Abstract
Tyzzer discovered the genus Cryptosporidium a century ago, and for almost 70 years cryptosporidiosis was regarded as an infrequent and insignificant infection that occurred in the intestines of vertebrates and caused little or no disease. Its association with gastrointestinal illness in humans and animals was recognized only in the early 1980s. Over the next 25 years, information was generated on the disease's epidemiology, biology, cultivation, taxonomy and development of molecular tools. Milestones include: (i) recognition in 1980 of cryptosporidiosis as an acute enteric disease; (ii) its emergence as a chronic opportunistic infection that complicates AIDS; (iii) acknowledgement of impact on the water industry once it was shown to be waterborne; and (iv) study of Cryptosporidium genomics.
Cryptosporidium – a harmless and incidental agent
Over 100 years have passed since Ernest Edward Tyzzer (1875–1965) first made his observations on the genus Cryptosporidium in 1907 [1] (Figure 1). His initial observations were followed by two publications in 1910 and 1912 [2,3]. Cryptosporidium was so named because of the absence of sporocysts within the oocysts, a characteristic of other coccidia. Tyzzer, a distinguished medical parasitologist at Harvard University in Boston, MA, published numerous papers in the scientific literature on many diverse topics until 1958 [4]. Astonishingly, these first three publications on Cryptosporidium have defined most of what we currently know about the biology and life history of Cryptosporidium muris and Cryptosporidium parvum, the two morphologically and biologically most distinct prototype species within the genus. Since the early 1900s, our understanding of the phylogeny of the genus Cryptosporidium has undergone several cycles of reversals, but our understanding of C. muris and C. parvum, so elegantly and precisely described by Tyzzer, survives intact to this day. Equipped with a light microscope, Tyzzer was able to delineate and characterize in minute and painstaking detail the morphology and sequence of the asexual and sexual life-cycle stages of this organism, which (at 2–5 μm) are barely visible under the light microscope. He identified an oocyst (questioned later by others [5]) and observed that unlike other coccidia, the oocyst sporulates while still attached to the host cell, creating conditions for autoinfection. With the help of electron microscopy, one amendment was made to the life cycle in 1986 that revealed a second generation of schizogony [6] and established the currently accepted life-cycle model. Also in 1986, electron microscopy and freeze fracture were used to make the second correction: the observation that Cryptosporidium parasites are intracellular [7]. Although Tyzzer could not observe the location of the parasite precisely without electron microscopy, he did conclude that Cryptosporidium, despite its extracellular location, obtains all its nutrients from the host cell through the ‘organ of attachment’ (later termed ‘feeder’ organelle) and, thus, is entirely parasitic in nature. This conclusion was confirmed by the annotation of the genome sequence in 2004. It was not until 1978 that the existence of oocysts was unequivocally confirmed and their excretion in feces became the key method of diagnosis of cryptosporidiosis [8].
Figure 1.
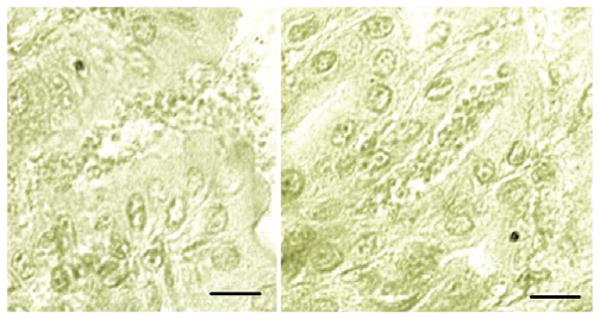
Micrographs of Tyzzer's histological sections of the stomach of an experimentally infected mouse. The information pertaining to this slide is copied from a handwritten catalogue card (# 1897), dated Jan 16, 1908, and entitled ‘Cryptosporidium feeding experiment’. The above micrographs were taken by Xiaochuan Feng, Tufts Cummings School of Veterinary Medicine, from an archive slide containing the gastric section of a mouse experimentally fed infected stomach contents and gastric mucus and sacrificed 14 days later. Note the small forms outpouring from an open pit onto the gastric surface (left) and a pit filled with parasite forms (right). There was no reference to fixative or stain used. Scale bar = 20 μm.
In the 70 years since its discovery, Cryptosporidium has been observed infrequently in the gastrointestinal tract of some 20, mostly healthy, animals in all four classes of vertebrates [9]. Notable observations during this period included the first reports in 1976 of cryptosporidiosis in humans [10]. In 1980, Bird and Smith (who reviewed all seven reported cases [11], of which six were immunocompromised patients) concluded that ‘Where immune mechanisms are functioning within normal limits and there is no other bowel disorder cryptosporidiosis does not appear to cause a problem and tentatively it can be regarded, therefore, as an opportunistic pathogenic parasite.’ This proved subsequently to be only partly true; although immunologically compromised individuals become chronically – and often fatally – infected, immunologically competent individuals frequently develop acute gastroenteritis [12]. Given how widespread and serious cryptosporidiosis turned out to be, it is curious that despite the discovery of cryptosporidiosis in 1907, it was not until the early 1980s that its clinical significance and widespread distribution were recognized.
Cryptosporidiosis – a serious and harmful agent of diarrhea
In a series of scientific papers published between 1980 and 1983, Tzipori et al. carried out extensive laboratory and field investigations on cryptosporidiosis. In crosstransmission experiments using Cryptosporidium isolates obtained from a calf, a lamb, a human and a deer, they infected newborn animals (mice, rats, guinea pigs, piglets, calves and lambs) and demonstrated that, contrary to the previously held view, mammalian Cryptosporidium isolates lack host specificity [13]. Consequently, these observations called into question the naming of species according to host origin and reflected the zoonotic nature of this parasite [14,15]. The subsequent emergence of the human species Cryptosporidium hominis some 20 years later, however, proved these observations to be only partly true (see below). The transmission experiments also opened the door to the first in vivo studies in laboratory rodents. Experimental infections enabled the first screening of anti-cryptosporidial drugs [16] and the study of disinfectants and extreme temperatures on oocyst viability [17]. Clinical and epidemiological investigations performed at the same time further showed that cryptosporidiosis was a common cause of serious and economically significant outbreaks of neonatal diarrhea in all ruminants, particularly in calves and lambs [18], and contributed to 4%–7% of sporadic cases of acute gastroenteritis in humans [19] (for a review of this work, see Ref. [20]).
Cryptosporidium in the HIV/AIDS era
The next significant milestone was the emergence of chronic and life-threatening cryptosporidiosis with HIV/AIDS in the early 1980s. The association with AIDS and the appearance of early clinical and epidemiological reports implicating cryptosporidiosis as a frequent cause of acute diarrhea in the general population firmly established that the infection was serious and widespread in humans. The first case of cryptosporidiosis in a homosexual man with AIDS was reported in 1982 [21] and by mid-1983, some 50 cases had been reported [22]. The infection in individuals with HIV/AIDS is persistent and life threatening and often involves infections of the hepatobiliary and the respiratory tracts in addition to the entire gastrointestinal tract. The link with AIDS was so strong that cryptosporidiosis became one of the defining features of the syndrome before the discovery of the causative virus. The associations with HIV/AIDS and malnutrition in children mean cryptosporidiosis remains to this day a serious, life-threatening condition that leads to intractable, often fatal, voluminous secretory diarrhea with profound weight loss and wasting, largely because of the lack of effective therapy and methods for control. The rate of infection among individuals with HIV/AIDS in the developed world, however, has subsided considerably between 1997 and 2007 because of the extensive use of anti-retroviral therapy. By restoring immune function, this therapy appears also to reduce the disease burden, but cryptosporidiosis remains a serious complication of HIV/AIDS and malnutrition in children in developing countries [23]. The incidence of cryptosporidiosis in humans ranges between 1% and 10%, depending on geography (it is more common in warmer climates), standard of hygiene (it is more common in developing countries), season, age (it is more common in children), proximity to farms and direct contact with farm animals [24]. In cattle, where the disease is economically significant, the infection probably occurs on all farms worldwide.
Cryptosporidium and the water industry
Although outbreaks of cryptosporidiosis owing to contaminated water have been recognized for some time, two events brought this link to the forefront. In 1989, after a waterborne outbreak of cryptosporidiosis in Swindon and Oxfordshire affected some 5000 people [25], the UK government established the Expert Group on Cryptosporidium in Water Supplies. In 1993, a major outbreak of cryptosporidiosis affecting >400 000 persons occurred in Milwaukee, WI, USA [26]. The magnitude of this outbreak, coupled with its association with treated drinking water, brought to public attention that Cryptosporidium-contaminated water was the most common source of outbreaks of this disease. Despite earlier reports of waterborne outbreaks of cryptosporidiosis, the magnitude of this outbreak highlighted the significance of drinking contaminated water as a major risk factor for contracting cryptosporidiosis in the USA. Over the ensuing decade a considerable research effort, funded primarily by government agencies in the USA and abroad, was devoted to this topic. This led to the initiation of risk-management studies including ‘Assessment of the risk of infection by Cryptosporidium or Giardia in drinking water from a surface water source’ [27]. Since 1992, methods have been developed for measuring oocyst concentration from large volumes of water [28]. Rapid, sensitive and specific polymerase chain reaction (PCR)-based tools for the identification, quantification and speciation of oocysts recovered from water also were developed [29,30]. The realization that Cryptosporidium oocysts are resistant to many chemical disinfectants [31] led to a search for methods that can inactivate oocysts without generating harmful byproducts. Much attention has focused on UV irradiation as an alternative method capable of inactivating waterborne oocysts [32]. However, control of surface-water contamination is being emphasized as a first measure to reduce the occurrence of waterborne oocysts. Regulations aimed at reducing the risk of exposure to waterborne oocysts have been put in place; for example, the Long Term 2 Enhanced Surface Water Treatment Rule in the USA and regulations in the UK requiring continuous monitoring for Cryptosporidium oocysts in drinking water. A treatment-based standard of one oocyst in 10 l has been adopted [33].
The evolving-species concept
The first observations of genetic heterogeneity among C. parvum (currently C. parvum and C. hominis) isolated from humans and livestock date back to 1992. Southern blotting of restriction-enzyme-digested genomic DNA [34], Western blotting [35] and isoenzyme profiles obtained from oocyst lysates [36] provided the first insights into the extent of heterogeneity in this species. Significantly, these studies showed for the first time that humans were infected with two types of Cryptosporidium parasites, one being apparently the same as that found in cattle and the other exclusively found in humans. Because of the large number of oocysts needed, Western blotting and isoenzyme analysis did not find wide application to typing of Cryptosporidium oocysts from field samples. In 1991, Mark Laxer was the first to apply PCR to the detection of Cryptosporidium oocysts [37]. Although the focus of this work was not the taxonomy of Cryptosporidium, but demonstrating the feasibility of detecting Cryptosporidium oocysts by PCR, it signaled the beginning of a rapid development of numerous PCR-based genotyping techniques, which together have shaped our understanding of the taxonomy of the genus in a fundamental way. Among these methods, PCR combined with restriction fragment length polymorphism (PCR-RFLP, which was first applied to Cryptosporidium typing by Awad-El-Kariem [38]) is the most popular. Papers describing such assays or their application to Cryptosporidium typing are too numerous to cite here. Other methods such as random amplification methods [39], sequencing [40], length polymorphisms of repetitive sequences [41] and conformational polymorphism detection methods [42] should also be mentioned in this context. The popularity of some PCR-RFLP assays, such as the one detecting an RsaI polymorphism in the Cryptosporidium oocyst wall protein (COWP) gene [43], and a species-specific assay targeting the small-subunit rRNA gene [44] is demonstrated by the fact that as of March 2007, these papers were quoted 151 and 138 times, respectively (source: ISI Web of Knowledge citation database).
The application of individual genetic markers, or multiple markers as part of multilocus typing schemes [45], confirmed the presence of two subgroups within C. parvum, which were variously named ‘human’ and ‘cattle,’ H and C or Type 1 and Type 2, respectively. These observations, subsequently confirmed in many laboratories, were significant in showing that humans are part of two distinct transmission cycles, one comprising ruminants and humans and the other exclusively comprising humans. Once it became apparent that RFLP alleles from multiple markers cosegregated into two distinct genotypic groups, it was a small step to the naming of a new species, C. hominis, proposed for C. parvum parasites exclusively infecting humans [46]. This proposal illustrated the difficulty in defining new species, a problem common to many microbes, which is rooted in the lack of defined criteria. In addition to the naming of C. hominis, several Cryptosporidium species have been named based on phenotypic and genotypic traits [47–50], whereas other genotypes have remained unnamed. In an attempt to put some order into the Cryptosporidium taxonomy, some authors now recognize 13 ‘valid’ species [51]. Oocyst size has been an important consideration for defining Cryptosporidium species, because oocysts of calf-infecting gastric species, such as C. andersoni, are larger than those of intestinal species found in the same host [52] and because oocyst size is considered a stable phenotype. Oocyst size, and morphology in general, have become less reliable for more recently named species, which instead rely primarily on genetic characteristics. Host specificity, another trait underlying the definition of Cryptosporidium species, also lacks the needed rigor, because many species have been successfully transmitted across different mammalian species [53,54] or were subsequently found to be infectious to different host species in Nature [55,56].
As the resolution of molecular methods increases, more diversity is uncovered. New genotypes defined on the basis of individual and multiple genetic markers have become apparent and raise new questions about Cryptosporidium taxonomy. Because the alternation of a sexual and asexual generation in the Cryptosporidium life cycle is thought to be obligatory, one could rely on the biological definition of species (i.e. defined on the basis of reproductive isolation) as an objective criterion. As is often the case with Cryptosporidium parasites, experiments that would be trivial with other organisms are difficult to perform. Crossing experiments in laboratory animals could, in theory, be used to assess the reproductive compatibility between species or genotypes and used as an objective measure of speciation. The feasibility of this approach was demonstrated with mixed C. parvum infection in mice [57], but applying this method to parasites that do not have common hosts is problematic. Moreover, as demonstrated in crossing experiments between C. parvum isolates [58], the lack of a mechanism for selecting recombinants and for propagating individual sporozoites makes crossing experiments a complicated and labor-intensive undertaking. In light of these limitations, the uncertainty in defining what is a species, or a variant within a species (genotype), is likely to be with us for the foreseeable future. An indication of how our thinking will continue to evolve is the recent identification of what appears to be new host-restricted populations within C. parvum infecting humans [59,60]. Although such observations run contrary to our desire to put order in a confusing system, the uncovering of new subgroups might be interpreted as a sign of rapid evolution of this parasite and its adaptation to different host species. Even though these observations are a taxonomist's nightmare, they make Cryptosporidium an interesting model for the study of parasite evolution.
In contrast to the extensive description of host-associated Cryptosporidium genotypes based on the application of genetic polymorphisms to oocyst and DNA samples recovered from various domestic and wildlife species, our understanding of host range and biological determinants of host specificity is superficial. The host range of a few species for which laboratory isolates are available has been tested in different animal models. This led to some surprising discoveries, perhaps the most interesting one being the observation that host range is not a stable phenotype. For instance, C. hominis, which was originally thought to be restricted to humans [46], can experimentally infect ruminants, piglets and immunosuppressed gerbils [53,54,61] and is rarely observed naturally in other species [56,62,63]. The extent to which host range could vary within a species also is unknown. These findings suggest that host specificity is not determined solely by receptor–ligand interaction at the level of the intestinal epithelium and that perhaps the microecology of the gut might play a significant part. It appears that any receptor-mediated restriction can be overridden with a large oocyst inoculum. The discovery that host specificity is maintained in monolayers of primary bovine and human cell lines [64], but not in transformed cell lines, might provide an interesting experimental system in which to investigate the molecular basis of host specificity.
Cryptosporidium in the postgenomic era
The publication of the complete sequence of the C. parvum genome [65] and the almost complete C. hominis genome sequence [66] represent a significant milestone in our understanding of these parasites. Before the completion of this effort, our understanding of the Cryptosporidium genome was rudimentary and based mainly on karyotype analyses with pulsed-field gel electrophoresis (PFGE) [67,68], a physical linkage map [69] and a survey of expressed sequences [70]. The availability of a complete genome sequence, and the easy access to this information through the cryptoDB.org database [71] (http://cryptodb.org/cryptodb/) represent a giant leap in our ability to conduct research. Examples of recent advances spawned by this information are new insights into the nucleotide metabolism of C. parvum, the discovery of horizontal gene transfer [72], the identification of regulatory sequences [73] and insights into oocyst-wall biogenesis [74].
Coinciding with the hundredth anniversary of the discovery of C. muris, the partial sequencing of the C. muris genome has been undertaken. The sequence from a species belonging to the gastric Cryptosporidium group, which has extensively diverged from the intestinal species, will enable comparative genome analyses between the three sequenced Cryptosporidium genomes. Until now, the power of such computational methods was limited by the high level of sequence similarity between the C. parvum and C. hominis genomes.
Some unresolved issues
Much has been learned about Cryptosporidium since 1980, but the genus remains enigmatic in ways that sets it apart from other pathogens. For instance, the intracellular, extra-cytoplasmic location is biologically unique, which might explain several characteristics including its resistance to anti-microbial agents.
The need for effective therapy
In the 25 years since the emergence of cryptosporidiosis as a significant disease in the human population and, more crucially, in individuals with HIV/AIDS and other immunodeficient individuals, little progress has been made with the development of effective treatments. Since 1987, the main focus has been directed toward evaluating antimicrobial agents developed against other Apicomplexa, including Plasmodium and Toxoplasma. Sadly, no serious attempts have been made by health agencies or the private sector to develop therapies specifically targeting Cryptosporidium, mostly because of the perception that the market for such drugs is too limited. This assessment was made despite the fact that cryptosporidiosis ranks among the most serious causes of a wide range of diarrheal illnesses globally. There is reason for optimism, however; recent developments, which include the sequencing of the genomes of C. parvum and C. hominis, have led to the identification of new molecular targets for drug development. In addition, the substantive number of chemical libraries available for drug discovery should facilitate the screening for effective drugs against cryptosporidiosis as well. Furthermore, the National Institutes of Health has placed cryptosporidiosis prominently among the Category B biothreat pathogens. Hopefully, this will counteract a loss of interest by some funding agencies resulting from the success of anti-retroviral therapies, which reduced the prevalence of chronic cryptosporidiosis considerably among individuals with HIV/AIDS and, consequently, reduced the incentive for drug development.
The need for better laboratory tools
Investigators working in the field of cryptosporidiosis still lack key tools available for studying other related pathogens. The inability to continually passage the parasite in cell culture and the inability to cryopreserve oocysts or intracellular stages are probably the most serious limitations. This limits access to endogenous stages of the life cycle, prevents the maintenance of well-characterized laboratory strains and limits the performance of meaningful comparative studies with multiple isolates. Parasite proteins of interest can only be expressed in surrogate hosts, such as Toxoplasma gondii [75], bacteria or yeast. Reports of successful completion of the parasite's life cycle in cell culture [76] or cell-free culture [77] have raised much interest, but are not widely adopted because of the difficulty in consistently reproducing these methods.
Concluding comments
Just as Tyzzer could not have predicted the dramatic expansion of our knowledge on Cryptosporidium parasites achieved to date, what the next century of research will bring is difficult to imagine. There is no shortage of research topics waiting to be tackled by inquisitive minds. Whether directly as a result of targeted research or (as is often the case) by serendipity, effective therapies are likely to become available in the near future. Access to endogenous forms and immortalization of strains in culture or by cryopreservation remain major challenges, which will require new ideas and new approaches. How soon these goals will be reached will depend largely on the extent of research support. Because the implementation of new water-treatment methods such as UV and ozone has reduced the threat of waterborne disease transmission and the AIDS epidemics in developed nations is contained, the focus will hopefully shift to developing nations, where cryptosporidiosis still contributes significantly to diarrhea and malnutrition.
References
- 1.Tyzzer EE. A sporozoon found in the peptic glands of the common mouse. Proc Soc Exp Biol Med. 1907;5:12–13. [Google Scholar]
- 2.Tyzzer EE. An extracellular coccidium, Cryptosporidium muris (gen. et sp. nov.), of the gastric glands of the common mouse. J Med Res. 1910;23:487–509. [PMC free article] [PubMed] [Google Scholar]
- 3.Tyzzer EE. Cryptosporidium parvum (sp. nov.), a coccidium found in the small intestine of the common mouse. Arch Protistenkd. 1912;26:394–412. [Google Scholar]
- 4.Weller TH. Ernest Edward Tyzzer 1875–1965. Biographical Memoirs. 1978;XLIX:353–373. [Google Scholar]
- 5.Vetterling JM, et al. Ultrastructure of Cryptosporidium wrairi from the guinea pig. J Protozool. 1971;18:248–260. doi: 10.1111/j.1550-7408.1971.tb03316.x. [DOI] [PubMed] [Google Scholar]
- 6.Current WL, Reese NC. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J Protozool. 1986;33:98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 7.Marcial MA, Madara JL. Cryptosporidium: cellular localization, structural analysis of absorptive cell-parasite membrane-membrane interactions in guinea pigs, and suggestion of protozoan transport by M cells. Gastroenterology. 1986;90:583–594. doi: 10.1016/0016-5085(86)91112-1. [DOI] [PubMed] [Google Scholar]
- 8.Pohlenz J, et al. Bovine cryptosporidiosis: a transmission and scanning electron microscopic study of some stages in the life cycle and of the host–parasite relationship. Vet Pathol. 1978;15:417–427. doi: 10.1177/030098587801500318. [DOI] [PubMed] [Google Scholar]
- 9.Levine ND. Some corrections of coccidian (Apicomplexa: Protozoa) nomenclature. J Parasitol. 1980;66:830–834. [PubMed] [Google Scholar]
- 10.Nime FA, et al. Acute enterocolitis in a human being infected with the protozoan Cryptosporidium. Gastroenterology. 1976;70:592–598. [PubMed] [Google Scholar]
- 11.Bird RG, Smith MD. Cryptosporidiosis in man: parasite life cycle and fine structural pathology. J Pathol. 1980;132:217–233. doi: 10.1002/path.1711320304. [DOI] [PubMed] [Google Scholar]
- 12.Navin TR, Juranek DD. Cryptosporidiosis: clinical, epidemiologic, and parasitologic review. Rev Infect Dis. 1984;6:313–327. doi: 10.1093/clinids/6.3.313. [DOI] [PubMed] [Google Scholar]
- 13.Tzipori S, et al. Cryptosporidium: evidence for a single-species genus. Infect Immun. 1980;30:884–886. doi: 10.1128/iai.30.3.884-886.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzipori S, et al. Vomiting and diarrhea associated with cryptosporidial infection. N Engl J Med. 1980;303:818. [PubMed] [Google Scholar]
- 15.Tzipori S, et al. Experimental infection of lambs with Cryptosporidium isolated from a human patient with diarrhoea. Gut. 1982;23:71–74. doi: 10.1136/gut.23.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzipori SR, et al. The therapeutic effect of 16 antimicrobial agents on Cryptosporidium infection in mice. Aust J Exp Biol Med Sci. 1982;60:187–190. doi: 10.1038/icb.1982.20. [DOI] [PubMed] [Google Scholar]
- 17.Fayer R, Leek RG. The effects of reducing conditions, medium, pH, temperature, and time on in vitro excystation of Cryptosporidium. J Protozool. 1984;31:567–569. doi: 10.1111/j.1550-7408.1984.tb05504.x. [DOI] [PubMed] [Google Scholar]
- 18.Tzipori S, et al. An outbreak of calf diarrhoea attributed to cryptosporidial infection. Vet Rec. 1980;107:579–580. [PubMed] [Google Scholar]
- 19.Tzipori S, et al. Cryptosporidiosis in hospital patients with gastroenteritis. Am J Trop Med Hyg. 1983;32:931–934. doi: 10.4269/ajtmh.1983.32.931. [DOI] [PubMed] [Google Scholar]
- 20.Tzipori S. Cryptosporidiosis in animals and humans. Microbiol Rev. 1983;47:84–96. doi: 10.1128/mr.47.1.84-96.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma P, Soave R. Three-step stool examination for cryptosporidiosis in 10 homosexual men with protracted watery diarrhea. J Infect Dis. 1983;147:824–828. doi: 10.1093/infdis/147.5.824. [DOI] [PubMed] [Google Scholar]
- 22.Ma P. Cryptosporidium and the enteropathy of immune deficiency. J Pediatr Gastroenterol Nutr. 1984;3:488–490. [PubMed] [Google Scholar]
- 23.Tumwine JK, et al. Cryptosporidiosis and microsporidiosis in ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- 24.Hunter PR, et al. Sporadic cryptosporidiosis case-control study with genotyping. Emerg Infect Dis. 2004;10:1241–1249. doi: 10.3201/eid1007.030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson AJ, et al. An outbreak of waterborne cryptosporidiosis in Swindon and Oxfordshire. Epidemiol Infect. 1991;107:485–495. doi: 10.1017/s0950268800049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mac Kenzie WR, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 27.Teunis PFM, et al. Assessment of the risk of infection by Cryptosporidium or Giardia in drinking water from a surface water source. Water Res. 1997;31:1333–1346. [Google Scholar]
- 28.Zuckerman U, Tzipori S. Portable continuous flow centrifugation and method 1623 for monitoring of waterborne protozoa from large volumes of various water matrices. J Appl Microbiol. 2006;100:1220–1227. doi: 10.1111/j.1365-2672.2006.02874.x. [DOI] [PubMed] [Google Scholar]
- 29.Nichols RA, et al. Molecular fingerprinting of Cryptosporidium oocysts isolated during water monitoring. Appl Environ Microbiol. 2006;72:5428–5435. doi: 10.1128/AEM.02906-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruecker NJ, et al. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl Environ Microbiol. 2005;71:8991–8994. doi: 10.1128/AEM.71.12.8991-8994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korich DG, et al. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl Environ Microbiol. 1990;56:1423–1428. doi: 10.1128/aem.56.5.1423-1428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rochelle PA, et al. The response of Cryptosporidium parvum to UV light. Trends Parasitol. 2005;21:81–87. doi: 10.1016/j.pt.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd A, Drury D. Continuous monitoring for Cryptosporidium–a novel approach to public health protection. Water Sci Technol. 2002;46:297–301. [PubMed] [Google Scholar]
- 34.Ortega YR, et al. Restriction fragment length polymorphism analysis of Cryptosporidium parvum isolates of bovine and human origin. J Protozool. 1991;38:40S–41S. [PubMed] [Google Scholar]
- 35.Nina JM, et al. Comparative study of the antigenic composition of oocyst isolates of Cryptosporidium parvum from different hosts. Parasite Immunol. 1992;14:227–232. doi: 10.1111/j.1365-3024.1992.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 36.Ogunkolade BW, et al. Isoenzyme variation within the genus Cryptosporidium. Parasitol Res. 1993;79:385–388. doi: 10.1007/BF00931827. [DOI] [PubMed] [Google Scholar]
- 37.Laxer MA, et al. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am J Trop Med Hyg. 1991;45:688–694. doi: 10.4269/ajtmh.1991.45.688. [DOI] [PubMed] [Google Scholar]
- 38.Awad-el-Kariem FM, et al. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1994;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 39.Morgan UM, et al. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 40.Strong WB, et al. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng X, et al. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl Environ Microbiol. 2000;66:3344–3349. doi: 10.1128/aem.66.8.3344-3349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasser RB, et al. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis. 2001;22:433–437. doi: 10.1002/1522-2683(200102)22:3<433::AID-ELPS433>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 43.Spano F, et al. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 44.Xiao L, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spano F, et al. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan-Ryan UM, et al. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J Eukaryot Microbiol. 2002;49:433–440. doi: 10.1111/j.1550-7408.2002.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 47.Ryan UM, et al. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa) J Parasitol. 2004;90:769–773. doi: 10.1645/GE-202R1. [DOI] [PubMed] [Google Scholar]
- 48.Lindsay DS, et al. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos Taurus. J Eukaryot Microbiol. 2000;47:91–95. doi: 10.1111/j.1550-7408.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 49.Morgan UM, et al. Cryptosporidium in cats–additional evidence for C. felis. Vet J. 1998;156:159–161. doi: 10.1016/s1090-0233(05)80047-4. [DOI] [PubMed] [Google Scholar]
- 50.Fayer R, et al. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus) J Parasitol. 2005;91:624–629. doi: 10.1645/GE-3435. [DOI] [PubMed] [Google Scholar]
- 51.Xiao L, et al. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Upton SJ, Current WL. The species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) infecting mammals. J Parasitol. 1985;71:625–629. [PubMed] [Google Scholar]
- 53.Akiyoshi DE, et al. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect Immun. 2002;70:5670–5675. doi: 10.1128/IAI.70.10.5670-5675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giles M, et al. Experimental infection of a lamb with Cryptosporidium parvum genotype 1. Vet Rec. 2001;149:523–525. doi: 10.1136/vr.149.17.523. [DOI] [PubMed] [Google Scholar]
- 55.Pavlasek I, Ryan U. The first finding of a natural infection of Cryptosporidium muris in a cat. Vet Parasitol. 2007;144:349–352. doi: 10.1016/j.vetpar.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Ryan UM, et al. Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl Environ Microbiol. 2005;71:4992–4997. doi: 10.1128/AEM.71.9.4992-4997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng X, et al. Experimental evidence for genetic recombination in the opportunistic pathogen Cryptosporidium parvum. Mol Biochem Parasitol. 2002;119:55–62. doi: 10.1016/s0166-6851(01)00393-0. [DOI] [PubMed] [Google Scholar]
- 58.Tanriverdi S, et al. Genetic crosses in the apicomplexan parasite Cryptosporidium parvum define recombination parameters. Mol Microbiol. 2007;63:1432–1439. doi: 10.1111/j.1365-2958.2007.05594.x. [DOI] [PubMed] [Google Scholar]
- 59.Mallon ME, et al. Multilocus genotyping of Cryptosporidium parvum Type 2: population genetics and sub-structuring. Infect Genet Evol. 2003;3:207–218. doi: 10.1016/s1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 60.Grinberg A, et al. Host-shaped segregation of the Cryptosporidium parvum multilocus genotype repertoire. Epidemiol Infect. 2008;136:273–278. doi: 10.1017/S0950268807008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baishanbo A, et al. Infectivity of Cryptosporidium hominis and Cryptosporidium parvum genotype 2 isolates in immunosuppressed Mongolian gerbils. Infect Immun. 2005;73:5252–5255. doi: 10.1128/IAI.73.8.5252-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith HV, et al. Natural Cryptosporidium hominis infections in Scottish cattle. Vet Rec. 2005;156:710–711. doi: 10.1136/vr.156.22.710. [DOI] [PubMed] [Google Scholar]
- 63.Tanriverdi S, et al. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol Biochem Parasitol. 2003;130:13–22. doi: 10.1016/s0166-6851(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 64.Hashim A, et al. Host cell tropism underlies species restriction of human and bovine Cryptosporidium parvum genotypes. Infect Immun. 2004;72:6125–6131. doi: 10.1128/IAI.72.10.6125-6131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abrahamsen MS, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 66.Xu P, et al. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 67.Mead JR, et al. Field inversion gel electrophoretic separation of Cryptosporidium spp. chromosome-sized DNA. J Parasitol. 1988;74:366–369. [PubMed] [Google Scholar]
- 68.Le Blancq SM, et al. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 69.Piper MB, et al. A HAPPY map of Cryptosporidium parvum. Genome Res. 1998;8:1299–1307. doi: 10.1101/gr.8.12.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strong WB, Nelson RG. Preliminary profile of the Cryptosporidium parvum genome: an expressed sequence tag and genome survey sequence analysis. Mol Biochem Parasitol. 2000;107:1–32. doi: 10.1016/s0166-6851(99)00225-x. [DOI] [PubMed] [Google Scholar]
- 71.Puiu D, et al. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 2004;32:D329–D331. doi: 10.1093/nar/gkh050. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Striepen B, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci USA. 2004;101:3154–3159. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mullapudi N, et al. Identification of putative cis-regulatory elements in Cryptosporidium parvum by de novo pattern finding. BMC Genomics. 2007;8:13. doi: 10.1186/1471-2164-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Templeton TJ, et al. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect Immun. 2004;72:980–987. doi: 10.1128/IAI.72.2.980-987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Connor RM, et al. Stable expression of Cryptosporidium parvum glycoprotein gp40/15 in Toxoplasma gondii. Mol Biochem Parasitol. 2007;152:149–158. doi: 10.1016/j.molbiopara.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hijjawi NS, et al. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int J Parasitol. 2001;31:1048–1055. doi: 10.1016/s0020-7519(01)00212-0. [DOI] [PubMed] [Google Scholar]
- 77.Hijjawi NS, et al. Complete development of Cryptosporidium parvum in host cell-free culture. Int J Parasitol. 2004;34:769–777. doi: 10.1016/j.ijpara.2004.04.001. [DOI] [PubMed] [Google Scholar]


