Abstract
Background
In China, hepatitis is a huge public health problem. Outbreaks of hepatitis A are the most frequent cause of acute hepatitis, and to date, few epidemiologic investigations or molecular surveillance studies have been performed.
Materials and methods
In 2006, two major outbreaks of hepatitis A occurred, one in Guigang City, southern China, and the other in Hetian City, northwestern China. Field and molecular epidemiologic investigations were conducted.
Results
In Guigang, a single outbreak occurred in a school; 35 patients and 25 asymptomatic individuals were infected with 1 strain of hepatitis A virus (HAV). A case-control study showed that contaminated water was the likely transmission source. In Hetian, the epidemic of hepatitis A consisted of sporadic, small outbreaks involving as many as 20 wild HAV strains. A molecular epidemiology approach allowed us to identify two groups infected by individual HAV strains. Further fieldwork and a case-control study showed that ice cream was the suspected transmission source in one group. Our molecular epidemiology study showed that genetic variability between the HAV strains isolated from Guigang and Hetian and previously reported HAV strains was at least 4.3%.
Conclusion
Contaminated water and suspected ice cream were associated with outbreaks of hepatitis A. Viral genetic analysis may advance field investigations in complex situations.
Keywords: Hepatitis A, Molecular epidemiology
Introduction
Hepatitis A, a widespread infectious disease, is transmitted by the fecal-oral route [1–4]. Contact with an infected person and ingestion of contaminated food or water are the major routes of infection [4]. Hepatitis A virus (HAV) infection is generally self-limited and clinical features range from a lack of symptoms to death from fulminant hepatitis [1]. In children younger than 6 years, up to 70% are asymptomatic; in older children and adults, fever, malaise, anorexia, nausea, abdominal discomfort, dark urine, and/or jaundice usually results. The incubation period ranges from 15 to 50 days, and symptoms usually last less than 2 months [1]. No evidence exists of chronic liver disease following infection [1].
Hepatitis A virus is an RNA virus that belongs to the Picornaviridae family; the virion is a nonenveloped, 7.5-kb, positive-stranded RNA virus. The genome can be divided into three distinct regions, and a single open-reading frame encodes all the viral proteins. The P1 region encodes the structural proteins (VP1, VP2, VP3, and VP4), and the P2 and P3 regions encode the non-structural proteins associated with viral replication [5–7]. The 5′-UTR contains an extensive secondary structure, required for cap-independent translation, and is covalently linked to the viral protein VPg [5]. As a translation terminator sequence, the 3′UTR has a poly(A) tract. Six HAV genotypes have been identified; three genotypes (I, II, and III) are of human origin [1, 8], and three (IV, V, and VI) are of simian origin. Genotypes have more than 15% nucleotide variation and subgenotypes have 7–7.5% nucleotide variation [8]. Despite genetic heterogeneity at the nucleotide level, only a single serotype of HAV exists [1].
Hepatitis A is a common infectious disease in many developing countries, and in 1988, 47 (0.015%) deaths were reported in the large epidemic in Shanghai, China, which involved more than 300,000 adolescents and young adults [1]. Over the past 20 years, several outbreaks have occurred each year in China, and the number of acute hepatitis A cases reported in the United States was 4,488 in 2005 [5]. Recently, even in developed countries, numerous hepatitis A outbreaks have been reported [2, 3, 9]. In November 2003, a large hepatitis A outbreak was identified among patrons of a single Pennsylvania restaurant; of 601 patients identified, 3 (0.5%) died [2]. Thus, hepatitis A is a major threat to human health. Surveillance regarding cases and the genetic characteristics are important in controlling and preventing hepatitis A.
In 2006, two major hepatitis A incidents occurred in China. One was an outbreak at a school in Guigang City located in the Guangxi Zhuang Autonomous Region (GXZAR) of southern China. The other was an epidemic involving multiple HAV strains in Hetian City, located in the Xinjiang Uygur Autonomous Region (XJUAR) of northwestern China.
To help control outbreaks and evaluate possible transmission sources and routes, specialists performed field investigations in the two areas; we conducted separate case-control studies in Hetian (XJUAR) and Guigang (GXZAR). We isolated viral RNA and studied the molecular characterization of HAV strains from the identified cases. In Hetian, we used a molecular epidemiology approach to identify separate patients infected by the same transmission route. We further compared viral genotypes, not only between these two areas but also with other reported outbreaks.
In this report, we summarize the studies on the two outbreaks, including epidemiologic investigations, molecular surveillance, the application of molecular epidemiology, and the genetic relatedness of HAV strains reported in the different regions.
Materials and methods
Case definition and case-control studies
The two outbreaks of hepatitis A in China described here occurred in 2006. One outbreak took place in southern China (GXZAR) and the other was in Hetian (XJUAR) in northwestern China, which involved multiple outbreaks of multiple wild HAV strains. A case patient was defined by the onset of acute illness with clinical symptoms (fever, malaise, anorexia, nausea, abdominal discomfort, dark urine, jaundice, etc.), consistent with hepatitis A, within the outbreak periods and serologic confirmation of acute infection with HAV (serum IgM antibody to HAV).
In Guigang, the first case was reported on May 16 and last case on June 28. All 35 patients and 59 random candidate students who had no symptoms consistent with acute hepatitis A (of these 59 students, none was positive for IgM antibodies to HAV) were interviewed for their contact histories with possible transmission sources and routes during the incubation period. No control subject had a history of hepatitis A or had received a hepatitis A vaccine.
Similarly, in Hetian, 71 hospitalized patients (age range: 1–33 years, 5 patients were older than 30 years; 26 females and 45 males, the Hetian Infectious Diseases Hospital) and 127 control individuals who had no symptoms consistent with hepatitis A were interviewed. Informed consents were obtained from all patients we studied.
Detection of HAV-specific IgM
IgM anti-HAV enzyme immunoassays were performed by using a Diagnostic Kit for IgM Antibody to HAV (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China) according to the manufacturer’s instructions. For each enzyme-linked immunosorbent assay (ELISA), undiluted serum (50 μL) was used [10, 11].
Virus samples, RNA extraction, RT-PCR, and nested PCR
Viral serum samples were collected from case patients in Guigang and Hetian. HAV RNA was extracted from a part (100 μL) of the serum sample by using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. The RNA pellet was redissolved in 10 μL of 10 mM dithiothreitol containing 5% (v/v) RNasin (40 units/μL; Promega, Madison, WI) and stored at −80°C until use.
Primer 3381 N (all numbers refer to the HM175 strain) [12], 5′-CCA TTT CAA GAG TCC ACA CAC T-3′ [1] was used for the RT-PCR reaction. The reaction mix contained 1 μL of RNA stock, 0.5 μL of RNasin (Promega), 1 μL of 100 mM dithiothreitol (Promega), 1 μL of 10 mM dNTP (Pharmacia, Piscataway, NJ), 2.5 μL of 10 μM primer solution, 1 μL (200 units) of Superscript II reverse transcriptase (Gibco/BRL, Grand Island, NY), and 4 μL of 5 × 1st Strand Synthesis buffer (Gibco/BRL). The reaction was incubated for 1 h at 42°C; 1 μL of RNase H (Gibco/BRL) and 1 μL of RNase T1 (Gibco/BRL) were then added and the reaction was incubated for 20 min at 37°C. The cDNA was then purified with the DNA Clean Sweep kit (Amresco, Solon, OH). The cDNA pellet was redissolved in 20 μL of double-distilled water and kept on ice until it was added to the following PCR reaction [13, 14].
The VP1/P2A region was selected for amplification of HAV RNA. The external primers were 2870P, 5′-GAC AGA TTC TAC ATT TGG ATT GGT-3′ (forward [1]), and 3381 N, 5′-CCA TTT CAA GAG TCC ACA CAC T-3′ (reverse [1]). The internal primers were 2896P, 5′-CTA TTC AGA TTG CAA ATA CAA T-3′ (forward [1]), and 3289 N, 5′-AAC TTC ATT ATT TCA TGC TCC T-3′ (reverse [1]). Reactions were performed in thin-walled PCR tubes (Stratagene, La Jolla, CA). Both PCR reactions were performed by using a Robocycler thermal cycler (Stratagene) with the following cycling parameters: denaturation at 95°C for 35 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s, for 25 cycles.
Nucleotide sequencing
In positive samples, nucleotide sequence analysis was undertaken directly on the purified second PCR products by using internal primers and ABI PRISM BigDye Terminator Cycle Sequencing Kit (Applied BioSystems, Foster City, CA) in an ABI PRISMA 3130 XL DNA Analyzer (Applied BioSystems).
Phylogenetic analysis
Phylogenetic analysis was performed with the MEGA4 Molecular Evolutionary Genetics Analysis software (http://www.megasoftware.net/mega4/). The genetic distance was calculated by using the uncorrected distance algorithm within the distances program in MEGA4. Final tree construction was based on the unweighted pair group method with arithmetic mean (UPGMA) algorithms.
Statistical analysis
Odds ratios (ORs) were used to measure the strength of associations in the case-control studies, and 95% confidence intervals (CIs) were calculated with the SAS software, using the Mantel–Haenszel statistic. P-values < 0.05 were deemed to be statistically significant.
Results
Descriptive epidemiology in Guigang
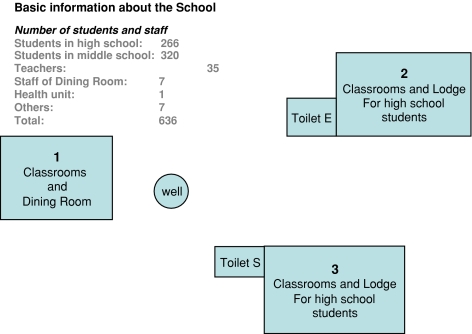
The outbreak occurred in a private school in Guigang. On May 16, 2006, the first hepatitis A case (a 17-year-old boy) in Guigang was reported to the GXZAR Center for Diseases Control and Prevention, and between then and the final case from the school reported on June 28, a total of 35 cases involving 25 males and 10 females were reported. No case was reported from outside this school in the city during the estimated incubation time. The ages of the infected students ranged from 13 to 19 years (average age = 15); all patients showed clinical symptoms consistent with hepatitis A and had elevated alanine aminotransferase (ALT). The GXZAR Center for Diseases Control and Prevention surveyed possible causes of the outbreak. The school consisted of 266 (190 male and 76 female) high school students, 320 (205 male and 115 female) middle school students, 35 teachers, 1 healthcare person, 7 staff in the dining room, and 7 male guards (Fig. 1). All students lived and studied on the independent school campus. After screening possible transmission factors for a fecal-oral disease at the school, investigators focused on the school dining room and the water supply system. The dining room was located in building 1 (Fig. 1). The antibody test for antihepatitis A IgM was negative for all seven staff in the dining room. The dining room used water from a well to prepare meals, and all students ate meals in the dining room. Two toilets were located close to the well, and the distances from toilets E and S to the well were 50 and 30 m, respectively (Fig. 1). Guigang is located in southern China, and the month of May is typically warm and rainy in this area. Investigators found evidence that water in the well was contaminated with ordure from the two toilets. Although we did not try to isolate HAV from the water supply, bacterial analysis showed that the water was contaminated with Escherichia coli (data not shown). Interviews with students revealed that some of them drank the water directly from this supply system. To determine the percentage of asymptomatic hepatitis A infections, we tested for IgM antibodies to hepatitis A in all 636 individuals at the school; 60 were found positive. Thus, in addition to the 35 patients, 25 asymptomatic individuals were confirmed. Investigation showed that the 60 individuals were infected with HAV during the same incubation time. Investigators corroborated that the water and other materials used for meal preparation and drinking had been sterilized. Together with the evidence that several children without illness also ate meals in the same dining room, we excluded the possibility that the dining room was a possible transmission source or route.
Fig. 1.
Schematic diagram of the school in Guigang. An independent school campus, surrounded by a wall. All students lived and studied here
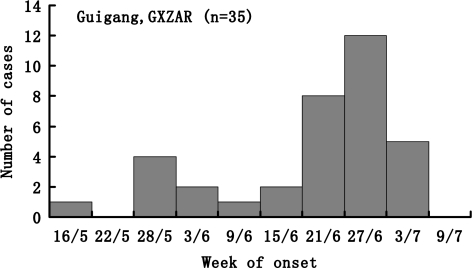
As shown in Fig. 2, it seemed that two peak dates of illness onset occurred. The first was around May 28 and the second was around June 27. Although illness onset in the same period and the case-control study showed that drinking unsterilized water was significantly associated with the hepatitis A transmission, we could not determine whether or how contact infection among children amplified this outbreak.
Fig. 2.
Cases of hepatitis A in Guigang, according to the date of onset. Bars represent students who were given a diagnosis of hepatitis A between May 16 and July 9, 2006
Descriptive epidemiology in Hetian
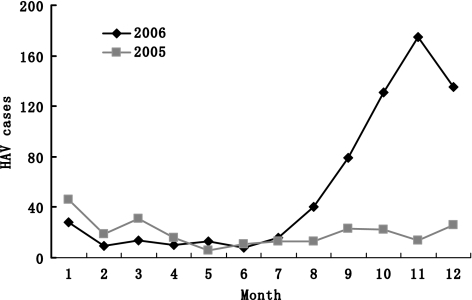
The XJUAR borders eight countries: Russia, Kazakhstan, Kirghizistan, Tajikistan, Pakistan, Mongolia, India, and Afghanistan. Hetian, an administration district of XJUAR consisting of seven counties and one city (Hetian City), had a total population of 1,743,813 in 2006. The total hepatitis A cases in this area was 240 (2005) and increased to 658 (2006) (Fig. 3). From epidemiologic survey of hepatitis A, every 7–8 years in XJUAR as many as approximately 74% of the children younger than 10 years were positive for anti-HAV IgG. Along with an increase in the population, the anti-HAV IgG rate in the population decreased to as low as 40%. So, after this accumulation of a larger susceptible population, new outbreaks are almost inevitable in XJUAR. In addition, hepatitis A cases in XJUAR typically increase annually between August and January. To analyze the epidemiology of hepatitis A in this district, we inspected Hetian City for possible transmission routes and sources. Nations in this area have different cultural traditions regarding diet, partying, and eating habits. August through January is a popular season for parties, and community activities increase. Inadequate sanitation and poor hygiene conditions favor the pollution of water and food, and similar to the other area, most patients were children and the rate in males was higher than in females.
Fig. 3.
The monthly hepatitis A survey in Hetian area. The total hepatitis A cases of each month in the year of 2005 and 2006. The cases increased within September to December of 2006, which represented the outbreaks in this area. Data was provided by local hepatitis survey system
In contrast to Guigang, it rarely rains in Hetian; water is typically supplied by deep wells and we found no evidence of contamination in the public water supply system. Together with some sporadic incidents, the increase in the incidence rate of hepatitis A was not likely due to a single source. Inspections showed that inadequate sanitation, poor hygiene conditions, contact infection, the existence of multiple HAV strains, and the accumulating population susceptible to hepatitis A were five major amplifying factors in the epidemiology of hepatitis A in Hetian.
Case-control studies
All 35 patients in Guigang, GXZAR, were enrolled in a case-control study; 59 random volunteer students who were free of this outbreak were used as controls. Of the 35 patients, 16 (46%) had a history of drinking unsterilized water (in China, people routinely drink boiled water; thus, unsterilized water means water that has not been boiled). Of the 59 control subjects, only 5 (8%) had a history of drinking unsterilized water, and drinking unsterilized water was significantly associated with hepatitis A (OR, 9.09 [95% CI, 2.93–28.21]); Table 1]. Thus, contaminated water was fixed as a source of the hepatitis A outbreaks in Guigang.
Table 1.
Association between drinking unsterilized water and hepatitis A in Guigang, GXZAR
| History of drinking unsterilized water | Case patients (n = 35) | Control subjects (n = 59) | OR (95% CI) |
|---|---|---|---|
| Yes | 16 | 5 | 9.09 (2.93–28.21) |
| No | 19 | 54 |
Abbreviations: CI confidence interval, OR odds ratio
Although the field investigation was complex and it was difficult to determine a clear transmission route or factor in Hetian, we did track some patients and performed a case-control study on the basis of evidence from molecular epidemiology. Our viral sequence analysis showed that seven patients in Hetian were infected with the same HAV; we then tried to establish a possible common transmission route. Interviews with all seven patients showed that six of them had eaten a certain ice cream during the estimated incubation time. One child had not eaten the suspect ice cream, but he had played with another child who was infected with same HAV and had an onset of illness 1 week earlier. We inspected the patients’ dwellings and selected 28 matching individuals who lived near the case patients, shared similar living conditions, and who were negative for IgM to HAV to perform a case-control study. Our findings showed that ingestion of the suspected ice cream was significantly associated with hepatitis A in those patients (OR, 36.00 [95% CI, 3.38–383.91]); Table 2].
Table 2.
Association between suspected ice cream and hepatitis A in seven infected patients
| Ingestion of suspected ice cream | Case patients (n = 7) | Control subjects (n = 28) | OR (95% CI) |
|---|---|---|---|
| Yes | 6 | 4 | 36.00 (3.38–383.91) |
| No | 1 | 24 |
Abbreviations: CI confidence interval, OR odds ratio
Viral sequencing analysis of HAV strains
To study the genotype(s) and genetic relatedness of the strains of HAVs isolated from the two areas, we performed a molecular epidemiology analysis. As the 168-nucleotide fragment of the VP1/2A junction was shown not to sufficiently differentiate genotypes or relatedness among HAV isolates, a 320-nucleotide fragment of the VP1/2A junction region (from 2935 to 3254, referring to strain HM175) was selected as the genotype-determining sequence [1].
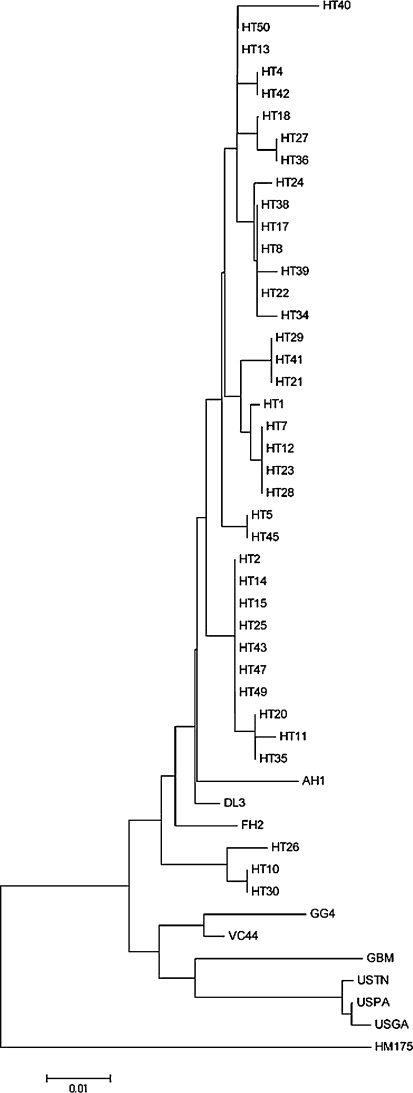
The HAV strains insolated in Guigang were demonstrated to be type 1A by genotype analysis. All 35 sequences showed 100% homology in the 320-nucleotide sequences of the VP1/2A region, which showed that this outbreak involved only one HAV strain. We selected only one strain to construct a phylogeny (Fig. 4, strain GG4). Of the 71 patient samples we collected in Hetian, 38 were successfully sequenced. Although all 38 viral sequences from Hetian belonged to the 1A genotype, the sequences showed considerable variability. On the basis of the 320 sequenced nucleotides, we found nearly 20 strains in Hetian during the same incubation period (Fig. 4). HT2, HT14, HT15, HT25, HT43, HT47, and HT49 were the same stain and formed the biggest infection cluster. H17, H12, H23, and H28 were another strain and formed the second biggest infection cluster (Fig. 4). This result showed that several wild HAV strains occurred in the Hetian area and that these HAV depots were the sources of hepatitis A outbreak in this region.
Fig. 4.
Phylogenetic analysis of the VP1/2A region of the strains isolated from Guigang and Hetian, and reported strains. Neighbor-joining phylogenetic trees of the VP1/2A region by using Kimura’s 2-parameter model. Strains are indicated at nodes. Bars denote genetic distances. HT indicates the strain isolated in Hetian, and GG4 refers to the strain isolated in Guigang. In Guigang, all 35 patients were infected by a single strain, and we selected GG4 to perform the phylogenetic analysis. AH1, AB020564 [15]; FH2, AB020568 [15]; VC44, AB253604; GBM, X75215 [16, 17]; HM175, M14707 [12], the DL3 sequence [11], and the strains isolated from the United States, USTN, USPA, and USGA
Molecular epidemiology of the hepatitis A outbreak in Hetian
To clarify whether patients might be linked to a suspected hepatitis A outbreak and to advance the investigation in the field, we tracked some patients who were infected with the same HAV strain. Patients HT2, HT14, HT15, HT25, HT43, HT47, and HT49 were infected with the same HAV strain and formed the biggest infection cluster. We then examined whether these patients shared a common source or route. We interviewed all seven patients (six children younger than 4 years and one 38-year-old adult). They had separate residences in Hetian City, although two children were neighbors. After screening for transmission routes, investigators found that six of the patients had a history of eating a certain brand of ice cream. We then inspected the small workshop where the ice cream was prepared. It had no authorization from the local health administration, and although the hygiene conditions were poor, we found no evidence that the material was contaminated with HAV at the time we made the inspection, and all the staffs were negative for IgM to HAV. We failed to confirm that HAV was present in the ice cream samples we took. We also found no other patient who was infected with the same HAV strain before or after the estimated incubation period.
Patients HT21, HT29, and HT41 were found to be infected with the same HAV strain. An investigation of these three patients showed that they were three visitors from eastern China, aged 33, 18, and 38 years, respectively. They had traveled to Hetian 3 weeks before the onset of illness and were living and eating in the same small hotel. Investigation of that lodging showed that inadequate sanitation measures (e.g., no refrigerator for meals) might have been risk factors for hepatitis A. However, the hotel staffs were negative for IgM to hepatitis A and only these three visitors stayed in this hotel during the estimated incubation time.
Although we were unable to confirm HAV in our suspected sources or routes, molecular epidemiology did help in finding links among patients coming from different regions. Our study showed that in the case of a complex situation, molecular epidemiology is a powerful approach to aid field investigations.
Genetic relatedness of HAV-reported strains
To study the distribution of HAV strains, we compared the HAV strains from these two incidents. In Guigang, all viral strains isolated belonged to one genotype, 1A, whereas in Hetian, our genetic study showed that nearly 20 wild viral strains occurred in the area during the estimated incubation period (Fig. 4). The genetic variability between the HAV strain isolated from Guigang and the HAV strains isolated from Hetian was more than 4.3%.
Although HAV displays a high degree of antigenic and genetic conservation and does not appear to accumulate the high frequency of genetic changes seen in many RNA viruses [1, 5], molecular surveillance on this widespread infectious disease is still important. We compared our sequences with those reported in the National Center for Biotechnology Information (NCBI) database (AH1, AB020564 [15]; FH2, AB020568 [15]; VC44, AB253604; GBM, X75215 [16, 17]; HM175, M14707 [12]), the DL3 sequence [11], and the strains isolated from the United States, which were involved in the outbreaks in Pennsylvania, Tennessee, and Georgia in 2003 (USTN, USPA, and USGA) [2, 9]. The genetic variability between the Chinese strains in our study and the strains isolated in the United States was at least 5% (Fig. 4). The GBM strain was close to the USTN, USPA, and USGA strains. The DL3 strain, isolated in Dalian, China, was close to the HT13 and HT50 strains, with a variability of 0.09%. The Japanese strain FH2 was close to HT27 and HT36, with a genetic variability of 1.3%. The GG4 strain was close to the Vietnamese strain VC44 strain (genetic variability of 1.9%), and the Japanese strain AH1 was close to the HT2 group (genetic variability of 1.9%). Our study showed that the Asian HAV strains were closer to each other than to the other reported sequences. Although our data were limited, we estimate that the prevalent HAV strains in China were domestic strains.
Discussion
Although HAV infection is generally self-limited, the rate at which patients die due to acute liver failure ranges from 0.015 to 0.5%. In the United States, 63% of the total viral hepatitis cases reported in 1998 were associated with hepatitis A [18], and the true incidence of hepatitis A is thought to be much higher because the disease often evolves without specific symptoms and consequently goes undetected.
In China, hepatitis is a huge public health problem. In fact, 9.8% of the Chinese populations are carriers of hepatitis B virus and 3.2% are infected with hepatitis C virus. The epidemic rate of hepatitis A in China is up to 80.9% and that of hepatitis E in China is 18.0% [19], so viral hepatitis poses a major problem. Above all, outbreaks of hepatitis A are the most frequent types of acute hepatitis. Although the Chinese government is planning to conduct vaccinations for hepatitis A, it will continue to be a major infectious disease for many years.
To date, few investigations on the epidemiology or molecular surveillance have been performed in China. In this study, we reported our efforts regarding two incidents that occurred in China in 2006. The outbreak in Guigang involved a single wild HAV strain. In Hetian, our samples were collected from the hospital; the epidemic consisted of many sporadic small outbreaks, which made it difficult to carry out a field investigation, the number of interviewed patients is small, and then the information got from these patients might not represent the whole situation in this area. It was also hard to verify transmission sources and routes. Although inadequate sanitation, poor hygiene conditions, contact infection, the existence of multiple HAV strains, and the accumulating population were believed to be the major factors to amplify the HAV epidemic in this area, there is possibly a unknown factor that was involved in the epidemic. Ongoing viral strain surveillance facilitated the rapid implementation of case and control measures. Incorporation of molecular epidemiologic methods into routine hepatitis A surveillance has improved the detection of hepatitis A outbreaks and increased our understanding of its epidemiology in China.
In XJUAR, hepatitis A is endemic. Wild HAV strains have accumulated for as long as the history of the Silk Road. Meanwhile, the national culture, inadequate sanitation, and poor hygiene have amplified the transmission of HAV in this region. In many regions of the world, including the United States, Japan, and China, HAV-related isolates tend to cluster, suggesting endemic spread [5]. A high degree of genetic conservation was shown during the infection period of an individual [20] or even among different isolates with common sources of infection [5, 21, 22]. In contrast, a higher degree of heterogeneity than previously reported has been found in strains isolated in South America [5, 23]. Recent studies suggest that a changing epidemiologic pattern in HAV infection throughout South America may result in more clinical cases in teenagers and adults and a greater potential for new outbreaks [24]. Whether this changing pattern in XJUAR is related to a higher genetic variability of HAV in that particular geographic region than previously expected or to changes in hygiene conditions is unclear. The evolutionary aspect is also vague, as is why these viruses were restricted to these regions. Much remains to be studied.
An extensive study of South American HAV strains revealed that the VP1 amino terminus contained more informative variable positions than the VP1/2A junction. Thus, the VP1/2A junction is more variable and can be used to distinguish one strain from another [25]. In our study, we used only 320 bp of the VP1/2A junction region to determine the HAV genotype.
In this study, although we either did not try or failed to isolate HAV from suspected samples, the case-control studies showed it to be related to the hepatitis A transmission route and our field investigation in Hetian was consistent with the molecular evidence.
In conclusion, we performed both field investigations and molecular epidemiology surveillance on two outbreaks of hepatitis A that occurred in China in 2006. Our study provides important information on the hepatitis A outbreaks.
Acknowledgments
We have no conflict of interest on this study. We thank the staff of the Chinese Center for Diseases Control and Prevention at GXZAR and XJUAR. Our research is supported by the National Natural Science Foundation of China (30671806) and grants from the Chinese Center for Disease Control and Prevention, Ministry of Health of the People’s Republic of China.
Abbreviations
- ALT
Alanine aminotransferase
- CI
Confidence interval
- GXZAR
Guangxi Zhuang Autonomous Region
- HAV
Hepatitis A virus
- OR
Odds ratio
- XJUAR
Xinjiang Uygur Autonomous Region
Contributor Information
Yue Wang, Phone: +86-10-63554474, FAX: +86-10-63510565, Email: euy-tokyo@umin.ac.jp.
Shengli Bi, Email: shengli_bi@163.com.
References
- 1.Nainan OV, Xia G, Vaughan G, Margolis HS. Diagnosis of hepatitis A virus infection: a molecular approach. Clin Microbiol Rev. 2006;19(1):63–79. doi: 10.1128/CMR.19.1.63-79.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler C, Vogt TM, Armstrong GL, Vaughan G, Weltman A, Nainan OV, et al. An outbreak of hepatitis A associated with green onions. N Engl J Med. 2005;353(9):890–897. doi: 10.1056/NEJMoa050855. [DOI] [PubMed] [Google Scholar]
- 3.Hutin YJ, Pool V, Cramer EH, Nainan OV, Weth J, Williams IT, et al. A multistate, foodborne outbreak of hepatitis A. National Hepatitis A Investigation Team. N Engl J Med. 1999;340(8):595–602. doi: 10.1056/NEJM199902253400802. [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Lemon SM, Hepatitis A. Virus: from discovery to vaccines. Hepatology. 2006;43(Suppl 1(2)):S164–S172. doi: 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- 5.Costa-Mattioli M, Di Napoli A, Ferre V, Billaudel S, Perez-Bercoff R, Cristina J. Genetic variability of hepatitis A virus. J Gen Virol. 2003;84(Pt 12):3191–3201. doi: 10.1099/vir.0.19532-0. [DOI] [PubMed] [Google Scholar]
- 6.Shimoike T. Structure and function of HAV genome (RNA) Nippon Rinsho. 2004;62(Suppl 8):428–432. [PubMed] [Google Scholar]
- 7.Shimoike T. Structure of HAV particle. Nippon Rinsho. 2004;62(Suppl 8):423–427. [PubMed] [Google Scholar]
- 8.Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, et al. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73(Pt 6):1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- 9.Amon JJ, Devasia R, Xia G, Nainan OV, Hall S, Lawson B, et al. Molecular epidemiology of foodborne hepatitis a outbreaks in the United States, 2003. J Infect Dis. 2005;192(8):1323–1330. doi: 10.1086/462425. [DOI] [PubMed] [Google Scholar]
- 10.Fan D, Wang C, Zhang Y, Li X, Wang X. The seroepidemiological investigation of HAV among the population aged from 1 to 15 years in HAV outbreak area in Yili Prefecture. Dis Surveill. 2006;21(10):526–528. [Google Scholar]
- 11.Cao J, Meng Q, Li X, Zhang L, Yuan J, Fan D, Wang C, et al. Genotyping of hepatitis A virus prevalent strains in Xinjiang Yili of China in 2005. Chin J Virol. 2007;23(2):110–114. [PubMed] [Google Scholar]
- 12.Cohen JI, Ticehurst JR, Purcell RH, Buckler-White A, Baroudy BM. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987;61(1):50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellier R, Bukh J, Emerson SU, Purcell RH. Amplification of the full-length hepatitis A virus genome by long reverse transcription-PCR and transcription of infectious RNA directly from the amplicon. Proc Natl Acad Sci USA 1996;93(9):4370–4373. Erratum in: Proc Natl Acad Sci USA 1997;94(19):10485 [DOI] [PMC free article] [PubMed]
- 14.Lu L, Ching KZ, Paula VS, Nakano T, Siegl G, Weitz M, et al. Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate) J Gen Virol. 2004;85(Pt 10):2943–2952. doi: 10.1099/vir.0.80304-0. [DOI] [PubMed] [Google Scholar]
- 15.Paula VS, Lu L, Niel C, Gaspar AM, Robertson BH. Genetic analysis of hepatitis A virus isolates from Brazil. J Med Virol. 2004;73(3):378–383. doi: 10.1002/jmv.20101. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara K, Yokosuka O, Fukai K, Imazeki F, Saisho H, Omata M. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J Hepatol. 2001;35(1):112–119. doi: 10.1016/S0168-8278(01)00074-5. [DOI] [PubMed] [Google Scholar]
- 17.Graff J, Normann A, Feinstone SM, Flehmig B. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J Virol. 1994;68(1):548–554. doi: 10.1128/jvi.68.1.548-554.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Available from www.cdc.gov/hepatitis/HAV/SurveillanceA.htm. Accessed 14 July 2007
- 19.Dai ZC, Qi GM. Viral Hepatitis in China, Seroepidemiological Survey in Chinese Population (Part One) 1992–1995. Beijing: Beijing Science and Technology Press; 1997. p. 39–58
- 20.Robertson BH, Averhoff F, Cromeans TL, Han X, Khoprasert B, Nainan OV, et al. Genetic relatedness of hepatitis A virus isolates during a community-wide outbreak. J Med Virol. 2000;62(2):144–150. doi: 10.1002/1096-9071(200010)62:2<144::AID-JMV4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Tallo T, Norder H, Tefanova V, Ott K, Ustina V, Prukk T, et al. Sequential changes in hepatitis A virus genotype distribution in Estonia during 1994 to 2001. J Med Virol. 2003;70(2):187–193. doi: 10.1002/jmv.10377. [DOI] [PubMed] [Google Scholar]
- 22.Diaz BI, Sariol CA, Normann A, Rodriguez L, Flehmig B. Genetic relatedness of Cuban HAV wild-type isolates. J Med Virol. 2001;64(2):96–103. doi: 10.1002/jmv.1023. [DOI] [PubMed] [Google Scholar]
- 23.Mbayed VA, Sookoian S, Alfonso V, Campos RH. Genetic characterization of hepatitis A virus isolates from Buenos Aires, Argentina. J Med Virol. 2002;68(2):168–174. doi: 10.1002/jmv.10194. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka J. Hepatitis A shifting epidemiology in Latin America. Vaccine. 2000;18(Suppl 1):S57–S60. doi: 10.1016/S0264-410X(99)00466-1. [DOI] [PubMed] [Google Scholar]
- 25.Costa-Mattioli M, Cristina J, Romero H, Perez-Bercof R, Casane D, Colina R, et al. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J Virol. 2002;76(18):9516–9525. doi: 10.1128/JVI.76.18.9516-9525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]