Abstract
Certain proteins contain subunits that enable their active translocation across the plasma membrane into cells. In the specific case of HIV-1, this subunit is the basic domain Tat49–57 (RKKRRQRRR). To establish the optimal structural requirements for this translocation process, and thereby to develop improved molecular transporters that could deliver agents into cells, a series of analogues of Tat49–57 were prepared and their cellular uptake into Jurkat cells was determined by flow cytometry. All truncated and alanine-substituted analogues exhibited diminished cellular uptake, suggesting that the cationic residues of Tat49–57 play a principal role in its uptake. Charge alone, however, is insufficient for transport as oligomers of several cationic amino acids (histidine, lysine, and ornithine) are less effective than Tat49–57 in cellular uptake. In contrast, a 9-mer of l-arginine (R9) was 20-fold more efficient than Tat49–57 at cellular uptake as determined by Michaelis–Menton kinetic analysis. The d-arginine oligomer (r9) exhibited an even greater uptake rate enhancement (>100-fold). Collectively, these studies suggest that the guanidinium groups of Tat49–57 play a greater role in facilitating cellular uptake than either charge or backbone structure. Based on this analysis, we designed and synthesized a class of polyguanidine peptoid derivatives. Remarkably, the subset of peptoid analogues containing a six-methylene spacer between the guanidine head group and backbone (N-hxg), exhibited significantly enhanced cellular uptake compared to Tat49–57 and even to r9. Overall, a transporter has been developed that is superior to Tat49–57, protease resistent, and more readily and economically prepared.
The bioavailability of drugs or molecular probes directed at intracellular receptors depends significantly on their being sufficiently polar for administration and distribution and sufficiently nonpolar for passive diffusion through the relatively nonpolar bilayer of the cell. As a consequence, although covering a broad range of structural diversity, most drugs are limited to a narrow range of physical properties. In addition, many promising drug candidates fail to advance clinically because they fall out of this range, being either too nonpolar for administration and distribution or too polar for passive cellular entry. Exceptions arise mainly through significant changes in formulation [e.g., poorly soluble taxol is formulated in ethanol:Cremophor EL (1)] or extensive tuning of physical properties [e.g., polar oligonucleotides modified with lipid groups (2)]. Several techniques have been developed to enable cellular uptake including drug incorporation into cationic liposomes (3), dendrimers (4), or siderophores (5).
In contrast to most drugs, certain naturally occurring macromolecules enter cells through an active transport mechanism. One important example is the nuclear transcription activator protein (Tat) encoded by HIV type 1 (HIV-1), a 101-aa protein that is required for viral replication (6, 7). Of particular interest for drug delivery is that exogenously added HIV-1 Tat efficiently crosses the plasma membranes of cells in an apparent energy-dependent fashion, localizes to the nucleus, and is functional, stimulating HIV-long terminal repeat-driven RNA synthesis (6–11). The sequence responsible for the cellular uptake of HIV-1 Tat consists of the highly basic region, amino acid residues 49–57 (RKKRRQRRR) (12–16). The detailed mechanism for the cellular uptake of HIV-1 Tat49–57 remains unknown.
HIV-1 Tat has been used to deliver functional biomolecules into cells. Although the entire protein can be used for this purpose, it is more efficient to use the truncated sequence containing only the basic residues required for transport, Tat49–57. Through covalent attachment to Tat-49–57, several proteins have been delivered into cells, including an inhibitor of human papillomavirus type 16 (HPV-16) (13), ovalbumin into the MHC class I pathway (17), the Cdk inhibitors p27Kip1 (18) and p16INK4a (19), and a caspase-3 protein (20). Tat49–57 has also been successfully used to deliver β-galactosidase in vivo into all tissues of the mouse including the brain (21). In addition to Tat49–57, several other short peptide sequences have been identified with membrane translocation activity, including those derived from Antennapedia (6, 22), fibroblast growth factor (23), Galparan (transportan) (24), and HSV-1 structural protein VP22 (25).
A structural analogy can be drawn between Tat49–57 and homopolymers [molecular weight (m.w.) = 4,000–200,000] of the cationic amino acids lysine (26), ornithine (27), and arginine (27) that are also able to enter cells. Polylysine (PL) has been used to efficiently deliver a range of biomolecules into cells including albumin and horseradish peroxidase (PL m.w. = 6700) (28), methotrexate (PL m.w. = 70,000) (29), oligonucleotides (PL m.w. = 14,000) (30), and adenovirus (PL m.w. = 20,500) (31). In addition, polylysine peptoid derivatives (32) have been used for gene delivery. Polyarginine (PA) has also been used to enhance the cellular uptake of tumor antigens (TA). The polyarginine-TA (PA m.w. = 100,000) conjugates are more efficiently translocated into cells than the corresponding polylysine-TA (PL m.w. = 94,000) conjugates by a factor of 10 as determined by fluorescent flow cytometry (33). However, problems related to toxicity, protein precipitation, and cost prevent such large cationic polymers from being broadly useful therapeutically. Because of the potential structural analogy to Tat49–57, we previously compared the cellular uptake of short oligomers of arginine, lysine, ornithine, and histidine. Interestingly, short oligomers of arginine were much more efficient at entering cells than the corresponding short oligomers of histidine, lysine, and ornithine (34).
Prompted by the potential broad value of using molecular transporters to enable or enhance drug delivery, we initiated a program aimed at elucidating the structural features of Tat49–57 that are required for its cellular entry. The second and more important goal of this study was to design and synthesize simpler and more effective molecular transporters for use in drug or probe delivery, a goal of broad fundamental and applied consequence.
Materials and Methods
General.
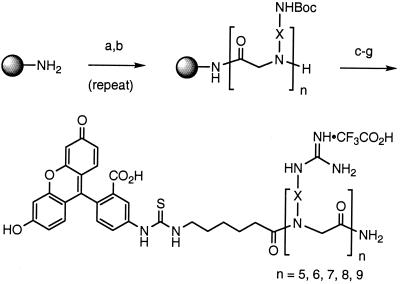
Rink amide resin and Boc2O were purchased from NovaBiochem. Diisopropylcarbodiimide, bromoacetic acid, fluorescein isothiocyanate (FITC-NCS), ethylenediamine, 1,3-diaminopropane, 1,4-diaminobutane, 1,6-diaminohexane, 1,8-diaminooctane, trans-1,6-diaminocyclohexane, and pyrazole-1-carboxamidine were all purchased from Aldrich. All solvents and other reagents were purchased from commercial sources and used without further purification. The mono-Boc amines were synthesized from the commercially available diamines by using a literature procedure (10 equiv of diamine and 1 equiv of Boc2O in chloroform followed by an aqueous work up to remove unreacted diamine) (35).
N-tert-butoxycarbonyl-1,6-trans-diaminocyclohexane.
Mp 159–161°C; 1H NMR (CDCl3) δ 4.35 (br s, 1H), 3.37 (br s, 1H), 2.61 (br s, 1H), 1.92–2.02 (m, 2H), 1.81–1.89 (m, 2H), 1.43 (s, 9H), 1.07–1.24 (m, 4H) ppm; 13C NMR (D6-DMSO) δ 154.9, 77.3, 49.7, 48.9, 35.1, 31.4, 28.3 ppm; ES-MS (M+1) calcd 215.17, found 215.22.
General Procedure for Peptide Synthesis.
Tat49–57 (RKKRRQRRR), truncated and alanine-substituted peptides derived from Tat49–57, Antennapedia43–58 (RQIKIWFQNRRMKWKK), and homopolymers of l-arginine (R5-R9) and d-arginine (r5-r9) were prepared with an automated peptide synthesizer (ABI433) by using standard solid-phase fluorenylmethoxycarbonyl (Fmoc) chemistry (36) with HATU as the peptide coupling reagent. The fluorescein moiety (Fl) was attached via an aminohexanoic acid spacer by treating a resin-bound peptide (1.0 mmol) with FITC (1.0 mmol) and diisopropyl ethyl amine (5 mmol) in dimethylformamide (DMF; 10 ml) for 12 h. Cleavage from the resin was achieved by using 95:5 trifluoroacetic acid (TFA)/triisopropylsilane. Removal of the solvent in vacuo gave a crude oil that was triturated with cold ether. The crude mixture thus obtained was centrifuged, the ether was removed by decantation, and the resulting orange solid was purified by RP-HPLC (H2O/CH3CN in 0.1% TFA). The products were isolated by lyophilization and characterized by electrospray mass spectrometry. The purity of the peptides was >95% as determined by analytical RP-HPLC (H2O/CH3CN in 0.1% TFA).
General Procedure for Peptoid Polyamine Synthesis.
Peptoids were synthesized manually by using a fritted glass apparatus and positive nitrogen pressure for mixing the resin following the literature procedure developed by Zuckermann (32, 37, 38). Treatment of Fmoc-substituted Rink amide resin (0.2 mmol) with 20% piperidine/DMF (5 ml) for 30 min (2×) gave the free resin-bound amine that was washed with DMF (3 × 5 ml). The resin was treated with a solution of bromoacetic acid (2.0 mmol) and diisopropylcarbodiimide (2.0 mmol) in DMF (5 ml) for 30 min. This procedure was repeated. The resin was then washed (3 × 5 ml DMF) and treated with a solution of mono-Boc diamine (8.0 mmol) in DMF (5 ml) for 12 h. These two steps were repeated until an oligomer of the required length was obtained (Note: the solution of mono-Boc diamine in DMF could be recycled without appreciable loss of yield). The resin was then treated with N-Fmoc-aminohexanoic acid (2.0 mmol) and DIC (2.0 mmol) in DMF for 1 h and this was repeated. The Fmoc moiety was then removed by treatment with 20% piperidine/DMF (5 ml) for 30 min. This step was repeated and the resin was washed with DMF (3 × 5 ml). The resin was then treated with FITC (0.2 mmol) and diisopropyl ethyl amine (2.0 mmol) in DMF (5 ml) for 12 h. The resin was then washed with DMF (3 × 5 ml) and dichloromethane (5 × 5 ml). Cleavage from the resin was achieved by using 95:5 TFA/triisopropylsilane (8 ml). Removal of the solvent in vacuo gave a crude oil that was triturated with cold ether (20 ml). The crude mixture thus obtained was centrifuged, the ether was removed by decantation, and the resulting orange solid was purified by RP-HPLC (H2O/CH3CN in 0.1% TFA). The products were isolated by lyophilization and characterized by electrospray mass spectrometry and in selected cases by 1H NMR spectroscopy.
General Procedure for Perguanidinylation of Peptoid Polyamines.
A solution of peptoid amine (0.1 mmol) dissolved in deionized water (5 ml) was treated with sodium carbonate (5 equiv per amine residue) and pyrazole-1-carboxamidine (5 equiv per amine residue) and heated at 50°C for 24–48 h. The crude mixture was then acidified with TFA (0.5 ml) and directly purified by RP-HPLC (H2O/CH3CN in 0.1% TFA). The products were characterized by electrospray mass spectrometry and isolated by lyophilization and further purified by RP-HPLC. The yield for the perguanidinylated peptoids was 60–70%, and their purity was >95% as determined by analytical RP-HPLC (H2O/CH3CN in 0.1% TFA).
Cellular Uptake Assay.
The arginine homopolymers and guanidine-substituted peptoids were each dissolved in PBS buffer (pH 7.2), and their concentration was determined by absorption of fluorescein at 490 nm (ɛ = 67,000). The accuracy of this method for determining transporter concentration was established by weighing selected samples and dissolving them in a known amount of PBS buffer. The concentrations determined by UV spectroscopy correlated with the amounts weighed out manually. Jurkat cells (human T cell line), murine B cells (CH27), or human PBL cells were grown in 10% FCS and DMEM and each of these were used for cellular uptake experiments. Varying amounts of arginine and oligomers of guanidine-substituted peptoids were added to approximately 3 × 106 cells in 2% FCS/PBS (combined total of 200 μl) and placed into microtiter plates (96-well) and incubated for varying amounts of time at 23°C or 4°C. The microtiter plates were centrifuged and the cells were isolated, washed with cold PBS (3 × 250 μl), incubated with 0.05% trypsin/0.53 mM EDTA at 37°C for 5 min, washed with cold PBS, and resuspended in PBS containing 0.1% propidium iodide. The cells were analyzed by using fluorescent flow cytometry (FACScan; Becton Dickinson) and cells staining with propidium iodide were excluded from the analysis. The data presented are the mean fluorescent signal for the 5,000 cells collected.
Inhibition of Cellular Uptake with Sodium Azide.
The assays were performed as previously described with the exception that the cells used were preincubated for 30 min with 0.5% sodium azide in 2% FCS/PBS buffer before the addition of fluorescent peptides and the cells were washed with 0.5% sodium azide in PBS buffer. All of the cellular uptake assays were run in parallel in the presence and absence of sodium azide.
Cellular Uptake Kinetics Assay.
The assays were performed as previously described except the cells were incubated for 0.5, 1, 2, and 4 min at 4°C in triplicate in 2% FCS/PBS (50 μl) in microtiter plates (96-well). The reactions were quenched by diluting the samples into 2% FCS/PBS (5 ml). The assays were then worked up and analyzed by fluorescent flow cytometry as previously described.
Results
Structure-Function Relationships of Fluorescently Labeled Peptides Derived from Tat49–57.‖
To determine the structural requirements for the cellular uptake of short arginine-rich peptides, a series of fluorescently labeled truncated analogues of Tat49–57 were synthesized by using standard solid-phase chemistry (36). A Fl was attached through an ahx acid spacer on the amino termini. The ability of these fluorescently labeled peptides to enter Jurkat cells was then analyzed by using flow cytometry (Fig. 1). Differentiation between cell surface binding and internalization was accomplished throughout by running a parallel set of assays in the presence and absence of sodium azide. Because sodium azide inhibits energy-dependent cellular uptake (39) but not cell surface binding, the difference in fluorescence between the two assays represents the amount of fluorescence resulting from internalization.
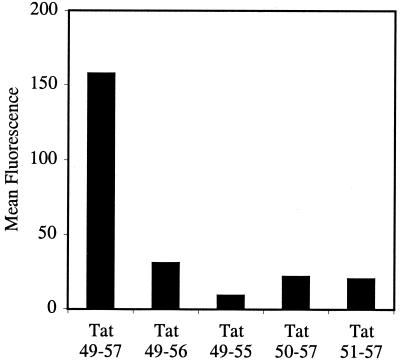
Figure 1.
FACS cellular uptake assay of truncated analogs of Tat49–57 (Fl-ahx-RKKRRQRRR): Tat49–56 (Fl-ahx-RKKRRQRR), Tat49–55 (Fl-ahx-RKKRRQR), Tat50–57 (Fl-ahx-KKRRQRRR), and Tat51–57 (Fl-ahx-KRRQRRR). Jurkat cells were incubated with varying concentrations (12.5 μM shown) of peptides for 10 min at 23°C.
Deletion of one arginine residue from either the amine terminus (Tat50–57) or the carboxyl terminus (Tat49–56) resulted in a significant (80%) loss of intracellular fluorescence relative to the parent sequence (Tat49–57). From the one amino acid truncated analogs, further deletion of R-56 from the carboxyl terminus (Tat49–55) resulted in an additional 60% loss of intracellular fluorescence, whereas deletion of K-50 from the amine terminus (Tat51–57) did not further diminish the amount of internalization. These results indicate that truncated analogs of Tat49–57 are significantly less effective at the transcellular delivery of fluorescein into Jurkat cells, and that the arginine residues appear to contribute more to cellular uptake than the lysine residues.
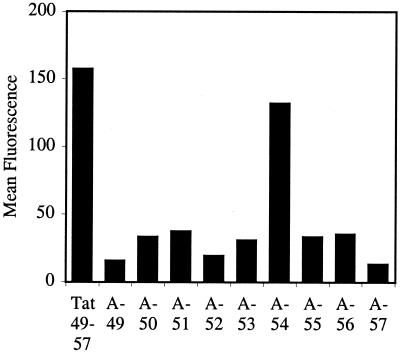
To determine the contribution of individual amino acid residues to cellular uptake, nine fluorescently labeled analogs containing alanine substitutions at each site of Tat49–57 were synthesized and assayed by flow cytometry (Fig. 2). Substitution of the noncharged glutamine residue of Tat49–57 with alanine (A-54) resulted in a modest decrease in cellular internalization. On the other hand, substitution of each of the cationic residues individually with alanine produced a 70–90% decrease in cellular uptake. In these cases, replacement of lysine (A-50, A-51) or arginine (A-49, A-52, A-55, A-56, A-57) residues with alanine had similar effects in reducing uptake.
Figure 2.
FACS cellular uptake assay of alanine-substituted analogs of Tat49–57: A-49 (Fl-ahx-AKKRRQRRR), A-50 (Fl-ahx-RAKRRQRRR), A-51 (Fl-ahx-RKARRQRRR), A-52 (Fl-ahx-RKKARQRRR), A-53 (Fl-ahx-RKKRAQRRR), A-54 (Fl-ahx-RKKRRARRR), A-55 (Fl-ahx-RKKRRQARR), A-56 (Fl-ahx-RKKRRQRAR), and A-57 (Fl-ahx-RKKRRQRRA). Jurkat cells were incubated with varying concentrations (12.5 μM shown) of peptides for 10 min at 23°C.
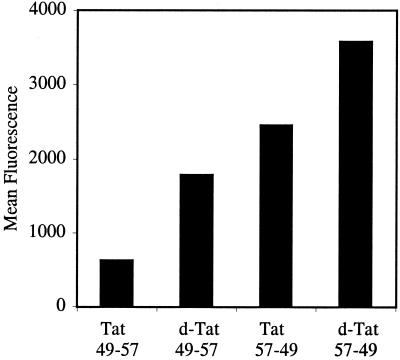
To determine whether the chirality of the transporter peptide was important, the corresponding d-isomer (d-Tat49–57) and retro-inverso isomers (l-Tat57–49 and d-Tat57–49) were synthesized and assayed by flow cytometry (Fig. 3). Importantly, all three analogs were more effective at entering Jurkat cells than Tat49–57. These results indicated that the chirality of the peptide backbone is not crucial for cellular uptake. Interestingly, the retro-l isomer (Tat57–49), which has three arginine residues located at the amine terminus instead of one arginine and two lysines, found in Tat49–57 demonstrated enhanced cellular uptake. Thus, residues at the amine terminus appear to be important and arginines are more effective than lysines for internalization. The improved cellular uptake of the unnatural d-peptides is most likely because of their increased stability to proteolysis in 2% FCS used in the assays. When serum was excluded, the d- and l-peptides were equivalent as expected.
Figure 3.
FACS cellular uptake assay of d- and retro-isomers of Tat49–57: d-Tat49–57 (Fl-ahx-rkkrrqrrr), Tat57–49 (Fl-ahx-RRRQRRKKR), and d-Tat57–49 (Fl-ahx-rrrqrrkkr). Jurkat cells were incubated with varying concentrations (12.5 μM shown) of peptides for 15 min at 23°C.
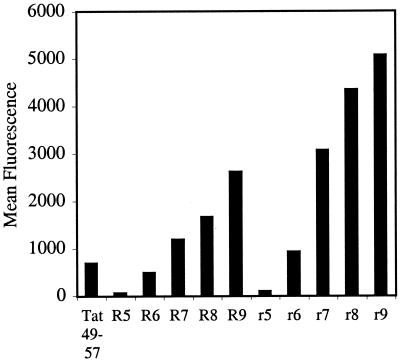
These initial results indicated that arginine content is primarily responsible for the cellular uptake of Tat49–57. Furthermore, these findings are consistent with our previous results showing that short oligomers of arginine are more effective at entering cells than the corresponding short oligomers of lysine, ornithine, and histidine (34). What had not been established was whether arginine homo-oligomers are more effective than Tat49–57. To address this point, Tat49–57 was compared with the l-arginine (R5-R9) and d-arginine (r5-r9) oligomers. Although Tat49–57 contains eight cationic residues, its cellular internalization was between that of R6 and R7 (Fig. 4), indicating that the presence of at least six arginine residues is an important factor for cellular uptake. Significantly, conjugates containing seven to nine arginine residues exhibited better uptake than Tat49–57.
Figure 4.
FACS cellular uptake of a series of arginine oligomers and Tat49–57: R5 (Fl-ahx-RRRRR), R6 (Fl-ahx-RRRRRR), R7 (Fl-ahx-RRRRRRR), R8 (Fl-ahx-RRRRRRRR), R9 (Fl-ahx-RRRRRRRRR), r5 (Fl-ahx-rrrrr), r6 (Fl-ahx-rrrrrr), r7 (Fl-ahx-rrrrrrr), r8 (Fl-ahx-rrrrrrrr), r9 (Fl-ahx-rrrrrrrrr). Jurkat cells were incubated with varying concentrations (12.5 μM shown) of peptides for 15 min at 23°C.
To quantitatively compare the ability of these arginine oligomers and Tat49–57 to enter cells, Michaelis–Menton kinetic analyses were performed. The rates of cellular uptake were determined after incubation (3°C) of the peptides in Jurkat cells for 30, 60, 120, and 240 s (Table 1). The resultant Km values revealed that R9 entered cells approximately 20-fold faster than Tat47–59. Significantly, the r9 transporter was 100-fold faster than Tat47–59 at entering cells. For comparison, Antennapedia43–58 was also analyzed and was shown to enter cells approximately 2-fold faster than Tat47–59, but significantly slower than r9 or R9.
Table 1.
Michaelis–Menton kinetics: Antennapedia43–58 (Fl-ahx-RQIKIWFQNRRMKWKK)
| Peptide | Km, μM | Vmax |
|---|---|---|
| Tat49–57 | 770 | 0.38 |
| Antennapedia43–58 | 427 | 0.41 |
| R9 | 44 | 0.37 |
| r9 | 7.6 | 0.38 |
Design and Synthesis of Peptidomimetic Analogs of Tat49–57.
Using the above structure–function relationships obtained with Tat47–59, we designed a series of polyguanidine peptoid derivatives that preserve the 1,4-backbone spacing of side chains of arginine oligomers but have an oligo-glycine backbone devoid of stereogenic centers. These peptoids incorporating arginine-like sides chains on the amide nitrogen were selected because of their expected resistance to proteolysis (40), and potential ease and more significantly lower cost of synthesis (37, 38). Furthermore, racemization, frequently encountered in peptide synthesis, is not a problem in peptoid synthesis and the “submonomer” (38) approach to peptoid synthesis allows for facile modification of side-chain spacers. Although the preparation of an oligourea (41) and peptoid-peptide hybrid (42) derivatives of Tat49–57 have been previously reported, their cellular uptake was not explicitly studied.
The desired peptoids were prepared by using the “submonomer” (38) approach to peptoids followed by attachment of a Fl through an aminohexanoic acid spacer onto the amine termini.∥ After cleavage from the solid-phase resin, the fluorescently labeled polyamine peptoids thus obtained were converted in good yields (60–70%) into polyguanidine peptoids by treatment with excess pyrazole-1-carboxamidine (43) and sodium carbonate (Scheme S1). Previously reported syntheses of peptoids containing isolated N-Arg units have relied on the synthesis of N-Arg monomers (5–7 steps) before peptoid synthesis and the use of specialized and expensive guanidine protecting groups (Pmc, Pbf) (44, 45). The compounds reported here represent examples of polyguanidinylated peptoids prepared by using a perguanidinylation step. This method provides easy access to polyguanidinylated compounds from the corresponding polyamines and is especially useful for the synthesis of perguanidinylated homooligomers. Furthermore, it eliminates the use of expensive protecting groups (Pbf, Pmc). An additional example of a perguanidinylation of a peptide substrate using a novel triflyl-substituted guanylating agent has recently been reported (46).
Scheme 1.
(a) BrCH2CO2H, DIC, DMF. (b) BocNH-X-NH2, DMF. (c) Fmoc-ahx-CO2H, DIC, DMF. (d) piperidine, DMF. (e) FITC-NCS, DIEA, DMF. (f) 95:5 TFA/TIS. (g) pyrazole-1-carboxamidine, aq. Na2CO3, 50°C. Abbreviations: N-etg, X = (CH2)2; N-arg, X = (CH2)3; N-btg, X = (CH2)4; N-hxg, X = (CH2)6; N-ocg, X = (CH2)8; N-chg, X = trans-1,4-cyclohexyl.
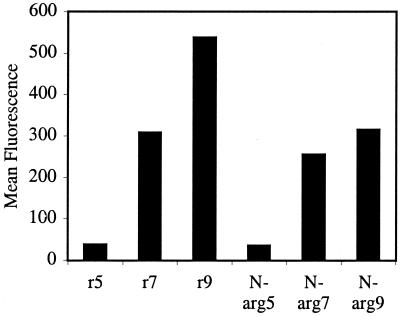
The cellular uptake of fluorescently labeled polyguanidine N-arg5,7,9 peptoids was compared with the corresponding d-arginine peptides r5,7,9 (similar proteolytic properties) by using Jurkat cells and flow cytometry. The amount of fluorescence measured inside the cells with N-arg5,7,9 was found to be proportional to the number of guanidine residues: N-arg9 > N-arg7 > N-arg5 (Fig. 5), analogous to that found for r5,7,9. Importantly, the N-arg5,7,9 peptoids showed only a slightly lower amount of cellular entry compared with the corresponding peptides, r5,7,9. Hence, it is clear from these results that the hydrogen bonding along the peptide backbone of Tat47–59 or arginine oligomers is not a required structural element for cellular uptake and oligomeric guanidine-substituted peptoids can be used in place of arginine-rich peptides as molecular transporters. The addition of sodium azide inhibited internalization demonstrating that the cellular uptake of peptoids was also energy-dependent.
Figure 5.
FACS cellular uptake of polyguanidine peptoids and d-arginine oligomers. Jurkat cells were incubated with varying concentrations (12.5 μM shown) of peptoids and peptides for 4 min at 23°C.
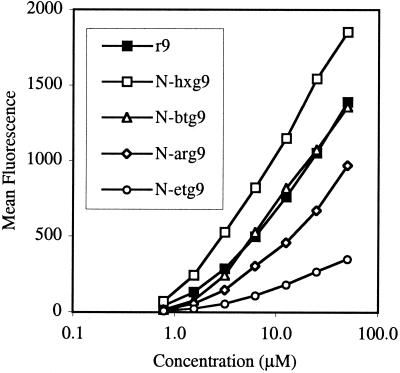
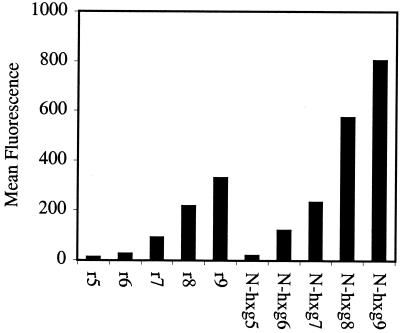
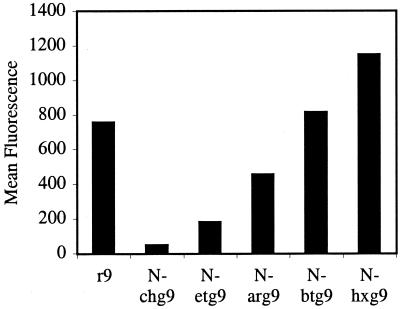
After establishing that the N-arg peptoids efficiently crossed cellular membranes, the effect of side chain length (number of methylenes) on cellular uptake was investigated. Significantly, for a given number of guanidine residues (5, 7, or 9), cellular uptake was proportional to side-chain length. Peptoids with longer side chains exhibited more efficient cellular uptake with N-hxg9 > N-btg9 > r9 > N-arg9 > N-etg9 (Fig. 6). Of special importance, the N-hxg peptoids showed remarkably high cellular uptake, even greater than the corresponding d-arginine oligomers. The cellular uptake of the corresponding heptamers and pentamers also showed the same relative trend. The longer side chains embodied in the N-hxg peptoids improved the cellular uptake to such an extent that the amount of internalization was comparable to the corresponding d-arginine oligomer containing one more guanidine residue (Fig. 7). For example, the N-hxg7 peptoid showed comparable cellular uptake to r8.
Figure 6.
FACS cellular uptake of d-arginine oligomers and polyguanidine peptoids. Jurkat cells were incubated with varying concentrations (12.5 μM shown) of fluorescently labeled peptoids and peptides for 4 min at 23°C.
Figure 7.
FACS cellular uptake of d-arginine oligomers and N-hxg peptoids. Jurkat cells were incubated with varying concentrations (6.3 μM shown) of fluorescently labeled peptoids and peptides for 4 min at 23°C.
To address whether the increase in cellular uptake was because of the hydrophobic nature or the flexibility of the side chains, a set of peptoids was synthesized containing cyclohexyl side chains, N-chg5,7,9 peptoids. These contain the same number of side-chain carbons as the N-hxg peptoids but possess different degrees of freedom. Interestingly, the N-chg peptoid showed much lower cellular uptake activity than all of the previously assayed peptoids, including the N-etg peptoids (Fig. 8). Therefore, the conformational flexibility and sterically unencumbered nature of the straight chain alkyl spacing groups is important for efficient cellular uptake.
Figure 8.
FACS cellular uptake of d-arginine oligomers and N-chg peptoids. Jurkat cells were incubated with varying concentrations (12.5 μM shown) of fluorescently labeled peptoids and peptides for 4 min at 23°C.
Discussion
The nona-peptide, Tat49–57, has been shown to translocate efficiently across plasma membranes (14). The goal of this research was to determine the structural basis for this activity and to use this information to develop simpler and more effective molecular transporters. Toward this end, truncated and alanine substituted derivatives of Tat49–57 conjugated to a fluorescein label were prepared. These derivatives exhibited greatly diminished cellular uptake compared with Tat49–57, indicating that all of the cationic residues of Tat49–57 are required for efficient cellular uptake. When compared with our previous studies on short oligomers of cationic peptides (34), these findings suggested that an oligomer of arginine might be superior to Tat49–57 and certainly more easily and cost-effectively prepared. Comparison of short arginine oligomers with Tat49–57 showed that members of the former were indeed more efficiently taken up into cells. This was quantified further by Michaelis–Menton kinetics analysis that showed that the R9 and r9 oligomers had Km values 20-fold and 100-fold greater than that found for Tat49–57.
Given the importance of the guanidino head group and the apparent insensitivity of the oligomer chirality revealed in our peptide studies, we designed and synthesized a series of polyguanidine peptoids. The peptoids N-arg5,7,9, incorporating the arginine side chain, exhibited comparable cellular uptake to the corresponding d-arginine peptides r5,7,9, indicating that the hydrogen bonding along the peptide backbone and backbone chirality are not essential for cellular uptake. This observation is consistent with molecular models of these peptoids, arginine oligomers, and Tat49–57, all of which have a deeply embedded backbone and a guanidinium dominated surface. Molecular models further reveal that these structural characteristics are retained in varying degree in oligomers with different alkyl spacers between the peptoid backbone and guanidino head groups. Accordingly, a series of peptoids incorporating 2- (N-etg), 4- (N-btg), and 6-atom (N-hxg) spacers between the backbone and side chain were prepared and compared for cellular uptake with the N-arg peptoids (3-atom spacers) and d-arginine oligomers. The length of the side chains had a dramatic effect on cellular entry. The amount of cellular uptake was proportional to the length of the side chain with N-hxg > N-btg > N-arg > N-etg. Cellular uptake was improved when the number of alkyl spacer units between the guanidine head group and the backbone was increased. Significantly, N-hxg9 was superior to r9, the latter being 100-fold better that Tat49–57. This result led us to prepare peptoid derivatives containing longer octyl spacers (N-ocg) between the guanidino groups and the backbone. Issues related to solubility prevented us from testing these compounds.
Because both perguanidinylated peptides and perguanidinylated peptoids efficiently enter cells, the guanidine head group (independent of backbone) is apparently a critical structural determinant of cellular uptake. However, the presence of several (over six) guanidine moieties on a molecular scaffold is not sufficient for active transport into cells as the N-chg peptoids did not efficiently translocate into cells. Thus, in addition to the guanidine head group, the conformational mobility of designed transporters also plays a role in cellular uptake.
In summary, this investigation identified a series of structural characteristics including sequence length, amino acid composition, and chirality that influence the ability of Tat49–57 to enter cells. These characteristics provided the blueprint for the design of a series of peptoids, of which 17 members were synthesized and assayed for cellular uptake. Significantly, the N-hxg9 transporter was found to be superior in cellular uptake to r9 which in turn was comparable to N-btg9. Hence, these peptoid transporters proved to be substantially better then Tat49–57. This research established that the peptide backbone and hydrogen bonding along that backbone are not required for cellular uptake, that the guanidino head group is superior to other cationic subunits, and most significantly, that an extension of the alkyl chain between the backbone and the head group provides superior transporters. In addition to better uptake performance, these peptoids offer several advantages over Tat49–57 including cost-effectiveness, ease of synthesis of analogs, and protease stability. These features along with their significant water solubility (>100 mg/ml) indicate that these peptoids could serve as effective transporters for the molecular delivery of drugs, drug candidates, and other agents into cells.
Acknowledgments
We thank Lee Wright and Chris Van Deusen for helpful discussions relating to the synthesis of the peptoid derivatives. Grants provided by the National Institutes of Health (CA 31841, CA 31845, AI 40968), National Institutes of Health fellowship to E.T.P. (CA 80344), and a Pharmacia graduate fellowship to K.P. are gratefully acknowledged.
Abbreviations
- DMF
dimethylformamide
- ahx
aminohexanoic
- Fl
fluorescein moiety
- m.w.
molecular weight
- PL
polylysine
- TFA
trifluoroacetic acid
- Fmoc
fluorenylmethoxycarbonyl
Footnotes
All synthetic peptides and peptoids contain an aminohexanoic (ahx) acid moiety attached to the N-terminal amino group with a fluorescein moiety (Fl) covalently linked to the amino group of the aminohexanoic acid spacer. The carboxyl terminus of every peptide and peptoid is a carboxamide.
References
- 1.Terwogt J M M, Nuijen B, Huinink W W T B, Beignen J H. Cancer Treat Rev. 1997;23:87–95. doi: 10.1016/s0305-7372(97)90022-0. [DOI] [PubMed] [Google Scholar]
- 2.Rait A, Pirollo K, Will D W, Peyman A, Rait V, Uhlmann E, Chang E H. Bioconjugate Chem. 2000;11:153–160. doi: 10.1021/bc990106n. [DOI] [PubMed] [Google Scholar]
- 3.Rui Y J, Wang S, Low P S, Thompson D H. J Am Chem Soc. 1998;120:11213–11218. [Google Scholar]
- 4.Kono K, Liu M, Fréchet J M J. Bioconjugate Chem. 1999;10:1115–1121. doi: 10.1021/bc990082k. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh A, Ghosh M, Niu C, Malouin F, Moellmann U, Miller M J. Chem Biol. 1996;3:1011–1019. doi: 10.1016/s1074-5521(96)90167-2. [DOI] [PubMed] [Google Scholar]
- 6.Lindgren M, Hällbrink M, Prochiantz A, Langel Ü. Trends Pharmacol Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 7.Jeang K-T, Xiao H, Rich E A. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 8.Green M, Loewenstein P M. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 9.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 10.Mann D A, Frankel A D. EMBO J. 1991;10:1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivés E, Charneau P, Van Rietschoten J, Rochat H, Bahraoui E. J Virol. 1994;68:3343–3353. doi: 10.1128/jvi.68.5.3343-3353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vivés E, Granier C, Prevot P, Leblue B. Lett Pept Sci. 1997;4:429–436. [Google Scholar]
- 13.Pepinsky R B, Androphy E J, Corina K, Brown R, Barsoum J. DNA Cell Biol. 1994;13:1011–1019. doi: 10.1089/dna.1994.13.1011. [DOI] [PubMed] [Google Scholar]
- 14.Vivés E, Brodin P, Lebleu B. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 15.Fawell S, Seery J, Daikh T, Moore C, Chen L L, Pepinsky B, Barsoum J. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson D C, Nichols E, Manger R, Woodle D, Barry M, Fritzberg A R. Biochem Biophys Res Commun. 1993;194:876–884. doi: 10.1006/bbrc.1993.1903. [DOI] [PubMed] [Google Scholar]
- 17.Kim D T, Mitchell D J, Brockstedt D G, Fong L, Nolan G P, Fathman C G, Engelman E G, Rothbard J B. J Immunol. 1997;159:1666–1668. [PubMed] [Google Scholar]
- 18.Nagahara H, Vocero-Akbani A M, Snyder E L, Ho A, Latham D G, Lissy N A, Becker-Hapak M, Ezhevsky S A, Dowdy S F. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 19.Gius D R, Ezhevsky S A, Becker-Hapak M, Nagahara N, Wei M C, Dowdy S F. Cancer Res. 1999;59:2577–2580. [PubMed] [Google Scholar]
- 20.Vocero-Akbani A M, Vander-Heyden N, Lissy N A, Ratner L, Dowdy S F. Nat Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 21.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 22.Derossi D, Chassaing G, Prochiantz A. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 23.Lin Y-Z, Yao S, Veach R A, Torgerson T R, Hawiger J. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 24.Pooga M, Hällbrink M, Zorko M, Langel U. FASEB J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 25.Elliott G, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 26.Ryser H J-P. Nature (London) 1967;215:934–936. doi: 10.1038/215934a0. [DOI] [PubMed] [Google Scholar]
- 27.Emi N, Kidoaki S, Yoshikawa K, Saito H. Biochem Biophys Res Commun. 1997;231:421–424. doi: 10.1006/bbrc.1997.6125. [DOI] [PubMed] [Google Scholar]
- 28.Shen W-C, Ryser H J-P. Proc Natl Acad Sci USA. 1978;75:1872–1876. doi: 10.1073/pnas.75.4.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryser H J-P, Shen W-C. Proc Natl Acad Sci USA. 1978;75:3867–3870. doi: 10.1073/pnas.75.8.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonetti J-P, Degols G, Lebleu B. Bioconjugate Chem. 1990;1:149–153. doi: 10.1021/bc00002a010. [DOI] [PubMed] [Google Scholar]
- 31.Mulders P, Pang S, Dannull J, Kaboo R, Hinkel A, Michel A, Tso C-L, Roth M, Belldegrun A. Cancer Res. 1998;58:956–961. [PubMed] [Google Scholar]
- 32.Murphy J E, Uno T, Hamer J D, Dwarki V, Zuckermann R N. Proc Natl Acad Sci USA. 1998;95:1517–1522. doi: 10.1073/pnas.95.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buschle M, Schmidt W, Zauner W, Mechtler K, Trska B, Kirlappos H, Birnstiel M L. Proc Natl Acad Sci USA. 1997;94:3256–3261. doi: 10.1073/pnas.94.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, D. J., Kim, D. T., Steinman, L. C., Fathman, C. G. & Rothbard, J. B. (2000) J. Peptide Res. 55, in press. [DOI] [PubMed]
- 35.Pons J-F, Fauchere J-L, Lamaty F, Molla A, Lazaro R. Eur. J. Org. Chem. 1998. 853–859. [Google Scholar]
- 36.Atherton E, Sheppard R C. Solid-Phase Peptide Synthesis. Oxford: IRL; 1989. [DOI] [PubMed] [Google Scholar]
- 37.Simon R J, Kania R S, Zuckermann R N, Huebner V D, Jewell D A, Banville S, Ng S, Wang L, Rosenberg S, Marlowe C K, et al. Proc Natl Acad Sci USA. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuckermann R N, Kerr J M, Kent S B H, Moos W H. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 39.Sandvig K, Olsnes S. J Biol Chem. 1982;257:7504–7513. [PubMed] [Google Scholar]
- 40.Miller S M, Simon R J, Ng S, Zuckermann R N, Kerr J M, Moos W H. Bioorg Med Chem Lett. 1994;4:2657–2662. [Google Scholar]
- 41.Tamilarasu N, Huq I, Rana T M. J Am Chem Soc. 1999;121:1597–1598. [Google Scholar]
- 42.Hamy F, Felder E R, Heizmann G, Lazdins J, Aboul-Ela F, Varani G, Karn J, Klimkait T. Proc Natl Acad Sci USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernatowicz M S, Wu Y L, Matsueda G R. J Org Chem. 1992;57:2497–2502. [Google Scholar]
- 44.Kruijtzer J A W, Hofmeyer L J F, Heerma W, Versluis C, Liskamp R M J. Chem Eur J. 1998;4:1570–1580. [Google Scholar]
- 45.Heizmann G, Felder E R. Peptide Res. 1994;7:328–332. [PubMed] [Google Scholar]
- 46.Feichtinger K, Sings H L, Baker T J, Matthews K, Goodman M. J Org Chem. 1998;63:8432–8439. [Google Scholar]