whereas it remains to be seen if some of the other proteins can associate with AS1411 in intact cells. In addition, it may be that some of the GRO-precipitated protein are not bound to GRO directly, but are instead associated in a complex with nucleolin [30,31]. Thus, although we continue to investigate the role of other proteins in the GRO mechanism, at present we believe that nucleolin is the primary molecular target of AS1411.
Cellular Responses to GRO29A and AS1411
Soon after identifying GRO29A as an antiproliferative agent, we began to examine the effects of this molecule on the metabolism and cycling of cancer cells [26]. By flow cytometry analysis of propidium iodide-stained cells, we observed an accumulation of GRO29A-treated cancer cells in the S phase of the cell cycle, which was apparent by 24 h and was very pronounced at 72 h after treatment. Induction of cell death with a similar time course was also indicated by the appearance of a sub-G1 peak. Both of these effects were specific for GRO29A compared to a much less active control GRO and for cancer cells compared to non-malignant cells. The effect on cell cycle was found to be independent of the starting phase of the cells and did not occur at a particular boundary, indicating that this is not a typical arrest of the cell cycle caused by checkpoint activation. Instead, the cells appeared to gradually accumulate in S phase over the course of several cycles, suggesting that DNA replication is arrested or severely hindered [26]. This postulate was confirmed by BrdU incorporation experiments, which showed a pronounced cessation of DNA synthesis in GRO29A-treated cells. In contrast, RNA and protein synthesis (assayed by BrU and 35S-methionine incorporation, respectively) were not inhibited [26]. When a series of six oligonucleotides were examined using an in vitro DNA replication assay, their inhibitory effects on DNA replication in this assay were correlated with their antiproliferative effects against cancer cells. In the in vitro tests, the inhibitory effects were related to the ability of oligonucleotides to block the unwinding activity of the exogenous helicase (SV40 large T antigen) required in this assay [26]. This suggests that inhibition of endogenous helicases may contribute to GRO activity in cancer cells, although this has yet to be confirmed. Whether there is a role for nucleolin in this effect is not yet known, although it is noteworthy that nucleolin is an important regulator of DNA replication [120,121].
A major area interest in the Bates' laboratory has been the mechanism of GRO-induced cell death. We noted in our 2001 publication [26] that although there was clear evidence of cell death in GRO29A-treated cells using both flow cytometry analysis and TUNEL staining, the morphology of treated cells (greatly enlarged cell size) was not consistent with apoptosis. Recently, we have presented evidence that AS1411, at least in some cell lines, can induce non-apoptotic cell death [122].
In the course of investigating the role of nucleolin and other associated proteins in the mechanism of GRO activity, we have also uncovered several other cellular responses to AS1411, including inhibition of NF-κB signaling [30] and induction of tumor suppressor gene expression [31], which are described in more detail below.
Possible Role of Cell Surface Nucleolin in AS1411 Activity
In trying to develop a model that might explain the unusual and tumor-selective effects of GROs, we tried to incorporate our knowledge of the distinguishing features of the drug molecule and its presumed target. One unusual property of the GROs, which distinguishes them from other types of oligonucleotide therapies, is their ability to exert potent effects on cultured cells in the absence of a transfection agent. This suggested to us that the target of GROs was a cell surface protein or that there was some mechanism that led to GROs being readily internalized into cancer cells (or both of these possibilities). Although in our original report [25], uptake of 5′-32P-labeled GRO did not appear to be significantly different than other sequences (possibly because cellular phosphoesterases cleave the 5′-radiolabel), subsequent studies using more appropriate methods (flow cytometry or confocal microscopy using fluorescenty-labeled oligonucleotides) have shown enhanced cancer cell uptake of AS1411 compared to non-G-rich sequences ([106] and Bates, unpublished).
At the time when we identified nucleolin as the GRO molecular target, very little was known about the specific role of this protein in cancer, although it was well established that high levels of nucleolar nucleolin (a major component of silver staining AgNOR proteins) were an indication of a high rate of cell proliferation and a poor clinical prognosis [123]. Nucleolin was generally considered at that time to be a predominantly nucleolar protein, although it was recognized that it could shuttle between the nucleus and cytoplasm [124]. However, we noted in our original paper that nucleolin was also present at high levels in the plasma membranes of cancer cells [25]. This observation was in accord with earlier reports from several other groups who had reported on the cell surface localization of nucleolin and its role as a cell surface receptor [125-130].
Based on this knowledge, we hypothesized that cell surface nucleolin is selectively expressed in cancer cells compared to normal cells, and that it mediates the binding and uptake of AS1411 in cancer cells. We predicted that most normal cells contain little or no surface nucleolin and would not be able to bind or take up AS1411, thus explaining its tumor selective effects (see Figure 5). By staining fixed but non-permeabilized cells with nucleolin antibody, we were able to show higher levels of surface and cytoplasmic nucleolin in several types of cultured cancer cells compared to Hs27 non-malignant fibroblasts [131]. Unfortunately, this method of detection did not provide uniform surface nucleolin staining in solid tumor tissues, so we have not yet been able to test our hypothesis using clinical specimens. Recently, another group reported on the tumor-selective expression of surface and cytoplasmic nucleolin in MCF7 breast cancer cells compared to MCF10A non-malignant breast cells [106] and in AML blasts compared to normal B cells [107].
Figure 5. Proposed Model for AS1411 Mechanism of Action.
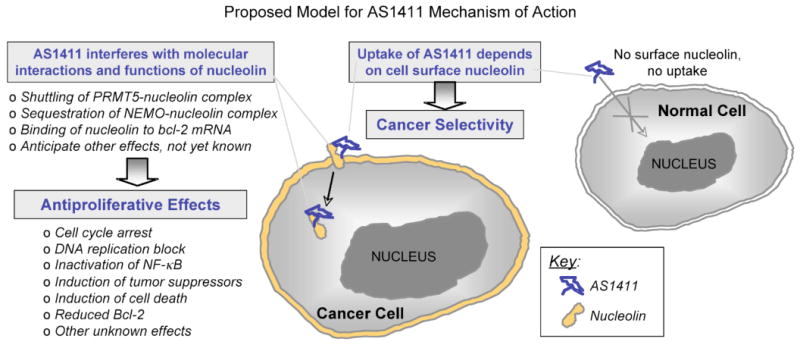
We propose that the cellular uptake of AS1411 is dependent upon cell surface nucleolin. Because cancer cells have high levels of surface nucleolin compared to normal cells, this would explain the preferential uptake and activity of AS1411 in malignant cells. We also propose that AS1411 causes pleiotropic antiproliferative effects because it interferes with some of the normal functions of nucleolin, which plays a role in many cancer-associated pathways. Some of the nucleolin complexes that are affected by AS1411 have already been identified, including those with PRMT5, NEMO and Bcl-2 mRNA (see text for details).
Since the time of our original observations, there have been dozens of additional reports describing the presence of nucleolin in the cytoplasm or on the cell surface (reviewed in [132]). The presence of nucleolin in the plasma membrane was initially controversial, since the protein contains no transmembrane domain and is transported to the surface via a non-classical mechanism [133], but there are now numerous studies providing additional compelling evidence for the presence of nucleolin on the cell surface [132-156]. Furthermore its involvement in mediating a wide range of pathways, including endocytosis [134-136], cellular adhesion [137-140], signal transduction [138-141], and as a receptor for various pathogens [144-149], has now been established. Perhaps most relevant in this case are several recent papers that provide strong support for the idea that surface nucleolin is a useful cancer target by demonstrating that it can mediate tumor-specific uptake and anti-tumor activity of targeted ligands in live animals. The first of these comes from Ruoslahti and colleagues who found that a tumor-homing peptide named F3, which was derived by phage display, works by targeting nucleolin protein [153]. They showed that nucleolin was expressed at high levels on the surface of cancer cells and tumor-associated angiogenic endothelial cells, and that a nucleolin monoclonal antibody was selectively taken up by tumors in a mouse model of breast cancer [154]. The second paper from Shi et al. [155] described cell surface nucleolin as a receptor that mediates the anti-angiogenic and anti-tumor effects of endostatin, an endogenous angiogenesis inhibitor, in a mouse model of melanoma. Most recently, a group led by Dr. Ara Hovanessian demonstrated that their nucleolin-binding pseudopeptide (HB-19) could also suppress tumor growth and angiogenesis in mouse models, while displaying no toxicity to normal cells [156]. In addition to these papers focused on the cell surface protein, there is mounting evidence that overexpression of nucleolin in general may play a direct role in the process of malignant transformation and progression [132, 157-162].
Possible Roles for Intracellular Nucleolin in AS1411 Activity
The next important questions we asked are how does binding of AS1411 affect nucleolin activity and how does that lead to cancer cell death? These turned out to be rather challenging questions, in large part because nucleolin is a remarkably multi-functional and dynamic protein [132,163-166]. It is known to interact with hundreds of other molecules (proteins, nucleic acids, peptides, nucleotides and sugars) and has been reported as a regulator of a similarly high number of cellular processes, including ribosome biogenesis, DNA replication, transcription, translation, chromatin remodeling, apoptosis, cytokinesis, protein trafficking and telomere maintenance [132,163-166]. We began by looking for obvious changes in nucleolin levels or stability that occurred specifically in AS1411-treated cells, but did not detect any. Therefore, we concluded that AS1411 binding was affecting the molecular interactions between nucleolin and its binding partners, which would lead to alterations in nucleolin-dependent pathways. In light of the characteristics of nucleolin noted above, we felt it would be impractical to examine each partner or function in turn. Consequently, in collaboration with other investigators at the Brown Cancer Center, we turned to proteomics analysis to allow us to identify candidate nucleolin-associated proteins that were affected by AS1411.
One of the first proteins we investigated was NEMO (also called IKKγ), a protein that was originally identified as a candidate GRO-associated protein after it was co-precipitated (with several other proteins, including nucleolin) from cancer cell extracts using a biotinylated version of AS1411 [30]. This protein was of interest to us because of its essential role in canonical NF-κB signaling, a pathway which is closely linked to the chemoresistance of cancer cells [167] and which had previously been reported to be inhibited by other types of G-rich oligonucleotides [33]. After confirming that our observation was not simply an in vitro artifact (by showing that NEMO from cancer cells treated with biotinylated AS1411 using streptavidin-coated beads), we demonstrated that the activity of the NEMO-containing IKK complex was markedly inhibited in AS1411-treated cells [30]. The IKK complex normally phosphorylates the cytoplasmic inhibitor, IκBα, thereby releasing NF-κB for translocation to the nucleus where it can activate target gene expression. Thus, we predicted that AS1411 would block activation of NF-κB and, as expected, we observed strong inhibition of both TNFα-stimulated and constitutive NF-κB signaling in a variety of AS1411-treated cancer cell lines [30]. Although we initially assumed that NEMO was bound directly to AS1411, our studies led us back to nucleolin as the likely mediator of the AS1411-NEMO association. In untreated or control-treated cells, a small amount of nucleolin-NEMO complex could be detected in the cytoplasm. However, in AS1411-treated cells, the levels of this complex were substantially enhanced suggesting that AS1411 can bind to the nucleolin-NEMO complex and stabilize it, thus sequestering NEMO activity and inhibiting the IKK-mediated activation of NF-κB [30]. The results suggest that inhibition of the pro-survival NF-κB pathway may contribute to the antiproliferative effects of AS1411. Furthermore, our paper provided the first evidence that nucleolin may play an important role in regulating this pathway.
Our next paper focused on protein arginine methyltransferase 5 (PRMT5), a protein that was identified by mass spectrometry in a screen for nucleolin-associated proteins [31]. Our strategy for this research was to immunoprecipitate nucleolin from prostate cancer cells that were untreated, control-treated or AS1411-treated. We utilized both immunoprecipitation of endogenous nucleolin and capture of FLAG-tagged nucleolin from cells that were transiently transfected with the appropriate plasmid. Immunoprecipitations were carried out using nuclear or cytoplasmic extracts prepared from control or AS1411-treated cells and then analyzed on one-dimensional SDS-polyacrylamide gels to identify protein bands of interest. PRMT5 was identified by MALDI-TOF fingerprinting and confirmed by western blotting as a specific nucleolin-associated protein. The levels of nucleolin-associated PRMT5 were found to be dramatically decreased in the nucleus of AS1411-treated cells, but increased in the cytoplasm, in a dose-dependent and time-dependent manner. This AS1411-induced translocation was apparently dependent on nucleolin because it was not observed in cells where nucleolin was downregulated by siRNA [31]. We also showed that the activity of PRMT5, which catalyzes the formation of symmetrically dimethylated arginine (sDMA), was similarly shifted from the nucleus to the cytoplasm, as assessed by levels of proteins recognized by an antibody specific for sDMA residues. In contrast, levels of asymmetric dimethylation of arginine (aDMA), which is catalyzed by another enzyme (primarily PRMT1) were unaffected by AS1411, indicating the specificity of this effect. One of the reasons why we focused on PRMT5 is that this gene is increasingly being recognized as an oncogene that functions in the nucleus by methylating histone arginines, leading to transcriptional repression of various tumor suppressor genes [168]. Since AS1411 causes a decrease in nuclear PRMT5, we reasoned that PRMT5 target genes should be de-repressed (i.e. re-expressed) in AS1411-treated cells. This was found to be the case with the tumor suppressor gene ST7 and cell cycle regulator cyclin E2, and it was confirmed by chromatin immunoprecipitation (ChIP) to be associated with decreased levels of PRMT5 at these gene promoters [31].
Thus, we had identified a second nucleolin complex whose function (and also location) was altered by AS1411. It seems likely that re-expression of genes that are silenced by PRMT5 may contribute to the antiproliferative effects of AS1411. Moreover, our work to this point has provided strong support that nucleolin, both at the surface and inside the cell, is the critical determinant of AS1411 activity. It has also suggested that the effects that we see on PRMT5 and NEMO are representative of a much more general mechanism for AS1411, in which binding of the GRO to nucleolin results in dysfunction or mislocalization of numerous nucleolin-containing complexes. Since nucleolin plays multiple roles in cancer cell survival, we predict that the end result will be a pleiotropic anticancer effect (see Figure 5). Support for this idea has come from our recent proteomic studies [169] showing that the majority of nucleolin-associated proteins we have identified by mass spectrometry techniques have perturbed nuclear-to-cytoplasmic ratios in AS1411-treated cells. Interestingly, some nucleolin complexes were translocated from nucleus to cytoplasm (like PRMT5), whereas others were increased in the nucleus and decreased in the cytoplasm, perhaps suggesting a general effect of AS1411 on nucleolin's nuclear-cytoplasmic shuttling function. Further validation of our model has come from a recent paper by Soundararajan et al. [106], who reported that AS1411 causes decreased binding of cytoplasmic nucleolin to Bcl-2 mRNA. Since nucleolin stabilizes this mRNA [157], the result is a decrease in levels of the anti-apoptotic Bcl-2 protein, which will presumably contribute to the anticancer effects of this molecule. Another group has reported that AS1411 can increase BMP-2 mRNA stability and protein levels, although this occurs only in mycoplasma-infected cells [170].
AS1411 Affects Only Certain Forms of Nucleolin
While most of the data seem to support our hypothesis that AS1411 affects the nuclear-cytoplasmic trafficking of nucleolin complexes, there was one major inconsistency: Why do we not see any changes in the nuclear/ cytoplasmic ratio of nucleolin itself? We believe that our recent studies [31] may have provided a compelling answer to that question. While examining the effects of AS1411 on the PRMT5-nucleolin complex, we wondered what proportion of the total PRMT5 existed in a complex with nucleolin, and conversely, how much of the total nucleolin was in a complex with PRMT5? To try to estimate this, we quantified bands from western blots in order to compare total precipitated PRMT5 or nucleolin with bands that represented nucleolin-precipitated PRMT5 or sDMA-modified nucleolin. This latter measure was used as a surrogate for PRMT5-associated nucleolin (since we had shown that PRMT5 can modify nucleolin by sDMA [31]) because the PRMT5 antibody was not very effective for immunoprecipitation. Although this was only a semi-quantitative method, it was clear that, whereas the majority (approximately 80%) of PRMT5 was precipitated with nucleolin, only a small proportion (approximately 5%) of the nucleolin was in a complex with PRMT5. This would explain why we could observe the nuclear-to-cytoplasmic shift of PRMT5 by simple western blotting of extracts, yet we were unable to observe any major change when examining levels of nucleolin. Furthermore, by specifically examining the sDMA-modified form of nucleolin, we were able to detect a nuclear-to-cytoplasmic translocation comparable to that seen with PRMT5 [31]. Therefore, we believe that only a small subset of the total nucleolin molecules in the cell is affected by AS1411. We have identified sDMA-modified nucleolin as one target, but it is not yet clear if there are other specific post-translationally modified forms of nucleolin that are also affected.
Comparison of AS1411 with other G-rich Oligonucleotides
It appears that we are one among a small number of groups who are developing GROs as potentially therapeutic agents, although, to the best of our knowledge, AS1411 is the only GRO to have yet been tested as an anticancer agent in human clinical trials. Interestingly, many of the sequences under development are also unmodified, or minimally modified, phosphodiester oligodeoxynucleotides that have been discovered through serendipitous observations of activities that are dependent on the presence of G-rich repeats or quadruplex formation. Other sequences being investigated include telomere homologs or “T-oligos” with (TTAGGG)n repeats [41-49,57,61], DZ13 [54-56], T40214 [50-53], G20 [58,59], “GT oligonucleotides” [21-24] and a c-myc promoter sequence [34]. There have been no systematic studies to compare the effects and mechanisms of these diverse G-rich sequences, but it is striking that there are some conserved activities among them. These include tumor-selective antiproliferative effects [23,26,41,45,46,48,59,106], cell cycle perturbations [26,44,45,52,59], non-apoptotic cell death [26,41,55,60], induction of DNA damage responses [45,49,57,61], inactivation of NF-κB signaling [30,33,47], Stat3 inhibition [41,50,51,60] and down-regulation of Bcl-2 protein [52,106]. On the other hand, in the few papers where oligonucleotides have been specifically compared, there are also observed differences between sequences [24,56,59].
Ongoing Studies and Future Directions
Development of AS1411 has been, and continues to be, an enormously collaborative effort involving a large number of scientists and physicians, principally from the University of Louisville and the Brown Cancer Center, as well as investigators at Antisoma and, previously, Aptamera. As AS1411 moves forward through Phase II clinical trials, we continue to address remaining questions about the activity and mechanism of action of this molecule. In collaboration with Antisoma, we are searching for biomarkers that can monitor response to AS1411 and predict which patients will be most responsive to the drug. With support from NCI and DOD, we continue to probe the detailed molecular mechanism of AS1411 and further investigate the fascinating biology of nucleolin. In projects led by Dr. John O. Trent at the University of Louisville, we are working to solve the structure of AS1411 (in collaboration with Drs. Andrew N. Lane, and J. Brad Chaires) and to develop small molecule inhibitors of nucleolin. In the future, we look forward to working with other colleagues to investigate the use of aptamers to achieve tumor-targeted delivery of therapeutic or imaging agents, and perhaps to investigate the role of other cell types (e.g. immune cells and endothelial cells) in mediating the effects of G-rich oligonucleotides. Our progress in all of these endeavors has been facilitated by the collegial atmosphere, strong leadership and state-of-the-art core facilities at the Brown Cancer Center. In particular, the alliance of several individuals at the Brown Cancer Center who have expertise in the biology, structure and biophysics of quadruplex nucleic acids, combined with the translational mindset of the institution, has enabled this exciting bench-to-bedside journey.
Acknowledgments
We wish to thank the current and past members of our research groups who have carried out much of the work described herein. We also thank our collaborators at the University of Louisville, including J. Brad Chaires, Andrew N. Lane, Hong Ye, Wolfgang Zacharias, William M. Pierce, Jr., Jon B. Klein, W. Glenn McGregor, A. Bennett Jenson, Vivek Sharma, John W. Eaton, Mariusz Z. Ratajczak, Janina Ratajczak, and all of the researchers in their laboratories who have worked on this project. In addition, we thank our collaborator, Alison Rodger, from the University of Warwick, England. We would like to acknowledgement the contributions of all those from Aptamera and Antisoma who have been involved in developing AS1411, particularly David Jones (Antisoma), Kerry M. Barnhart (formerly of Aptamera) and the late Lloyd Kelland (formerly of Antisoma), our primary contacts at these companies. Aptamera funded the IND-directed pre-clinical studies and early clinical trials of AS1411, and Antisoma sponsored part of the Phase I clinical trial and are funding all ongoing clinical studies of AS1411. The laboratory studies described in this review were funded in part by: the Department of Defense Prostate Cancer Research Program (DAMD17-01-1-0067 and W81XWH-04-1-0183 to PJB; DAMD17-98-1-8583 to DMM); the National Institutes of Health (R01 CA122383, R21 CA91115 and R21 CA104230 to PJB; R01 CA113735 to JOT); and grants to PJB from the Kentucky Lung Cancer Research Program and the Komen Breast Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement: Some of the authors (PJB, JOT, DMM) are inventors of patented or patent-pending technologies related to AS1411, G-rich oligonucleotides and nucleolin. Some of the authors (PJB, JOT, DMM) are shareholders in Antisoma, the company that is now sponsoring the development of AS1411. In addition, PJB is presently a recipient of grant support from Antisoma on projects related to AS1411. DMM is also a co-founder of Advanced Cancer Therapeutics, Inc. and a member of the scientific advisory boards of Appoimmune, Inc. and Transmed Oncology, Inc. The other authors (DAL, SDT) have declared no conflicts of interest.
References
- 1.Gleave ME, Monia BP. Antisense therapy for cancer. Nat Rev Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 2.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gewirtz AM. On future's doorstep: RNA interference and the pharmacopeia of tomorrow. J Clin Invest. 2007;117:3612–3614. doi: 10.1172/JCI34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomita N, Ogihara T, Morishita R. Transcription factors as molecular targets: molecular mechanisms of decoy ODN and their design. Curr Drug Targets. 2003;4:603–608. doi: 10.2174/1389450033490803. [DOI] [PubMed] [Google Scholar]
- 6.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–83. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 7.Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 9.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 10.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaswen P, Stampfer MR, Ghosh K, Cohen JS. Effects of sequence of thioated oligonucleotides on cultured human mammary epithelial cells. Antisense Res Dev. 1993;3:67–77. doi: 10.1089/ard.1993.3.67. [DOI] [PubMed] [Google Scholar]
- 12.Maltese JY, Sharma HW, Vassilev L, Narayanan R. Sequence context of antisense RelA/NF-kappa B phosphorothioates determines specificity. Nucleic Acids Res. 1995;23:1146–1151. doi: 10.1093/nar/23.7.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White JR, Gordon-Smith EC, Rutherford TR. Phosphorothioate-capped antisense oligonucleotides to Ras GAP inhibit cell proliferation and trigger apoptosis but fail to downregulate GAP gene expression. Biochem Biophys Res Commun. 1996;227:118–124. doi: 10.1006/bbrc.1996.1476. [DOI] [PubMed] [Google Scholar]
- 14.Burgess TL, Fisher EF, Ross SL, Bready JV, Qian YX, Bayewitch LA, Cohen AM, Herrera CJ, Hu SS, Kramer TB, et al. The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc Natl Acad Sci U S A. 1995;92:4051–4055. doi: 10.1073/pnas.92.9.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benimetskaya L, Berton M, Kolbanovsky A, Benimetsky S, Stein CA. Formation of a G-tetrad and higher order structures correlates with biological activity of the RelA (NF-kappaB p65) ‘antisense’ oligodeoxynucleotide. Nucleic Acids Res. 1997;25:2648–2656. doi: 10.1093/nar/25.13.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Chen HJ, Sun J, Benimetskaya L, Schwartz A, Cannon P, Stein CA, Rabbani LE. A comparison of guanosine-quartet inhibitory effects versus cytidine homopolymer inhibitory effects on rat neointimal formation. Antisense Nucleic Acid Drug Dev. 1998;8:227–236. doi: 10.1089/oli.1.1998.8.227. [DOI] [PubMed] [Google Scholar]
- 17.Saijo Y, Uchiyama B, Abe T, Satoh K, Nukiwa T. Contiguous four-guanosine sequence in c-myc antisense phosphorothioate oligonucleotides inhibits cell growth on human lung cancer cells: possible involvement of cell adhesion inhibition. Jpn J Cancer Res. 1997;88:26–33. doi: 10.1111/j.1349-7006.1997.tb00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramanathan M, Lantz M, MacGregor RD, Huey B, Tam S, Li Y, Garovoy MR, Hunt CA. Inhibition of interferon-gamma-induced major histocompatibility complex class I expression by certain oligodeoxynucleotides. Transplantation. 1994;57:612–615. [PubMed] [Google Scholar]
- 19.Ramanathan M, Lantz M, MacGregor RD, Garovoy MR, Hunt CA. Characterization of the oligodeoxynucleotide-mediated inhibition of interferon-gamma-induced major histocompatibility complex class I and intercellular adhesion molecule-1. J Biol Chem. 1994;269:24564–24574. [PubMed] [Google Scholar]
- 20.Balasubramanian V, Nguyen LT, Balasubramanian SV, Ramanathan M. Interferon-gamma-inhibitory oligodeoxynucleotides alter the conformation of interferon-gamma. Mol Pharmacol. 1998;53:926–932. [PubMed] [Google Scholar]
- 21.Scaggiante B, Morassutti C, Dapas B, Tolazzi G, Ustulin F, Quadrifoglio F. Human cancer cell lines growth inhibition by GTn oligodeoxyribonucleotides recognizing single-stranded DNA-binding proteins. Eur J Biochem. 1998;252:207–215. doi: 10.1046/j.1432-1327.1998.2520207.x. [DOI] [PubMed] [Google Scholar]
- 22.Morassutti C, Dapas B, Scaggiante B, Paroni G, Xodo L, Quadrifoglio F. Effect of oligomer length and base substitutions on the cytotoxic activity and specific nuclear protein recognition of GTn oligonucleotides in the human leukemic CCRF-CEM cell line. Nucleosides Nucleotides. 1999;18:1711–1716. doi: 10.1080/07328319908044830. [DOI] [PubMed] [Google Scholar]
- 23.Dapas B, Tell G, Scaloni A, Pines A, Ferrara L, Quadrifoglio F, Scaggiante B. Identification of different isoforms of eEF1A in the nuclear fraction of human T-lymphoblastic cancer cell line specifically binding to aptameric cytotoxic GT oligomers. Eur J Biochem. 2003;270:3251–3262. doi: 10.1046/j.1432-1033.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- 24.Scaggiante B, Dapas B, Grassi G, Manzini G. Interaction of G-rich GT oligonucleotides with nuclear-associated eEF1A is correlated with their antiproliferative effect in haematopoietic human cancer cell lines. FEBS J. 2006;273:1350–1361. doi: 10.1111/j.1742-4658.2006.05143.x. [DOI] [PubMed] [Google Scholar]
- 25.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J Biol Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Hamhouyia F, Thomas SD, Burke TJ, Girvan AC, McGregor WG, Trent JO, Miller DM, Bates PJ. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J Biol Chem. 2001;276:43221–43230. doi: 10.1074/jbc.M104446200. [DOI] [PubMed] [Google Scholar]
- 27.Dapić V, Bates PJ, Trent JO, Rodger A, Thomas SD, Miller DM. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry. 2002;41:3676–3685. doi: 10.1021/bi0119520. [DOI] [PubMed] [Google Scholar]
- 28.Dapić V, Abdomerović V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMicken HW, Bates PJ, Chen Y. Antiproliferative activity of G-quartet-containing oligonucleotides generated by a novel single-stranded DNA expression system. Cancer Gene Ther. 2003;10:867–869. doi: 10.1038/sj.cgt.7700652. [DOI] [PubMed] [Google Scholar]
- 30.Girvan AC, Teng Y, Casson LK, Thomas SD, Jüliger S, Ball MW, Klein JB, Pierce WM, Jr, Barve SS, Bates PJ. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 31.Teng Y, Girvan AC, Casson LK, Pierce WM, Jr, Qian M, Thomas SD, Bates PJ. AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin. Cancer Res. 2007;67:10491–10500. doi: 10.1158/0008-5472.CAN-06-4206. [DOI] [PubMed] [Google Scholar]
- 32.Tam RC, Wu-Pong S, Pai B, Lim C, Chan A, Thomas DF, Milovanovic T, Bard J, Middletonm PJ. Increased potency of an aptameric G-rich oligonucleotide is associated with novel functional properties of phosphorothioate linkages. Antisense Nucleic Acid Drug Dev. 1999;9:289–300. doi: 10.1089/oli.1.1999.9.289. [DOI] [PubMed] [Google Scholar]
- 33.Shen W, Waldschmidt M, Zhao X, Ratliff T, Krieg AM. Antitumor mechanisms of oligodeoxynucleotides with CpG and polyG motifs in murine prostate cancer cells: decrease of NF-kappaB and AP-1 binding activities and induction of apoptosis. Antisense Nucleic Acid Drug Dev. 2002;12:155–164. doi: 10.1089/108729002760220752. [DOI] [PubMed] [Google Scholar]
- 34.Simonsson T, Henriksson M. c-myc Suppression in Burkitt's lymphoma cells. Biochem Biophys Res Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
- 35.Anselmet A, Mayat E, Wietek S, Layer PG, Payrastre B, Massoulié J. Non-antisense cellular responses to oligonucleotides. FEBS Lett. 2002;510:175–180. doi: 10.1016/s0014-5793(01)03248-3. [DOI] [PubMed] [Google Scholar]
- 36.Akhtar S, Dunnion D, Poyner D, Ackroyd J, Bibby M, Double J. Sequence and chemistry requirements for a novel aptameric oligonucleotide inhibitor of EGF receptor tyrosine kinase activity. Biochem Pharmacol. 2002;63:2187–2195. doi: 10.1016/s0006-2952(02)00985-1. [DOI] [PubMed] [Google Scholar]
- 37.Cogoi S, Ballico M, Bonora GM, Xodo LE. Antiproliferative activity of a triplex-forming oligonucleotide recognizing a Ki-ras polypurine/polypyrimidine motif correlates with protein binding. Cancer Gene Ther. 2004;11:465–476. doi: 10.1038/sj.cgt.7700722. [DOI] [PubMed] [Google Scholar]
- 38.Cogoi S, Quadrifoglio F, Xodo LE. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- 39.Filion MC, Filion B, Roy J, Ménard S, Reader S, Phillips NC. Development of immunomodulatory six base-length non-CpG motif oligonucleotides for cancer vaccination. Vaccine. 2004;22:2480–2488. doi: 10.1016/j.vaccine.2003.11.072. [DOI] [PubMed] [Google Scholar]
- 40.Filion MC, Saha N, Gueddi M, Phillips NC. Development of short non-CpG phosphodiester oligonucleotides as immune stimulatory agents. Vaccine. 2003;21:983–989. doi: 10.1016/s0264-410x(02)00549-2. [DOI] [PubMed] [Google Scholar]
- 41.Aoki H, Iwado E, Eller MS, Kondo Y, Fujiwara K, Li GZ, Hess KR, Siwak DR, Sawaya R, Mills GB, Gilchrest BA, Kondo S. Telomere 3′ overhang-specific DNA oligonucleotides induce autophagy in malignant glioma cells. FASEB J. 2007;21:2918–2930. doi: 10.1096/fj.06-6941com. [DOI] [PubMed] [Google Scholar]
- 42.Eller MS, Puri N, Hadshiew IM, Venna SS, Gilchrest BA. Induction of apoptosis by telomere 3′ overhang-specific DNA. Exp Cell Res. 2002;276:185–193. doi: 10.1006/excr.2002.5531. [DOI] [PubMed] [Google Scholar]
- 43.Li GZ, Eller MS, Firoozabadi R, Gilchrest BA. Evidence that exposure of the telomere 3′ overhang sequence induces senescence. Proc Natl Acad Sci U S A. 2003;100:527–531. doi: 10.1073/pnas.0235444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eller MS, Li GZ, Firoozabadi R, Puri N, Gilchrest BA. Induction of a p95/Nbs1-mediated S phase checkpoint by telomere 3′ overhang specific DNA. FASEB J. 2003;17:152–162. doi: 10.1096/fj.02-0197com. [DOI] [PubMed] [Google Scholar]
- 45.Puri N, Eller MS, Byers HR, Dykstra S, Kubera J, Gilchrest BA. Telomere-based DNA damage responses: a new approach to melanoma. FASEB J. 2004;18:1373–1381. doi: 10.1096/fj.04-1774com. [DOI] [PubMed] [Google Scholar]
- 46.Li GZ, Eller MS, Hanna K, Gilchrest BA. Signaling pathway requirements for induction of senescence by telomere homolog oligonucleotides. Exp Cell Res. 2004;301:189–200. doi: 10.1016/j.yexcr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 47.Marwaha V, Chen YH, Helms E, Arad S, Inoue H, Bord E, Kishore R, Sarkissian RD, Gilchrest BA, Goukassian DA. T-oligo treatment decreases constitutive and UVB-induced COX-2 levels through p53- and NFkappaB-dependent repression of the COX-2 promoter. J Biol Chem. 2005;280:32379–32388. doi: 10.1074/jbc.M503245200. [DOI] [PubMed] [Google Scholar]
- 48.Yaar M, Eller MS, Panova I, Kubera J, Wee LH, Cowan KH, Gilchrest BA. Telomeric DNA induces apoptosis and senescence of human breast carcinoma cells. Breast Cancer Res. 2007;9:R13. doi: 10.1186/bcr1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohashi N, Yaar M, Eller MS, Truzzi F, Gilchrest BA. Features that determine telomere homolog oligonucleotide-induced therapeutic DNA damage-like responses in cancer cells. J Cell Physiol. 2007;210:582–595. doi: 10.1002/jcp.20848. Erratum in: J Cell Physiol. 2185, 285. [DOI] [PubMed] [Google Scholar]
- 50.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 51.Jing N, Zhu Q, Yuan P, Li Y, Mao L, Tweardy DJ. Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: a potential novel therapy for head and neck cancer. Mol Cancer Ther. 2006;5:279–286. doi: 10.1158/1535-7163.MCT-05-0302. [DOI] [PubMed] [Google Scholar]
- 52.Weerasinghe P, Garcia GE, Zhu Q, Yuan P, Feng L, Mao L, Jing N. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol. 2007;31:129–136. [PubMed] [Google Scholar]
- 53.Weerasinghe P, Li Y, Guan Y, Zhang R, Tweardy DJ, Jing N. T40214/PEI complex: a potent therapeutics for prostate cancer that targets STAT3 signaling. Prostate. 2008;68:1430–1442. doi: 10.1002/pros.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivory L, Tucker C, King A, Lai A, Goodchild A, Witherington C, Gozar MM, Birkett DJ. The DNAzymes Rs6, Dz13, and DzF have potent biologic effects independent of catalytic activity. Oligonucleotides. 2006;16:297–312. doi: 10.1089/oli.2006.16.297. [DOI] [PubMed] [Google Scholar]
- 55.Gozar MM, Goodchild A, Passioura T, King A, Lai A, Witherington C, Rivory L. Dz13, a DNAzyme targeting c-jun, induces off-target cytotoxicity in endothelial cells with features of nonapoptotic programmed cell death. Oligonucleotides. 2008;18:257–268. doi: 10.1089/oli.2008.0139. [DOI] [PubMed] [Google Scholar]
- 56.Goodchild A, King A, Gozar MM, Passioura T, Tucker C, Rivory L. Cytotoxic G-rich oligodeoxynucleotides: putative protein targets and required sequence motif. Nucleic Acids Res. 2007;35:4562–4572. doi: 10.1093/nar/gkm465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi H, Lin CP, Fu X, Wood LM, Liu AA, Tsai YC, Chen Y, Barbieri CM, Pilch DS, Liu LF. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 58.Skogen M, Roth J, Yerkes S, Parekh-Olmedo H, Kmiec E. Short G-rich oligonucleotides as a potential therapeutic for Huntington's Disease. BMC Neurosci. 2006;7:65. doi: 10.1186/1471-2202-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz TR, Vasta CA, Bauer TL, Parekh-Olmedo H, Kmiec EB. G-rich oligonucleotides alter cell cycle progression and induce apoptosis specifically in OE19 esophageal tumor cells. Oligonucleotides. 2008;18:51–63. doi: 10.1089/oli.2007.0109. [DOI] [PubMed] [Google Scholar]
- 60.Yokoyama T, Kondo Y, Kondo S. Roles of mTOR and STAT3 in autophagy induced by telomere 3′ overhang-specific DNA oligonucleotides. Autophagy. 2007;3:496–498. doi: 10.4161/auto.4602. [DOI] [PubMed] [Google Scholar]
- 61.Tsolou A, Passos JF, Nelson G, Arai Y, Zglinicki T. ssDNA fragments induce cell senescence by telomere uncapping. Exp Gerontol. 2008;43:892–899. doi: 10.1016/j.exger.2008.08.043. [DOI] [PubMed] [Google Scholar]
- 62.Matsugami A, Mashima T, Nishikawa F, Murakami K, Nishikawa S, Noda K, Yokoyama T, Katahira M. Structural analysis of r(GGA)4 found in RNA aptamer for bovine prion protein. Nucleic Acids Symp Ser. 2008;52:179–180. doi: 10.1093/nass/nrn091. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida W, Mochizuki E, Takase M, Hasegawa H, Morita Y, Yamazaki H, Sode K, Ikebukuro K. Selection of DNA aptamers against insulin and construction of an aptameric enzyme subunit for insulin sensing. Biosens Bioelectron. 2008 doi: 10.1016/j.bios.2008.06.016. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 64.Li T, Li B, Dong S. Aptamer-based label-free method for hemin recognition and DNA assay by capillary electrophoresis with chemiluminescence detection. Anal Bioanal Chem. 2007;389:887–893. doi: 10.1007/s00216-007-1487-5. [DOI] [PubMed] [Google Scholar]
- 65.Lévesque D, Beaudoin JD, Roy S, Perreault JP. In vitro selection and characterization of RNA aptamers binding thyroxine hormone. Biochem J. 2007;403:129–138. doi: 10.1042/BJ20061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Homann M, Lorger M, Engstler M, Zacharias M, Göringer HU. Serum-stable RNA aptamers to an invariant surface domain of live African trypanosomes. Comb Chem High Throughput Screen. 2006;9:491–499. doi: 10.2174/138620706777935324. [DOI] [PubMed] [Google Scholar]
- 67.Phan AT, Kuryavyi V, Ma JB, Faure A, Andréola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc Natl Acad Sci U S A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori T, Oguro A, Ohtsu T, Nakamura Y. RNA aptamers selected against the receptor activator of NF-kappaB acquire general affinity to proteins of the tumor necrosis factor receptor family. Nucleic Acids Res. 2004;32:6120–6128. doi: 10.1093/nar/gkh949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wen JD, Gray DM. The Ff gene 5 single-stranded DNA-binding protein binds to the transiently folded form of an intramolecular G-quadruplex. Biochemistry. 2002;41:11438–11448. doi: 10.1021/bi020276e. [DOI] [PubMed] [Google Scholar]
- 70.Wilson C, Szostak JW. Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem Biol. 1998;5:609–617. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 71.Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D. High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor. I J Immunol. 1996;157:221–230. [PubMed] [Google Scholar]
- 72.Lauhon CT, Szostak JW. RNA aptamers that bind flavin and nicotinamide redox cofactors. J Am Chem Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 73.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 74.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci U S A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SW, Song MK, Baek KH, Park Y, Kim JK, Lee CH, Cheong HK, Cheong C, Sung YC. Effects of a hexameric deoxyriboguanosine run conjugation into CpG oligodeoxynucleotides on their immunostimulatory potentials. J Immunol. 2000;165:3631–3639. doi: 10.4049/jimmunol.165.7.3631. [DOI] [PubMed] [Google Scholar]
- 76.Dalpke AH, Zimmermann S, Albrecht I, Heeg K. Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology. 2002;106:102–112. doi: 10.1046/j.1365-2567.2002.01410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu CC, Castro JE, Motta M, Cottam HB, Kyburz D, Kipps TJ, Corr M, Carson DA. Selection of oligonucleotide aptamers with enhanced uptake and activation of human leukemia B cells. Hum Gene Ther. 2003;14:849–860. doi: 10.1089/104303403765701141. [DOI] [PubMed] [Google Scholar]
- 78.Bartz H, Mendoza Y, Gebker M, Fischborn T, Heeg K, Dalpke A. Poly-guanosine strings improve cellular uptake and stimulatory activity of phosphodiester CpG oligonucleotides in human leukocytes. Vaccine. 2004;23:148–155. doi: 10.1016/j.vaccine.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 79.Wu CC, Lee J, Raz E, Corr M, Carson DA. Necessity of oligonucleotide aggregation for toll-like receptor 9 activation. J Biol Chem. 2004;279:33071–33078. doi: 10.1074/jbc.M311662200. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Z, Guo K, Schluesener HJ. The immunostimulatory activity of CpG oligonucleotides on microglial N9 cells is affected by a polyguanosine motif. J Neuroimmunol. 2005;161:68–77. doi: 10.1016/j.jneuroim.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z, Weinschenk T, Schluesener HJ. Uptake, intracellular distribution, and novel binding proteins of immunostimulatory CpG oligodeoxynucleotides in microglial cells. J Neuroimmunol. 2005;160:32–40. doi: 10.1016/j.jneuroim.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 82.Stein CA. The experimental use of antisense oligonucleotides: a guide for the perplexed. J Clin Invest. 2001;108:641–644. doi: 10.1172/JCI13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agrawal S, Iadarola PL, Temsamani J, Zhao Q, Shaw DR. Effect of G-rich sequences on the synthesis, purification, hybridization, cell uptake and hemolytic activity of oligonucleotides. Bioorgan Med Chem Lett. 1996;6:2219–2224. [Google Scholar]
- 84.Biyani M, Nishigaki K. Structural characterization of ultra-stable higher-ordered aggregates generated by novel guanine-rich DNA sequences. Gene. 2005;364:130–138. doi: 10.1016/j.gene.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 85.Shida T, Suda M, Sekiguchi J. Quadruplex formation of oligonucleotides containing G clusters and their nuclease resistance. Nucleic Acids Symp Ser. 1997;37:129–130. [PubMed] [Google Scholar]
- 86.Bishop JS, Guy-Caffey JK, Ojwang JO, Smith SR, Hogan ME, Cossum PA, Rando RF, Chaudhary N. Intramolecular G-quartet motifs confer nuclease resistance to a potent anti-HIV oligonucleotide. J Biol Chem. 1996;271:5698–56703. doi: 10.1074/jbc.271.10.5698. [DOI] [PubMed] [Google Scholar]
- 87.Cao Z, Huang CC, Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal Chem. 2006;78:1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 88.Srividya S, Roy RP, Basu SK, Mukhopadhyay A. Selective activation of antitumor activity of macrophages by the delivery of muramyl dipeptide using a novel polynucleotide-based carrier recognized by scavenger receptors. Biochem Biophys Res Commun. 2000;268:772–777. doi: 10.1006/bbrc.2000.2216. [DOI] [PubMed] [Google Scholar]
- 89.Prasad V, Hashim S, Mukhopadhyay A, Basu SK, Roy RP. Oligonucleotides tethered to a short polyguanylic acid stretch are targeted to macrophages: enhanced antiviral activity of a vesicular stomatitis virus-specific antisense oligonucleotide. Antimicrob Agents Chemother. 1999;43:2689–2696. doi: 10.1128/aac.43.11.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hughes JA, Avrutskaya AV, Juliano RL. Influence of base composition on membrane binding and cellular uptake of 10-mer phosphorothioate oligonucleotides in Chinese hamster ovary (CHRC5) cells. Antisense Res Dev. 1994;4:211–215. doi: 10.1089/ard.1994.4.211. [DOI] [PubMed] [Google Scholar]
- 91.Vlassov VV, Balakireva LA, Yakubov LA. Transport of oligonucleotides across natural and model membranes. Biochim Biophys Acta. 1994;1197:95–108. doi: 10.1016/0304-4157(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 92.Beltinger C, Saragovi HU, Smith RM, LeSauteur L, Shah N, DeDionisio L, Christensen L, Raible A, Jarett L, Gewirtz AM. Binding, uptake, and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J Clin Invest. 1995;95:1814–1823. doi: 10.1172/JCI117860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basner-Tschakarjan E, Mirmohammadsadegh A, Baer A, Hengge UR. Uptake and trafficking of DNA in keratinocytes: evidence for DNA-binding proteins. Gene Ther. 2004;11:765–774. doi: 10.1038/sj.gt.3302221. [DOI] [PubMed] [Google Scholar]
- 94.Corrias S, Cheng YC. Interaction of human plasma membrane proteins and oligodeoxynucleotides. Biochem Pharmacol. 1998;55:1221–1227. doi: 10.1016/s0006-2952(97)00595-9. [DOI] [PubMed] [Google Scholar]
- 95.Biessen EA, Vietsch H, Kuiper J, Bijsterbosch MK, Berkel TJ. Liver uptake of phosphodiester oligodeoxynucleotides is mediated by scavenger receptors. Mol Pharmacol. 1998;53:262–269. doi: 10.1124/mol.53.2.262. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki K, Doi T, Imanishi T, Kodama T, Tanaka T. Oligonucleotide aggregates bind to the macrophage scavenger receptor. Eur J Biochem. 1999;260:855–860. doi: 10.1046/j.1432-1327.1999.00233.x. [DOI] [PubMed] [Google Scholar]
- 97.Wyatt JR, Vickers TA, Roberson JL, Buckheit RW, Jr, Klimkait T, DeBaets E, Davis PW, Rayner B, Imbach JL, Ecker DJ. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc Natl Acad Sci U S A. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ojwang J, Elbaggari A, Marshall HB, Jayaraman K, McGrath MS, Rando RF. Inhibition of human immunodeficiency virus type 1 activity in vitro by oligonucleotides composed entirely of guanosine and thymidine. J Acquir Immune Defic Syndr. 1994;7:560–570. [PubMed] [Google Scholar]
- 99.Ojwang J, Okleberry KM, Marshall HB, Vu HM, Huffman JH, Rando RF. Inhibition of Friend murine leukemia virus activity by guanosine/thymidine oligonucleotides. Antiviral Res. 1994;25:27–41. doi: 10.1016/0166-3542(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 100.Fennewald SM, Mustain S, Ojwang J, Rando RF. Inhibition of herpes simplex virus in culture by oligonucleotides composed entirely of deoxyguanosine and thymidine. Antiviral Res. 1995;26:37–54. doi: 10.1016/0166-3542(94)00064-f. [DOI] [PubMed] [Google Scholar]
- 101.Kaur H, Jaso-Friedmann L, Leary JH, 3rd, Evans DL. Single-base oligodeoxyguanosine-binding proteins on nonspecific cytotoxic cells: identification of a new class of pattern-recognition receptors. Scand J Immunol. 2004;60:238–248. doi: 10.1111/j.0300-9475.2004.01455.x. [DOI] [PubMed] [Google Scholar]
- 102.Lang R, Hultner L, Lipford GB, Wagner H, Heeg K. Guanosine-rich oligodeoxynucleotides induce proliferation of macrophage progenitors in cultures of murine bone marrow cells. Eur J Immunol. 1999;29:3496–3506. doi: 10.1002/(SICI)1521-4141(199911)29:11<3496::AID-IMMU3496>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 103.Patole PS, Zecher D, Pawar RD, Gröne HJ, Schlöndorff D, Anders HJ. G-rich DNA suppresses systemic lupus. J Am Soc Nephrol. 2005;16:3273–3280. doi: 10.1681/ASN.2005060658. [DOI] [PubMed] [Google Scholar]
- 104.Trieu A, Roberts TL, Dunn JA, Sweet MJ, Stacey KJ. DNA motifs suppressing TLR9 responses. Crit Rev Immunol. 2006;26:527–544. doi: 10.1615/critrevimmunol.v26.i6.50. [DOI] [PubMed] [Google Scholar]
- 105.Kochetkova M, Iversen PO, Lopez AF, Shannon MF. Deoxyribonucleic acid triplex formation inhibits granulocyte macrophage colony-stimulating factor gene expression and suppresses growth in juvenile myelomonocytic leukemic cells. J Clin Invest. 1997;99:3000–3008. doi: 10.1172/JCI119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 107.Shah K, Djeha H, Richie C, McGeever G, Ahmad T, Green C, Courtenay-Luck N, Otake Y, Fernandes D. AS1411, a novel DNA aptamer as a potential treatment of acute myelogenous leukaemia (AML) Blood. 2006;108:564A–565A. Meeting Abstract. [Google Scholar]
- 108.Barnhart KM, Laber DA, Bates PJ, Trent JO, Miller DM. AGRO100: The translation from lab to clinic of a tumor-targeted nucleic acid aptamer. J Clin Oncol. 2004;22:226S–226S. Meeting Abstract. [Google Scholar]
- 109.Laber DA, Choudry MA, Taft BS, Bhupalam L, Sharma VR, Hendler FJ, Barnhart KM. A phase I study of AGRO100 in advanced cancer. J Clin Oncol. 2004;22:3112. Meeting Abstract. [Google Scholar]
- 110.Laber DA, Sharma VR, Bhupalam L, Taft B, Hendler FJ, Barnhart KM. Update on the first phase I study of AGRO100 in advanced cancer. J Clin Oncol. 2005;23:3064. Meeting Abstract. [Google Scholar]
- 111.Laber D, Bates P, Trent J, Barnhart K, Taft B, Miller D. Long term clinical response in renal cell carcinoma patients treated with quadruplex forming oligonucleotides. Clin Cancer Res. 2005;11:9088S. Meeting Abstract. [Google Scholar]
- 112.Miller DM, Laber DA, Bates PJ, Trent JO, Taft BS, Kloecker GH, Acton G. Extended phase I study of AS1411 in renal and non-small cell lung cancers. Ann Oncol. 2006;17:147–148. Meeting Abstract. [Google Scholar]
- 113.Laber DA, Taft BS, Kloecker GH, et al. Pharmacokinetics of the anti-nucleolin aptamer AS1411 in a phase I study. Mol Cancer Ther. 2007;6:3574S–3574S. Meeting Abstract. [Google Scholar]
- 114.Chen W, Sridharan V, Soundararajan S, Otake Y, Stuart R, Jones D, Fernandes D. Activity and mechanism of action of AS1411 in acute myeloid leukemia cells. Blood. 2007;110:479A–479A. Meeting Abstract. [Google Scholar]
- 115.Dickinson LA, Kohwi-Shigematsu T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol Cell Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 118.Dempsey LA, Sun H, Hanakahi LA, Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 119.Girvan AC. University of Louisville Electronic Theses and Dissertations. 2006. Mechanistic studies of guanine-rich oligonucleotides (Doctoral Dissertation) [Google Scholar]
- 120.Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M, Borowiec JA. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iliakis G, Krieg T, Guan J, Wang Y, Leeper D. Evidence for an S-phase checkpoint regulating DNA replication after heat shock: a review. Int J Hyperthermia. 2004;20:240–249. doi: 10.1080/02656730310001656379. [DOI] [PubMed] [Google Scholar]
- 122.Soleimani-Meigooni DN, Mi Y, Thomas SD, Jenson AB, Bates PJ. Uncoupling of cell growth and division: An unusual mechanism of non-apoptotic cell death in leukemia cells treated with novel anticancer aptamer, AS1411. Proc AACR 2007 (Meeting Abstract).2007. [Google Scholar]
- 123.Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- 124.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1996;56(3):379–390. doi: 10.1016/0092-8674(89)90241-9. 1989 Feb 10. [DOI] [PubMed] [Google Scholar]
- 125.Semenkovich CF, Ostlund RE, Jr, Olson MO, Yang JW. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry. 1990;29:9708–9713. doi: 10.1021/bi00493a028. [DOI] [PubMed] [Google Scholar]
- 126.Jordan P, Heid H, Kinzel V, Kübler D. Major cell surface-located protein substrates of an ecto-protein kinase are homologs of known nuclear proteins. Biochemistry. 1994;33:14696–14706. doi: 10.1021/bi00253a007. [DOI] [PubMed] [Google Scholar]
- 127.Kibbey MC, Johnson B, Petryshyn R, Jucker M, Kleinman HK. A 110-kD nuclear shuttling protein, nucleolin, binds to the neurite-promoting IKVAV site of laminin-1. J Neurosci Res. 1995;42:314–322. doi: 10.1002/jnr.490420305. [DOI] [PubMed] [Google Scholar]
- 128.Krantz S, Salazar R, Brandt R, Kellermann J, Lottspeich F. Purification and partial amino acid sequencing of a fructosyllysine-specific binding protein from cell membranes of the monocyte-like cell line U937. Biochim Biophys Acta. 1995;1266:109–112. doi: 10.1016/0167-4889(95)00028-q. [DOI] [PubMed] [Google Scholar]
- 129.Deng JS, Ballou B, Hofmeister JK. Internalization of anti-nucleolin antibody into viable HEp-2 cells. Mol Biol Rep. 1996;23:191–195. doi: 10.1007/BF00351168. [DOI] [PubMed] [Google Scholar]
- 130.Callebaut C, Blanco J, Benkirane N, Krust B, Jacotot E, Guichard G, Seddiki N, Svab J, Dam E, Muller S, Briand JP, Hovanessian AG. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J Biol Chem. 1998;273:21988–21697. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- 131.Bates PJ, Miller DM, Trent JO, Xu X. Method for the diagnosis and prognosis of malignant diseases. United States Patent 7,357,928 2008
- 132.Storck S, Shukla M, Dimitrov S, Bouvet P. Functions of the histone chaperone nucleolin in diseases. Subcell Biochem. 2007;41:125–144. doi: 10.1007/1-4020-5466-1_7. [DOI] [PubMed] [Google Scholar]
- 133.Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, Krust B. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261:312–28. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- 134.Legrand D, Vigie K, Said EA, Elass E, Masson M, Slomianny MC, Carpentier M, Briand JP, Mazurier J, Hovanessian AG. Surface nucleolin participates in both the binding and endocytosis of lactoferrin in target cells. Eur J Biochem. 2004;271:303–317. doi: 10.1046/j.1432-1033.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- 135.Chen X, Kube DM, Cooper MJ, Davis PB. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 2008;16:333–342. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- 136.Dumler I, Stepanova V, Jerke U, Mayboroda OA, Vogel F, Bouvet P, Tkachuk V, Haller H, Gulba DC. Urokinase-induced mitogenesis is mediated by casein kinase 2 and nucleolin. Curr Biol. 1999;9:1468–1476. doi: 10.1016/s0960-9822(00)80116-5. [DOI] [PubMed] [Google Scholar]
- 137.Harms G, Kraft R, Grelle G, Volz B, Dernedde J, Tauber R. Identification of nucleolin as a new L-selectin ligand. Biochem J. 2001;360:531–538. doi: 10.1042/0264-6021:3600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reyes-Reyes EM, Akiyama SK. Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp Cell Res. 2008;314:2212–2223. doi: 10.1016/j.yexcr.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tate A, Isotani S, Bradley MJ, Sikes RA, Davis R, Chung LW, Edlund M. Met-Independent Hepatocyte Growth Factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer. 2006;6:197. doi: 10.1186/1471-2407-6-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Larrucea S, Gonzalez-Rubio C, Cambronero R, Ballou B, Bonay P, Lopez-Granados E, Bouvet P, Fontan G, Fresno M, Lopez-Trascasa M. Cellular adhesion mediated by factor J, a complement inhibitor. Evidence for nucleolin involvement J Biol Chem. 1998;273:31718–31725. doi: 10.1074/jbc.273.48.31718. [DOI] [PubMed] [Google Scholar]
- 141.Sorokina EA, Wesson JA, Kleinman JG. An acidic peptide sequence of nucleolin-related protein can mediate the attachment of calcium oxalate to renal tubule cells. J Am Soc Nephrol. 2004;15:2057–2065. doi: 10.1097/01.ASN.0000133024.83256.C8. [DOI] [PubMed] [Google Scholar]
- 142.Joo EJ, ten Dam GB, van Kuppevelt TH, Toida T, Linhardt RJ, Kim YS. Nucleolin: acharan sulfate-binding protein on the surface of cancer cells. Glycobiology. 2005;15:1–9. doi: 10.1093/glycob/cwh132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carpentier M, Morelle W, Coddeville B, Pons A, Masson M, Mazurier J, Legrand D. Nucleolin undergoes partial N- and O-glycosylations in the extranuclear cell compartment. Biochemistry. 2005;44:5804–5815. doi: 10.1021/bi047831s. [DOI] [PubMed] [Google Scholar]
- 144.Sinclair JF, O'Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002;277:2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- 145.de Verdugo UR, Selinka HC, Huber M, Kramer B, Kellermann J, Hofschneider PH, Kandolf R. Characterization of a 100-kilodalton binding protein for the six serotypes of coxsackie B viruses. J Virol. 1995;69:6751–6757. doi: 10.1128/jvi.69.11.6751-6757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nisole S, Said EA, Mische C, Prevost MC, Krust B, Bouvet P, Bianco A, Briand JP, Hovanessian AG. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J Biol Chem. 2002;277:20877–20886. doi: 10.1074/jbc.M110024200. [DOI] [PubMed] [Google Scholar]
- 147.Said EA, Krust B, Nisole S, Svab J, Briand JP, Hovanessian AG. The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J Biol Chem. 2002;277:37492–37502. doi: 10.1074/jbc.M201194200. [DOI] [PubMed] [Google Scholar]
- 148.Bose S, Basu M, Banerjee AK. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. J Virol. 2004;78:8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barel M, Hovanessian AG, Meibom K, Briand JP, Dupuis M, Charbit A. A novel receptor - ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol. 2008;8:145. doi: 10.1186/1471-2180-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shibata Y, Muramatsu T, Hirai M, Inui T, Kimura T, Saito H, McCormick LM, Bu G, Kadomatsu K. Nuclear targeting by the growth factor midkine. Mol Cell Biol. 2002;22:6788–6796. doi: 10.1128/MCB.22.19.6788-6796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alete DE, Weeks ME, Hovanession AG, Hawadle M, Stoker AW. Cell surface nucleolin on developing muscle is a potential ligand for the axonal receptor protein tyrosine phosphatase-sigma. FEBS J. 2006;273:4668–4681. doi: 10.1111/j.1742-4658.2006.05471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hirano K, Miki Y, Hirai Y, Sato R, Itoh T, Hayashi A, Yamanaka M, Eda S, Beppu M. A multifunctional shuttling protein nucleolin is a macrophage receptor for apoptotic cells. J Biol Chem. 2005;280:39284–39293. doi: 10.1074/jbc.M505275200. [DOI] [PubMed] [Google Scholar]
- 153.Huang Y, Shi H, Zhou H, Song X, Yuan S, Luo Y. The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood. 2006;107:3564–3571. doi: 10.1182/blood-2005-07-2961. [DOI] [PubMed] [Google Scholar]
- 154.Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J Cell Biol. 2003;163:871–878. doi: 10.1083/jcb.200304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shi H, Huang Y, Zhou H, Song X, Yuan S, Fu Y, Luo Y. Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood. 2007;110:2899–2906. doi: 10.1182/blood-2007-01-064428. [DOI] [PubMed] [Google Scholar]
- 156.Destouches D, El Khoury D, Hamma-Kourbali Y, Krust B, Albanese P, Katsoris P, Guichard G, Briand JP, Courty J, Hovanessian AG. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS ONE. 2008;3:e2518. doi: 10.1371/journal.pone.0002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Di Segni A, Farin K, Pinkas-Kramarski R. Identification of nucleolin as new ErbB receptors- interacting protein. PLoS ONE. 2008;3:e2310. doi: 10.1371/journal.pone.0002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Otake Y, Soundararajan S, Sengupta TK, Kio EA, Smith JC, Pineda-Roman M, Stuart RK, Spicer EK, Fernandes DJ. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood. 2007;109:3069–3075. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grinstein E, Shan Y, Karawajew L, Snijders PJ, Meijer CJ, Royer HD, Wernet P. Cell cycle-controlled interaction of nucleolin with the retinoblastoma protein and cancerous cell transformation. J Biol Chem. 2006;281:22223–22235. doi: 10.1074/jbc.M513335200. [DOI] [PubMed] [Google Scholar]
- 160.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]; Cell. 123:536–537. Erratum in: [Google Scholar]
- 161.Fu Z, Fenselau C. Proteomic evidence for roles for nucleolin and poly[ADP-ribosyl] transferase in drug resistance. J Proteome Res. 2005;4:1583–1591. doi: 10.1021/pr0501158. [DOI] [PubMed] [Google Scholar]
- 162.Grinstein E, Wernet P, Snijders PJ, Rösl F, Weinert I, Jia W, Kraft R, Schewe C, Schwabe M, Hauptmann S, Dietel M, Meijer CJ, Royer HD. Nucleolin as activator of human papillomavirus type 18 oncogene transcription in cervical cancer. J Exp Med. 2002;196:1067–1078. doi: 10.1084/jem.20011053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 164.Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 165.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 166.Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- 167.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 168.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Teng Y, Pierce WM, Bates PJ. Proteomic studies of nucleolin complexes in prostate cancer cells treated with novel anticancer agent, AS1411. Proc AACR 2007 (Meeting Abstract).2007. [Google Scholar]
- 170.Jiang S, Zhang S, Langenfeld J, Lo SC, Rogers MB. Mycoplasma infection transforms normal lung cells and induces bone morphogenetic protein 2 expression by post-transcriptional mechanisms. J Cell Biochem. 2008;104:580–894. doi: 10.1002/jcb.21647. [DOI] [PubMed] [Google Scholar]
- 171.Ritchie C, Doran B, Shah K, Rowlinson-Busza G, Courtenay-Luck N, Jones D. Combination of the aptamer AS1411 with paclitaxel or Ara-C produces synergistic inhibition of cancer cell growth. Proc AACR 2007 (Meeting Abstract).2007. [Google Scholar]


