Abstract
Objective
We compared eight genotypic interpretation methods to determine whether the method used would affect the rates of reported transmitted drug resistance.
Design
Retrospective cohort study.
Methods
For the International AIDS Society-USA method we classified a mutation as resistant if it was a ‘major’ resistance-associated mutation. For the Stanford algorithm, we classified a mutation as resistant if the score was at least 60 (Stanford 60), and alternatively, if the score was at least 30 (Stanford 30). For Agence Nationale de Recherches sur le SIDA and Rega, we interpreted resistance as either ‘intermediate resistance’ or ‘resistance’ (ANRS 1 and Rega 1), and ‘resistance’ only (ANRS 2 and Rega 2). We also used the calibrated population resistance algorithm. We then determined the rates of transmitted drug resistance within the Acute Infection Early Disease Research Program cohort (n = 1311) enrolled between March 1995 and August 2006 using each method; agreement was assessed using kappa coefficients.
Results
Differences in estimated rates of transmitted drug resistance using International AIDS Society-USA, calibrated population resistance, Stanford 30, ANRS 1, Rega 1 and Rega 2 methods were mostly minor for resistance to protease and non-nucleoside reverse transcriptase inhibitors (1% range) and more pronounced for nucleoside reverse transcriptase inhibitors (5% range). For these methods kappa agreement was substantial or almost perfect across all drug classes. The Stanford 60 was most conservative.
Conclusions
The persistent high rates of transmitted drug resistance support the need for continued genotypic surveillance. The currently available interpretation algorithms can be used for this purpose.
Keywords: algorithms, HIV, prevalence, transmitted drug resistance
Introduction
The transmission of drug resistant HIV-1 represents a public health concern by potentially reducing the number of active antiretroviral medications available to infected persons and can increase the risk of treatment failure, particularly in resource limited settings in which assays for drug resistance are not readily available [1,2].
Accurate surveillance of transmitted drug resistant (TDR) HIV-1 can inform public health prevention and treatment strategies [3,4]. Such surveillance has led to recommendations for obtaining genotype testing at baseline for recently infected or newly diagnosed antiretroviral naïve individuals [5–7]. There are various algorithms that can be used to infer HIV-1 susceptibility to antiretroviral drugs from genotypic data, and differences in interpretation among algorithms have been investigated previously to assess protease inhibitor susceptibility using both genotypic and phenotypic data [8], TDR among treatment naïve cohorts [4,9] and to predict virologic success of antiretroviral therapy using calculated genotypic susceptibility scores [10,11].
To address the differences among various interpretation methods for the epidemiologic surveillance of TDR HIV-1, we used genotype sequence data from a large primary HIV-1 infection cohort to systematically compare five algorithms (and eight methods) commonly used to determine the presence of drug resistance in a genotype. One of them is the recently published calibrated population resistance (CPR) algorithm that was specifically developed for the epidemiologic surveillance of TDR HIV-1 [4]. The other four are the International AIDS Society-USA (IAS-USA) [12], the Stanford [13], the Agence Nationale de Recherches sur le SIDA (ANRS) [14] and the Rega [15] algorithms. Each of the last three algorithms was considered in two versions (including and excluding an intermediate level in the definition of resistance). The latter three algorithms are accessible online through the Stanford drug resistance database website (http://hivdb.stanford.edu/).
Methods
To address whether the interpretation method used would affect the rates of reported TDR, we compared the eight methods in the setting of a large primary infection cohort, the Acute Infection and Early Disease Research Program (AIEDRP). This is a retrospective study of genotype data (1311 pol sequences) collected at baseline by sites of the AIEDRP network located in Sydney, Australia; Baltimore, Maryland; Denver, Colorado; Los Angeles, California; Montreal, Canada; New York, New York; Seattle, Washington; San Diego, California; and San Francisco, California.
The TDR rates to the antiretroviral drug classes, nucleoside reverse transcriptase inhibitors (NRTI), non-nucleoside reverse transcriptase inhibitors (NNRTI) and protease inhibitors as well as to ‘any’ of these three classes were studied. We interpreted the prevalence of TDR HIV-1 within the AIEDRP network for time period March 1995 to August 2006 using eight methods based on five publicly available, commonly used algorithms. For the Stanford HIV drug resistance algorithm (version 4.2.0), a drug-specific penalty score is assigned a priori to each annotated drug resistance mutation; the sequence is scanned for the presence of such mutations and the sum of scores for each detected mutation determines the overall resistance score. We classified a sequence as resistant to a given drug if the computed score was either at least 60 (Stanford 60) or at least 30 (Stanford 30). For the IAS-USA 2005 method, we classified a mutation as conferring resistance if it was defined as a ‘major’ resistance-associated mutation [12]. Accordingly, a ‘major’ resistance-associated mutation should be selected by the presence of an antiretroviral medication and lead to an alteration in medication binding or decrease in antiretroviral activity [12]. Since ritonavir was removed from the IAS-USA 2006 list [16] and the majority of our participants were enrolled during the time when ritonavir was being used as an antiretroviral medication, rather than as a pharmacoenhancer, we chose to use the IAS-USA 2005 drug resistance interpretation list instead of the IAS-USA 2006 list. Also, as the minor mutations listed in the IAS-USA interpretation by definition should not have significant effect on phenotypic resistance to antiretroviral medications, they were not considered in the definition of resistance for the purposes of this study. We also estimated the prevalence of TDR using the CPR algorithm [4]. Drug resistance was inferred from sequence data by class of antiretroviral medication based on 31 protease inhibitors, 31 NRTI and 18 NNRTI resistance surveillance mutations. The ANRS (version 2006.07) [14] and Rega (version 6.4.1) [15] algorithms report the drug resistance at three levels: susceptible, resistant, and an intermediate level, which is described differently for each algorithm but which we refer to here as intermediate. We adopted two protocols for classifying a sequence as resistant: whether it was classified as either ‘intermediately resistant’ or ‘resistant’ (ANRS 1 and Rega 1), or strictly ‘resistant’ (ANRS 2 and Rega 2).
We compared the level of agreement between algorithms using two quantitative measures. Firstly, we computed the observed agreement proportion, defined as O = (N++ + N−−)/N; where N is the total number of sequences and, N++ and N−− are the numbers of cases where both algorithms agreed (either in positive or negative classification). Secondly, we used Cohen’s kappa coefficient [17], defined as the ratio kappa = (O–C)/ (1–C), where O is the observed agreement proportion as above, and C is the probability that two tests agree by chance, based on the observed positive and negative classifications of each test. Kappa is greater than zero if the observed agreement exceeds the proportion expected by chance, and reaches its maximum value of one for two tests with perfect agreement. Tests of kappa equal to zero were performed with Bonferroni adjustment for multiple comparisons by dividing α = 0.05 by the total number of tests (0.05/112 = 0.0004).
Written informed consent was obtained from all patients and the human experimentation guidelines of the U.S. Department of Health and Human Services and the individual institutions were followed in conducting this research.
Patient characteristics
Between March 1995 and August 2006, the participating sites of the AIEDRP network collected 1311 baseline genotypes to assess the rate of TDR HIV-1 [18]. All study participants were recently diagnosed with primary HIV-1 infection and were antiretroviral naïve at the time of enrollment. Study participants were mostly male (94.7%), non-Hispanic white (70.1%), reported sex with men as the most common HIV-1 risk factor (>90%), and were infected with clade B HIV-1 (97.5%). The median age of the participants at enrollment was 35 years (range 15.6–66 years).
Results
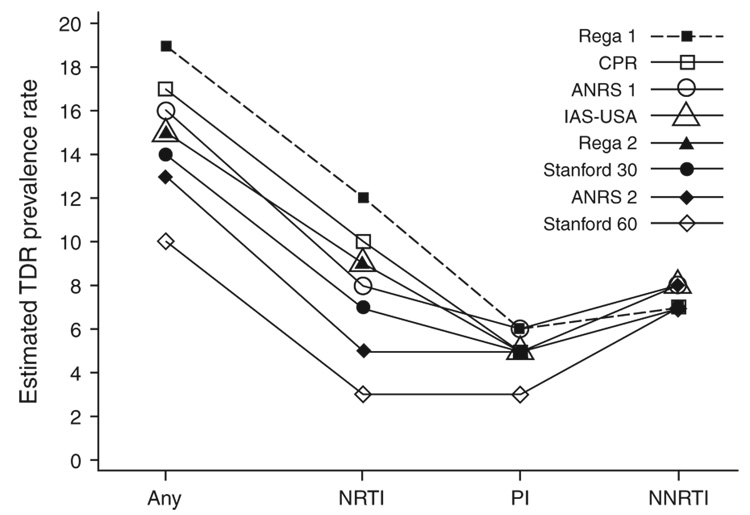
The estimated prevalence of ‘any’ TDR HIV-1 and TDR by antiretroviral drug classes using eight interpretation methods (IAS-USA, CPR, Stanford 30, Stanford 60, ANRS 1, ANRS 2, Rega 1, and Rega 2) is presented in Fig. 1. For the majority of methods the rates differed only slightly for the protease inhibitors and NNRTI drug classes. When considering NRTI and any resistance the TDR rates estimated by the most conservative and least conservative methods differed substantially. On the basis of the estimated prevalence rates for NRTI and any resistance, the eight methods could be rank-ordered and classified into five groups. The Stanford 60 method was the most conservative. It provided similar NNRTI TDR rates to the other seven methods, but the TDR rates for NRTI, protease inhibitors and ‘any’ were clearly lower. For NRTI and any TDR, the discrepancy was as large as 9% between the least conservative method (Rega 1) and Stanford 60. The ANRS 2 was the second most conservative approach, providing similar NNRTI and protease inhibitor TDR rates when compared with the other six methods, but the TDR rates for NRTI and ‘any’ were notably lower. For NRTI and any TDR, the discrepancy was as large as 7% between ANRS 2 and the least conservative method (Rega 1). Stanford 30, Rega 2, and IAS-USA provided similar TDR rates, which were 7–9% for NRTI, 7–8% for NNRTI, 5% for protease inhibitors and 14–15% for any drug resistance. The interpretations by ANRS 1 and CPR were also similar and the discrepancy between the two methods was in the 1–2% range. The least conservative method, Rega 1, provided the highest TDR rates except for NNRTI. As the Stanford 60, ANRS 1 and Rega 1 methods were designed to designate resistance mutations as a subset of their corresponding less conservative versions (Stanford 30, ANRS 2 and Rega 2), all rates estimated by Stanford 30, ANRS 1 and Rega 1 are equal to or higher than those estimated by Stanford 60, ANRS 2 and Rega 2 (Fig. 1). The observed proportion of overall agreement (number of subjects with agreement over total number of subjects) among the eight methods was, nevertheless, very high, with a range of 0.910–1.00.
Fig. 1. Estimated NRTI, protease inhibitors, NNRTI and ‘any’ transmitted drug resistance prevalence rates using IAS-USA, CPR, Stanford 30, Stanford 60, ANRS 1, ANRS 2, Rega 1 and Rega 2.
ANRS, Agence Nationale de Recherches sur le SIDA; CPR, calibrated population resistance; IAS-USA, International AIDS Society-USA; NNRTI, non-nucleoside reverse transcriptase inhibitors; NRTI, nucleoside reverse transcriptase inhibitors, PI, protease inhibitors, TDR, transmitted drug resistance.
The chance-corrected agreement was almost perfect among the Stanford 30, IAS-USA, CPR, Rega 1, Rega 2, and ANRS 1 methods for NNRTI, protease inhibitors and ‘any’ TDR prevalence with kappa ranging from 0.81 to 0.99 [19]. The agreement for NRTI among these six methods was substantial or almost perfect (range of kappa 0.75–0.93). The agreement among these six, Stanford 60 and ANRS 2 depended on the class of antiretroviral medication. Almost perfect agreement was maintained for NNRTI TDR (range of kappa 0.91–1.00) and the agreement remained substantial for ‘any’ resistance (range of kappa 0.63–0.89). For protease inhibitors resistance, except for the kappa coefficient of 0.54 between Stanford 60 and Rega 1 and 0.57 between Stanford 60 and ANRS 1, the agreement among other methods was still substantial with kappa ranging from 0.63 to 0.98. The discrepancy was the largest for the estimates of NRTI resistance. The agreement between ANRS 2 and the other seven methods was substantial (range of kappa 0.61–0.78) except for Rega 1, with kappa equal to 0.57. The agreement estimated by kappa between Stanford 60 and ANRS 2 was 0.74 but the agreement between Stanford 60 and the other six methods was either moderate or close to moderate (range of kappa 0.39–0.57) (Table 1). All observed agreement was significantly greater than chance agreement (P<0.001 after Bonferroni correction).
Table 1.
Kappa coefficients for agreement of Stanford 60, Stanford 30, IAS-USA, CPR, Rega 1, Rega 2, ANRS 1 and ANRS 2 in NRTI. NNRTI, protease inhibitors and any transmitted drug resistance.
| Any | NRTI | PI | NNRTI | |
|---|---|---|---|---|
| Rega 1/ANRS 1 | 0.86 | 0.82 | 0.84 | 0.97 |
| Rega 1/CPR | 0.91 | 0.93 | 0.83 | 0.96 |
| ANRS 1/CPR | 0.88 | 0.85 | 0.86 | 0.99 |
| Rega 1/IAS-USA | 0.85 | 0.84 | 0.88 | 0.94 |
| Rega 1/Rega 2 | 0.86 | 0.85 | 0.83 | 0.99 |
| Rega 1/Stanford 30 | 0.81 | 0.75 | 0.84 | 0.96 |
| ANRS 1/IAS-USA | 0.83 | 0.77 | 0.91 | 0.91 |
| ANRS 1/Rega 2 | 0.86 | 0.81 | 0.84 | 0.97 |
| ANRS 1/Stanford 30 | 0.88 | 0.87 | 0.87 | 0.95 |
| CPR/IAS-USA | 0.89 | 0.89 | 0.94 | 0.92 |
| CPR/Rega 2 | 0.84 | 0.78 | 0.92 | 0.96 |
| CPR/Stanford 30 | 0.87 | 0.80 | 0.97 | 0.96 |
| IAS-USA/Rega 2 | 0.85 | 0.82 | 0.90 | 0.93 |
| IAS-USA/Stanford 30 | 0.88 | 0.86 | 0.95 | 0.92 |
| Stanford 30/Rega 2 | 0.93 | 0.90 | 0.95 | 0.97 |
| ANRS 2/Rega 1 | 0.78 | 0.57 | 0.88 | 0.97 |
| ANRS 2/CPR | 0.82 | 0.61 | 0.93 | 0.99 |
| ANRS 2/ANRS 1 | 0.88 | 0.73 | 0.93 | 1 |
| ANRS 2/IAS-USA | 0.83 | 0.66 | 0.98 | 0.91 |
| ANRS 2/Rega 2 | 0.86 | 0.70 | 0.91 | 0.97 |
| ANRS 2/Stanford 30 | 0.89 | 0.78 | 0.94 | 0.95 |
| Stanford 60/Rega 1 | 0.63 | 0.39 | 0.54 | 0.98 |
| Stanford 60/CPR | 0.69 | 0.42 | 0.66 | 0.97 |
| Stanford 60/ANRS 1 | 0.70 | 0.50 | 0.57 | 0.96 |
| Stanford 60/IAS-USA | 0.72 | 0.48 | 0.63 | 0.94 |
| Stanford 60/Rega 2 | 0.76 | 0.50 | 0.69 | 0.99 |
| Stanford 60/Stanford 30 | 0.80 | 0.57 | 0.68 | 0.98 |
| Stanford 60/ANRS 2 | 0.81 | 0.74 | 0.63 | 0.96 |
P - values for testing kappa coefficient equals to zero are all less than 0.001 after Bonferroni adjustment for all 112 comparisons. Kappa greater than 0.80 represents almost perfect agreement, 0.61–0.80 represents substantial agreement and 0.41–0.60 represents moderate agreement [19]. ANRS, Agence Nationale de Recherches sur le SIDA; CPR, calibrated population resistance; IAS-USA, International AIDS Society-USA; NNRTI, nonnucleoside reverse transcriptase inhibitors; NRTI nucleoside reverse transcriptase inhibitors, PI, protease inhibitors.
Conclusion
The epidemiologic surveillance of TDR HIV-1 can be important for the allocation of prevention, education and treatment resources [3], and there has been some debate about what available method should be used to determine the rates of TDR HIV-1, especially for comparisons between studies and populations [4,20]. This study represents the most comprehensive comparison of widely used genotype interpretation algorithms for the largest well characterized cohort of individuals with primary HIV-1 infection (n = 1311 baseline genotypes). As previously reported [18], the prevalence rate of TDR to any class of antiretroviral medication was between 10 and 19% and the resistance to NRTI antiretroviral medications was the most common class of TDR HIV-1 (Fig. 1).
Differences in the interpretation of algorithms may be important for the clinical management of an individual patient, the epidemiologic surveillance of TDR HIV-1 in a population in which options for antiretroviral therapy are specific and limited, or the surveillance of populations that have been heavily exposed to complex and different antiretroviral treatments. The rates of TDR HIV-1 found in this very large primary infection cohort study did not differ substantially with the method that was used to interpret genotypic data for protease inhibitors and NNRTI resistance except when using the most conservative method (Stanford 60). For NRTI resistance, the rates of TDR HIV-1 were comparable when the two most conservative methods (Stanford 60 and ANRS 2) and the least conservative method (Rega 1) are excluded. This may be because the different algorithms classify roughly the same mutations as conferring major and relevant resistance, and differences between the algorithms are relatively few and mostly involve relatively infrequent mutations. A limitation of the study is that the cohort used is predominantly white and represents predominantly men who have sex with men [18]. Different results might have been obtained for populations infected with different subtypes of HIV and having different HIV risk factors; therefore, it will be important to perform similar comparative analyses in other populations.
Additionally, the currently available algorithms may actually underestimate the extent to which selective drug pressures have shaped the transmitted viral population, as enrichment of ‘secondary’ mutations, which may have been selected by the treatment of a donor partner, are not often included [12,16,21,22]. Although not strictly defining drug susceptibility for an individual or for a population, using less conservative algorithms may thus give a more accurate picture of the extent to which drug pressure has influenced transmitted virus. On the basis of these analyses, both Rega 1 and CPR algorithms seem particularly well suited for this purpose, but are closely followed by the algorithms of ANRS 1, IAS-USA, Rega 2 and Stanford 30. Taken together, we conclude that the persistent high rates of TDR support the need for continued surveillance and that the currently available algorithms can be used for this surveillance.
Acknowledgements
We are grateful to all the participants, investigators and staff of the AIEDRP network whose unwavering generosity and dedication allowed us to conduct these investigations. In particular, we would like to thank Drs. Thomas Campbell, Simon Frost, Sergei Kosakovsky Pond and Elizabeth Connick for their scientific insight and contribution to these investigations. We would also like to thank Laureen Copfer for her administrative assistance.
Financial support: Data in this study were collected by sites participating in the Acute Infection and Early Disease Research Program (AIEDRP). AIEDRP is funded by the National Institute of Allergy and Infectious Diseases and NIH grants 5K23AI055276, AI27670, AI38858, AI43638, AI43752, AI29164, AI47745, Al41532, MH62512, AI55356, AI047033, AI041534, 1P01 AI057127, AI41531, AI41535, AI57005, RR024143, RR00051, and AI57167, the UCSD Center for AIDS Research (AI36214), the Colorado Center for AIDS Research (AI054907), and the Research Center for AIDS and HIV-1 Infection of the San Diego Veterans Affairs Healthcare System (10-92-035) with the contribution of Fonds de la recherché en santé du Québec (FRSQ).
References
- 1.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 2.Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 3.Truong HH, Grant RM, McFarland W, Kellogg T, Kent C, Louie B, et al. Routine surveillance for the detection of acute and recent HIV infections and transmission of antiretroviral resistance. AIDS. 2006;20:2193–2197. doi: 10.1097/01.aids.0000252059.85236.af. [DOI] [PubMed] [Google Scholar]
- 4.Shafer RW, Rhee SY, Pillay D, Miller V, Sandstrom P, Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammer SM, Saag MS, Schechter M, Montaner JSG, Schooley RT, Jacobsen DM, et al. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society-USA panel. JAMA. 2006;296:827–843. doi: 10.1001/jama.296.7.827. [DOI] [PubMed] [Google Scholar]
- 6.Smith D, Moini N, Pesano R, Cachay E, Aiem H, Lie Y, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44:456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 7.Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2006 http://aidsinfo.nih.gov/
- 8.Ross L, Boulme R, Fusco G, Scarsella A, Florance A. Comparison of HIV type 1 protease inhibitor susceptibility results in viral samples analyzed by phenotypic drug resistance assays and by six resistance algorithms: an analysis of a subpopulation of the CHORUS cohort. AIDS Res Hum Retroviruses. 2005;21:696–701. doi: 10.1089/aid.2005.21.696. [DOI] [PubMed] [Google Scholar]
- 9.Masquelier B, Bhaskaran K, Pillay D, Gifford R, Balestre E, Jorgensen LB, et al. Prevalence of transmitted HIV-1 drug resistance and the role of resistance algorithms: data from seroconverters in the CASCADE collaboration from 1987 to 2003. J Acquir Immune Defic Syndr. 2005;40:505–511. doi: 10.1097/01.qai.0000186361.42834.61. [DOI] [PubMed] [Google Scholar]
- 10.De Luca A, Cozzi-Lepri A, Perno CF, Balotta C, Di Giambenedetto S, Poggio A, et al. Variability in the interpretation of transmitted genotypic HIV-1 drug resistance and prediction of virological outcomes of the initial HAART by distinct systems. Antivir Ther. 2004;9:743–752. [PubMed] [Google Scholar]
- 11.De Luca A, Vendittelli M, Baldini F, Di Giambenedetto S, Trotta MP, Cingolani A, et al. Construction, training and clinical validation of an interpretation system for genotypic HIV-1 drug resistance based on fuzzy rules revised by virological outcomes. Antivir Ther. 2004;9:583–593. [PubMed] [Google Scholar]
- 12.Johnson VA, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: 2005. Top HIV Med. 2005;13:51–57. [PubMed] [Google Scholar]
- 13.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meynard JL, Vray M, Morand-Joubert L, Race E, Descamps D, Peytavin G, et al. Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: a randomized trial. AIDS. 2002;16:727–736. doi: 10.1097/00002030-200203290-00008. [DOI] [PubMed] [Google Scholar]
- 15.Van Laethem K, De Luca A, Antinori A, Cingolani A, Perna CF, Vandamme AM. A genotypic drug resistance interpretation algorithm that significantly predicts therapy response in HIV-1-infected patients. Antivir Ther. 2002;7:123–129. [PubMed] [Google Scholar]
- 16.Johnson VA, Brun-Vezinet F, Clotet B, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: Fall 2006. Top HIV Med. 2006;14:125–130. [PubMed] [Google Scholar]
- 17.Fleiss JH, Levin B, Paik MC. Statistical Methods for Rates and Proportions. 3rd ed. New Jersey: Wiley; 2003. [Google Scholar]
- 18.Little S, May S, Hecht F, Markowitz M, Daar ES, Kaldor J, et al. Increase in transmitted NNRTI drug resistance among recently HIV-infected patients from North America and Australia. Antiviral Ther. 2006;11:S110. [Google Scholar]
- 19.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 20.Snoeck J, Kantor R, Shafer RW, Van Laethem K, Deforche K, Carvalho AP, et al. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother. 2006;50:694–701. doi: 10.1128/AAC.50.2.694-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J Virol. 2004;78:7545–7552. doi: 10.1128/JVI.78.14.7545-7552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc Natl Acad Sci U S A. 2001;98:13907–13912. doi: 10.1073/pnas.241300698. [DOI] [PMC free article] [PubMed] [Google Scholar]