Abstract
Induction of wild-type p53 in the ECV-304 bladder carcinoma cell line by infection with a p53 recombinant adenovirus (Ad5CMV-p53) resulted in extensive apoptosis and eventual death of nearly all of the cells. As a strategy to determine the molecular events important to p53-mediated apoptosis in these transformed cells, ECV-304 cells were selected for resistance to p53 by repeated infections with Ad5CMV-p53. We compared the expression of 5,730 genes in p53-resistant (DECV) and p53-sensitive ECV-304 cells by reverse transcription–PCR, Northern blotting, and DNA microarray analysis. The expression of 480 genes differed by 2-fold or more between the two p53-infected cell lines. A number of potential targets for p53 were identified that play roles in cell cycle regulation, DNA repair, redox control, cell adhesion, apoptosis, and differentiation. Proline oxidase, a mitochondrial enzyme involved in the proline/pyrroline-5-carboxylate redox cycle, was up-regulated by p53 in ECV but not in DECV cells. Pyrroline-5-carboxylate (P5C), a proline-derived metabolite generated by proline oxidase, inhibited the proliferation and survival of ECV-304 and DECV cells and induced apoptosis in both cell lines. A recombinant proline oxidase protein tagged with a green fluorescent protein at the amino terminus localized to mitochondria and induced apoptosis in p53-null H1299 non-small cell lung carcinoma cells. The results directly implicate proline oxidase and the proline/P5C pathway in p53-induced growth suppression and apoptosis.
The p53 tumor suppressor protein plays multiple roles in cell cycle control (1–3), differentiation (4), genomic stability (5, 6), angiogenesis (7, 8), and apoptosis (9–13). After damage to DNA, p53 is induced and activated to generate a late G1 block, thus allowing DNA repair to proceed with fidelity (5, 6, 14–16). Mutations that inactivate the p53 gene product are frequently found in human cancers (17, 18), which has generated a concept that p53 mutations impair cell cycle control, allowing further mutations to accumulate increasing the risk for cancer. The p53 protein functions as a sequence-specific DNA-binding factor and can activate genes whose promoters contain a p53 response element (reviewed in refs. 19 and 20). Genes involved in the regulation of the cell cycle, apoptosis, and DNA repair are transactivated by p53 (19–21).
To identify other genes involved in p53-mediated apoptosis of transformed cells, we selected for ECV-304 bladder carcinoma cells that exhibited resistance to apoptosis induced by p53. Proline oxidase, an enzyme involved in the proline/pyrroline-5-carboxylate (proline/P5C) redox pathway, was identified as a p53-induced gene in ECV-304 tumor cells. Both P5C and a recombinant proline oxidase protein exhibited the ability to inhibit cell growth and to induce apoptosis, suggesting that the proline oxidase pathway may play a role in p53-mediated growth inhibition and apoptosis.
Materials and Methods
Cell Lines and Cell Culture.
Recent investigation by the American Type Culture Collection using DNA profiling has revealed that the ECV-304 cell line is genetically similar to the T24 bladder carcinoma cell line (see http://www.atcc.org). The DECV cell line was generated from ECV-304 by selection for resistance to p53-mediated apoptosis (22). ECV-304 and DECV cells were maintained and propagated in DMEM containing 10% FCS.
Construction of a Recombinant Green Fluorescent Proline Oxidase Expression Vector.
Attempts to clone the complete proline oxidase cDNA proved unsuccessful, potentially because of complex DNA structure at the 5′ portion of the cDNA or the mRNA used to generate cDNA. However, cDNA encoding amino acids 186–516 of proline oxidase/dehydrogenase I was obtained by reverse transcription–PCR (RT-PCR) and cloned in the frame of the pEGFP-C2 (CLONTECH) green fluorescent protein (GFP) fusion expression vector.
Recombinant Adenoviruses.
The construction and generation of Ad5CMV-p53 and Ad5CMV-galactosidase have been described (23). The GFP recombinant adenovirus was constructed by using the pAdTrack vector, which contains GFP, and generating recombinant adenoviruses using the pAdeasy-1 vector as described (24).
Generation of ECV-304 Cells Resistant to p53-Mediated Apoptosis.
ECV-304 cells resistant to p53-mediated apoptosis were generated by repeated infections with Ad5CMV-p53 as described (22). The details of the cell behavior differences between DECVs and the parental ECV-304 cells are reported elsewhere (22).
RNA Preparation and Analysis.
For RT-PCR and Northern analyses, total cellular RNA was extracted with TRI-REAGENT (Molecular Research Center, Cincinnati) according to the suggested manufacturer's protocol. RNA was harvested at 12 h after infection with recombinant p53 adenovirus when signs of apoptosis were minimally visible in the ECV-304 cells. After analysis of quality by formaldehyde agarose gel electrophoresis, the total RNA was then used in RT-PCR assays or extracted further to isolate poly(A)+RNA.
DNA Microarray Analysis.
To isolate poly(A)+RNA to generate fluorescent DNA probes in DNA microarray hybridization, total RNA isolated with TRI-REAGENT was extracted four times with phenol-chloroform, ethanol-precipitated, and resuspended in diethyl pyrocarbonate-treated water to give a final concentration of 0.5 mg/ml. Contaminating DNA was removed by incubating the RNA mixture with DNase I (50 units/0.5 mg RNA) at 37°C for 1 h. The reaction was terminated by adding 100 μl of 0.1 M EDTA (pH 8.0), and the RNA was extracted twice with phenol-chloroform. The RNA was ethanol-precipitated, washed once with 80% ethanol, and resuspended in 2 ml of diethyl pyrocarbonate-treated water. Poly(A)+RNA was isolated from the total RNA, using Oligotex resin in a batchwise protocol (Qiagen, Chatsworth, CA). The poly(A)+RNA from p53-induced ECV-304 and p53-induced DECV cells was used to generate carbocyanine 5- and carbocyanine 3-labeled cDNA, respectively, for competitive hybridization of a DNA microarray grid containing 5,730 sequences of known genes and expressed sequence tags (UniGEM Human V screening services; Genome Systems, St. Louis).
Northern Blotting.
Total cellular RNA was isolated as described above, run on a 1% agarose minigel (4 μg per gel lane) containing 2.2 M formaldehyde, and transferred to a nitrocellulose membrane (Schleicher & Schuell). Membranes were dried at room temperature, and RNA was cross-linked by UV irradiation in a Stratalinker (Stratagene). Hybridization was performed in ExpressHyb hybridization buffer (CLONTECH) containing 1 × 106 cpm/ml of 32P-labeled cDNA probe at 65°C for 24 h. Blots were washed at 65°C for 15 min in 1× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.1% SDS followed by a wash in 0.1× SSC and 0.1% SDS for 15 min.
RT-PCR.
Total RNA was extracted as described above, and cDNA was synthesized according to the Advantage RT-for-PCR procedure (CLONTECH). The primers for PCR were used at an excess concentration of 500 ng in a 50-μl final reaction volume. PCR was performed by using AmpliTaq DNA polymerase under conditions described in the CLONTECH Advantage RT-for-PCR kit (94°C for 45 s; 60°C for 45 s; 72°C for 2 min; 25 cycles; 7 min final extension at 72°C). The PCR were performed in the linear range of the reaction to obtain a semiquantitative result. The products generated in each PCR were cloned into the TA cloning vector system (Invitrogen) and sequenced to confirm the appropriate identity of the cDNAs.
Flow Cytometry [Fluorescence-Activated Cell Sorter (FACS)].
The green fluorescent apoptotic cells expressing GFP or GFP-proline oxidase were quantified as the proportion of cells that contained a DNA content of less than 2 M (subG1 DNA content), as described for analysis of p53-mediated apoptosis (22, 25). H1299 cell monolayers (3–5 × 106 total cells) were trypsinized, washed twice in PBS at 4°C, and fixed in 70% ethanol as described (22).
Cell Proliferation Assays.
Cells (1 × 105) were seeded into wells of a 24-well culture plate and incubated with or without recombinant adenoviruses or amino acids for the times indicated. WST-1 cell proliferation reagent (Roche Molecular Biochemicals) was added directly to the supernatant (10 μl/200 μl growth medium), and the cell cultures were returned to the incubator for 1–2 h. The absorbency of the formazan product was then determined at 460 nm.
Isolation of Mitochondria.
Mitochondria were isolated by centrifugation through a sucrose cushion as described (26). The mitochondrial pellet was washed twice in hypertonic lysis buffer and suspended in 200 μl of the same buffer, and 2 μl was solubilized in SDS gel loading buffer for analysis of GFP-proline oxidase by immunoblotting.
Confocal Microscopy.
H1299 cells were transfected for 24 h with GFP-proline oxidase, typsinized as described above, and reseeded onto glass coverslips precoated with collagen. After 24 h, medium was removed and the cell monolayers on the coverslips were rinsed once with basal DMEM lacking FCS. The coverslips were rinsed once in basal medium and overlaid with basal medium containing 300 nM MitoTracker Red mitochondrial stain (Molecular Probes). The cells on the coverslips were then incubated at 37°C for 30 min, mounted on glass slides, and sealed with fingernail polish, and the excited green GFP-proline oxidase and red MitoTracker fluorescence were viewed under a Zeiss Ultima Z confocal fluorescence microscope.
Results
Generation of a p53-Resistant Derivative of ECV-304 Cells.
ECV-304 cells express low levels of wild-type p53 and, when induced to express high levels of the p53 protein by adenovirus-mediated gene transfer, undergo extensive apoptosis, resulting in the eventual death of almost all of the cells within 36 h (27). We generated a derivative line of ECV-304 cells that is resistant to p53-mediated apoptosis (differentiated ECVs or DECVs) as a strategy to study the relevant molecular events in the apoptosis of these transformed cells. The DECV cells were generated by repeated selection of ECV-304 cells that were resistant to apoptosis induced by the Ad5CMV-p53 adenovirus (22). The parental ECV-304 cells but not DECV cells undergo extensive apoptosis after infection with p53 recombinant adenovirus. The same amount of p53 protein can be induced by adenovirus infection in the DECV and ECV-304 cell lines, but apoptotic death only occurs in a significant amount in the parental ECV-304 cells. Immunofluorescent localization experiments revealed a similar intracellular distribution of p53 in the two cell lines, with a predominance of p53 observed in the nucleus (22).
Molecular Characterization of p53-Induced DECV and ECV-304 Cells.
In an initial attempt to identify the molecular basis for the resistance of DECV cells to p53-mediated apoptosis, the expression of apoptosis-associated genes was examined by semiquantitative RT-PCR in ECV-304 and DECV cells up-regulated for wild-type p53, by infection with Ad5CMV-p53. No differences in the expression of Bcl-2, Bak, Bad, Waf-1, and Bax-α genes were observed between DECV and ECV-304 cells up-regulated for p53 (data not shown). The absence of any significant changes in apoptotic gene expression between DECV and ECV-304 cells prompted us to perform a DNA microarray analysis that included sequences of a total of 5,730 known genes and expressed sequence tag sequences. RNA was harvested at 12 h after infection with recombinant adenovirus, a time when signs of apoptosis are minimally visible in the ECV-304 cells, and used to generated carbocyanine 3- and carbocyanine 5-labeled cDNA for competitive hybridization on a DNA microarray grid. Apoptosis occurs rapidly thereafter in ECV-304 cells, with as many as 30–40% of the cells in apoptosis after 16–20 h of p53 up-regulation. Approximately 8% of the 5,730 genes examined by DNA microarray analysis exhibited 2-fold or greater changes in expression between p53-infected DECV and ECV-304 cells. Table 1 is a partial list of genes that were detected to be differentially expressed between p53-induced ECV-304 and DECV.
Table 1.
Differential gene expression detected by DNA microarray analysis
| Gene | Function | Change in expression* |
|---|---|---|
| Cyclin A1 | Associates with cdk2 protein kinase at S and G2-M phases (39) | 6.5 |
| PISSLRE | CDC2-related protein kinase essential for cell proliferation and acts at G2-M phase (40) | −2.0 |
| HLH 1R21 | Inhibitor of DNA binding (41), member of Id helix–loop–helix family of negative regulators of differentiation | 4.6 |
| Protocadherin-42 | Cell–cell adhesion and communication (42) | −8.0 |
| Apaf-1 | Activates caspase-9 (43) | 2.9 |
| XRCC9 | DNA postreplication repair or cell cycle checkpoint control (44) | −2.1 |
| E2F | Positive regulator for entry into S phase (45) | −3.2 |
| BTG2 | Involved in p53-dependent cell cycle control and the cellular response to DNA damage (46) | 2.4 |
| PIG-3 | p53-induced apoptosis-associated gene (29) | 2.5 |
| PIG-11 | p53-induced apoptosis-associated gene (29) | 3.6 |
A positive number indicates that a gene was expressed at higher levels in ECV-304 cells than in DECV, and a negative value indicates higher expression in DECV.
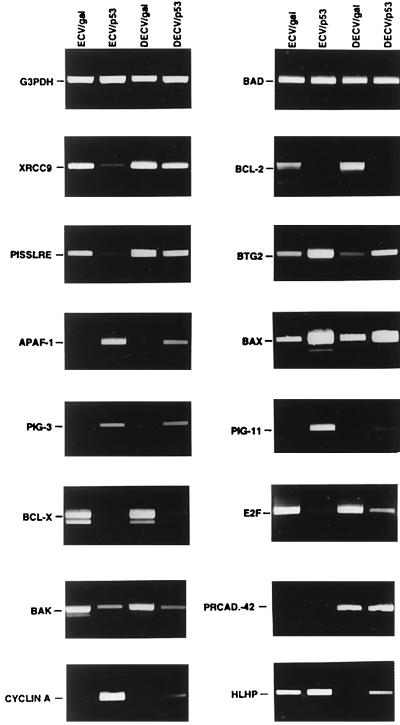
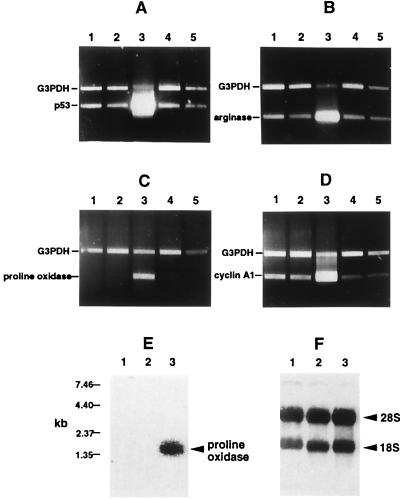
RT-PCR was performed to confirm the differential expression of some of the genes identified by DNA microarray. Analysis of glyceraldehyde 3-phosphate dehydrogenase and Bad transcript levels, which show no changes in expression in p53-infected ECV-304 and DECV cells, were used as controls to ensure that equivalent amounts of cDNA were included in each assay (Fig. 1). In all cases, p53 either repressed or induced differentially the expression of the genes that were analyzed (Fig. 1). Cyclin A1, Pig-11, Baxδ, BTG2, Pig-3, and Apaf-1 were observed to be induced more highly by p53 in ECV-304 cells than in DECV cells, whereas XRCC9, PISSLRE, and E2F were repressed by p53 in ECV-304 cells to greater degrees than in DECV cells. The Bcl-2, Bak, and Bcl-XL anti-apoptotic genes were dramatically repressed by p53 in both ECV-304 and DECV cells. The patterns of expression of these genes are specific, inasmuch as no changes in the expression of glyceraldehyde 3-phosphate dehydrogenase or Bad occurred in response to induction of p53. Northern blotting for expression of cyclin A1 generated results similar to the microarray and RT-PCR data by showing a much greater induction of cyclin A1 by p53 in ECV-304 cells than in DECV cells (data not shown) and thus validated the results of the DNA microarray and RT-PCR analyses. The induction of arginase II, proline oxidase, and cyclin A1 was specific to the up-regulation of p53, inasmuch as no increases in expression of those genes were detected by RT-PCR in control GFP adenovirus-infected cells or cells treated with P5C (Fig. 2 B–D).
Figure 1.
Differential regulation of gene expression by p53 in ECV-304 and DECV cells. ECV-304 and DECV cells were infected with control recombinant β-galactosidase adenovirus (ECV/gal; DECV/gal) or recombinant p53 adenovirus (ECV/p53; DECV/p53) for 12 h. Total RNA was extracted and subjected to RT-PCR to analyze the expression of XRCC9, Bcl-2, PISSLRE, BTG2, Apaf-1, Bax, Pig-3, Pig-11, BclXL, E2F, Bak, protocadherin-42 (PRCAD-42), cyclin A1 (CYCLIN A), and HLH 1R21 (HLHP). Glyceraldehyde 3-phosphate dehydrogenase and Bad, which were not influenced by p53, were used as expression controls to ensure that equivalent amounts of cDNA were included in each assay.
Figure 2.
Induction of proline oxidase and arginase II in ECV-304 cells up-regulated for p53. (A–D) RT-PCR was performed to analyze p53 (A), arginase II (B), proline oxidase (C), and cyclin A1 (D) in mock-infected (lane 1), GFP recombinant adenovirus-infected (lane 2), p53 recombinant adenovirus-infected (lane 3), DMSO-treated (lane 4), and P5C-treated (lane 5) ECV-304 cells. (E) Northern blot analysis was performed on total RNA isolated from mock-infected (lane 1), GFP-infected (lane 2), and p53-infected (lane 3) ECV-304 cells using a radiolabeled proline oxidase cDNA probe and shows induction of proline oxidase mRNA only in p53-induced cells. (F) After capillary transfer from the agarose gel to a nylon membrane, total RNA was stained with methylene blue before hybridization with the proline oxidase probe and shows equivalent amounts of quality total RNA in each gel lane.
Up-Regulation of Enzymes Involved in the Proline/P5C Redox Cycle in p53-Induced Cells.
Two enzymes involved in a redox pathway were observed to be up-regulated more than 5-fold by DNA microarray analysis in p53-induced ECV-304 cells than in p53-induced DECV cells (Table 1). Proline oxidase, a mitochondrial enzyme, is involved in the transfer of redox potential across the mitochondrial membrane through the proline/delta-P5C (proline-P5C) pathway (28). Proline oxidase has also been previously identified as a p53-induced gene (PIG6) in a colorectal cell line (29). Arginase II, a cytoplasmic and mitochondrial enzyme, may be connected to the proline-P5C pathway by converting arginine to ornithine, which can be used by ornithine aminotransferase to produce P5C (30). RT-PCR confirmed that proline oxidase and arginase II were induced specifically in cells up-regulated for p53 but not in mock-infected cells or in cells infected with GFP control adenovirus (Fig. 2 B and C, respectively). The two enzymes were also confirmed by RT-PCR to be differentially expressed in ECV-304 and DECV cells up-regulated for p53 (data not shown). Northern blotting also showed induction of proline oxidase in p53-induced cells but not in GFP-infected or mock-infected cells (Fig. 2E).
Proline oxidase catalyzes the conversion of proline to P5C, which has been reported to function as a metabolic signal regulated by humoral factors to coordinate the metabolism of amino acids and ribonucleotides (28). Both proline and P5C have been shown to be transferred from cell to cell and function as intercellular communicators through the transfer of oxidizing and reducing potential. We thus were interested in determining the effect of P5C on cell growth and whether it was capable of inducing apoptosis. P5C was purchased commercially as a dinitrophenylhydrazone conjugate, which was solubilized in DMSO and added directly to the cell culture media. Dinitrophenylhydrazine by itself produced little or no toxicity in any of the cell types that we investigated, at concentrations as high as 800 μM and for the time duration used in this study (data not shown). P5C that was acid-extracted and purified from the 2,4-dinitrophenylhydrazine (DNP) was found not to be as efficient in entering cells as the dinitrophenylhydrazone conjugate (DNP-P5C) and exhibited much less of an effect on cell growth than the DNP-P5C compound. We believe that the DNP moiety facilitated the entry of P5C into cells.
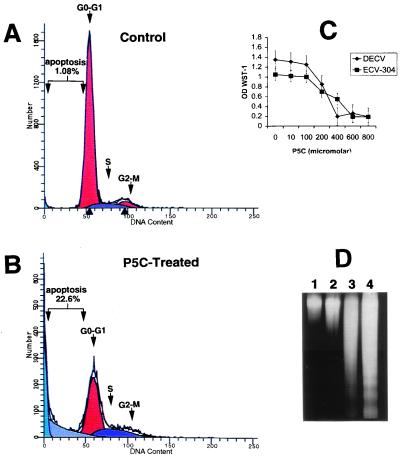
The effect of the DMSO-solubilized DNP-P5C compound on the growth of several transformed and normal cells was investigated. DNP-P5C was observed to inhibit the proliferation and survival of DECV and ECV-304 cells (Fig. 3C). The growth of H358 non-small cell lung carcinoma, human umbilical vein endothelial cells, and normal fibroblast cells was also inhibited by DNP-P5C (data not shown). The inhibitory effect was maximal at doses of 400 μM for most of the different cell types, with human umbilical vein endothelial cells being the most sensitive. Several amino acids showed no effect on cell number at doses similar to and much higher than that of P5C (data not shown), indicating that the inhibition of growth was specific for the P5C metabolite.
Figure 3.
P5C inhibits growth and induces apoptosis in p53-sensitive and p53-resistant cells. (A and B) FACS was performed on ECV-304 cells treated with DMSO (A) and on cells treated with P5C (B) for 16 h. (C) P5C inhibited the proliferation and reduced the survival of DECV and ECV-304 cells. (D) Internucleosomal DNA fragmentation analysis supported the FACS analysis in showing DNA fragmentation ladders characteristic of apoptosis. DMSO-treated ECV-304 and DECV are shown in lanes 1 and 2, and ECV-304 and DECV cells treated with 600 μM P5C for 36 h are shown in lanes 3 and 4, respectively.
Flow cytometry (FACS) was used to determine whether P5C influenced apoptosis in ECV-304 cells. Apoptotic cells were quantified as the proportion of cells that had a DNA content of less than 2 N (sub-G1 DNA content), as previously described for analysis of cells undergoing p53-mediated apoptosis (22, 25). FACS scan analysis and internucleosomal fragmentation assays indicated that 22% of the ECV-304 cell population was undergoing apoptosis after 48 h of treatment with DNP-P5C (Fig. 3B). In contrast, approximately 1% of the ECV-304 cells were undergoing apoptosis with the added DMSO vehicle control (Fig. 3A). Internucleosomal DNA fragmentation analysis confirmed the detection of apoptosis in the FACS analysis by revealing the characteristic DNA ladder in DNP-P5C-treated ECV-304 and DECV cells, along with a considerable amount of a DNA smear (Fig. 3D).
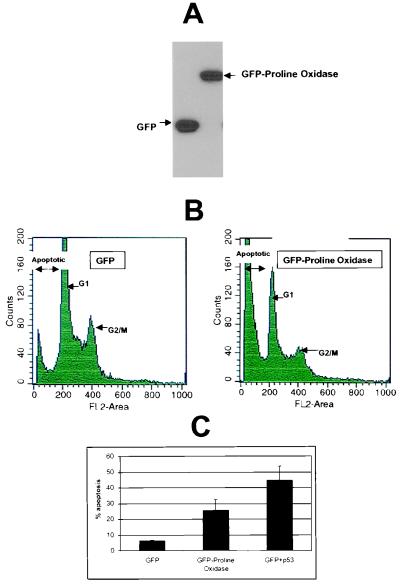
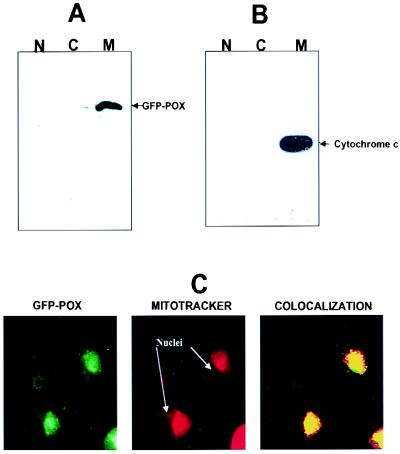
To determine whether proline oxidase could directly mediate apoptosis in the absence of p53, H1299 cells devoid of endogenous p53 were transiently transfected with a recombinant hybrid protein containing GFP fused to amino acids 186–516 of proline oxidase. Expression of GFP and the GFP-proline oxidase proteins is shown in Fig. 4A. H1299 cells transiently transfected with the GFP-proline oxidase expression vector underwent significantly higher rates of apoptosis than cells expressing only GFP (Fig. 4 B and C). Levels of apoptosis in GFP-proline oxidase-expressing cells were not as high as those in cells up-regulated for p53 (Fig. 4C). Subcellular fractionation revealed that GFP-proline oxidase localized predominantly in the mitochondria (Fig. 5A, lane M), with only trace amounts found in the nuclear (Fig. 5A, lane N) and cytoplasmic (Fig. 5A, lane C) compartments. Cytochrome c, a mitochondrial marker, confirmed that mitochondria had been purified, because it was found most predominantly in the mitochondrial fraction (Fig. 5B, lane M). Confocal microscopy confirmed the subcellular fractionation results by showing the colocalization of GFP-proline oxidase with mitochondria, which were concentrated at perinuclear areas in the cytoplasm of H1299 cells (Fig. 5C). MitoTracker Red was used to localize mitochondria, and the colocalization of the excitated green GFP-proline oxidase with MitoTracker Red resulted in a yellow color in the confocal imaging (Fig. 5C, Colocalization). We conclude that a proline oxidase molecule localizes to the mitochondria and is capable of inducing apoptosis in a transformed human cell line.
Figure 4.
A recombinant GFP-proline oxidase induces apoptosis in H1299 non-small cell lung carcinoma cells. (A) H1299 cells were transfected with GFP (lane 1) or GFP-proline oxidase (lane 2) expression vector for 24 h and immunoblotted with a polyclonal GFP antibody. (B) H1299 cells at 40–50% confluence were transfected with GFP or GFP-proline oxidase and subjected to FACS analyses 72 h later. (C) Quantitation of apoptosis occurring in cells transfected with GFP, GFP-proline oxidase fusion protein, or a combination of GFP + p53. The data values shown were compiled from four independent FACS scans.
Figure 5.
Recombinant GFP-proline oxidase localizes to mitochondria. Cells were subfractionated into nuclei (N), cytoplasmic/membrane (C), and mitochondrial (M) fractions. (A and B) A portion of each subcellular fraction (5 μg protein) was solubilized in SDS gel loading buffer and immunoblotted with a polyclonal GFP antibody (A) or a monoclonal cytochrome c antibody (B). (C) Confocal microscopy was used to visualize the localization of GFP-proline oxidase in transfected H1299 cells (GFP-POX; green fluorescence). These same cells were counterstained with MitoTracker Red (red fluorescence) to visualize the localized concentration of mitochondria in the cytoplasm. The right panel shows the simultaneous excitation of GFP-proline oxidase and MitoTracker Red in the same field as that shown in the left and middle panels, which yielded a yellowish-red fluorescence at areas of colocalization of GFP-POX and MitoTracker Red. Filtered control analyses indicated that there was no contamination of the red fluorescence window by the green fluorescence signal or of the green fluorescence window by the red fluorescence signal.
Discussion
We have shown that induction of wild-type p53 by adenovirus-mediated gene transfer leads to rapid and extensive apoptosis in ECV-304 cells (27). To identify molecular events relevant to p53-mediated apoptosis in ECV-304 cells, we established a variant of ECV-304 (DECV) that is resistant to p53-mediated apoptosis. The DECV cells appear to represent a more differentiated, less tumorigenic variant of ECV-304 cells, based on the formation of lattice- and cyst-like structures in culture, the lower efficiency of growth in soft agar, and the demonstration of contact inhibition (22).
Serial analysis of gene expression in a p53-induced colorectal cell line identified several p53-regulated genes involved in control over the redox status of cells (29). Two of those redox regulators, PIG3 (quinone oxidoreductase homolog) and PIG6 (proline oxidase), were also up-regulated by p53 in the sensitive ECV-304 but not in the resistant DECV cells. We identified several other redox regulators that were differentially expressed in p53-sensitive and p53-resistant endothelial cells. These included arginase II, γ-glutamyl transferase-2, and NADH ubiquinone dehydrogenase Fe-S protein-5. The differential induction of proline oxidase by p53 between ECV-304 and DECV cells and the ability of a recombinant proline oxidase protein to induce apoptosis are observations that are of particular interest, because this enzyme is involved in the transfer of reducing equivalents between the cytoplasm and mitochondria. Proline oxidase catalyzes the conversion of proline to P5C and transfers electrons into the mitochondrial electron transport with an intervening flavoprotein (31). These interconversions have been postulated to shuttle redox between cellular compartments, transfer electrons from NADPH to NAD+, couple the oxidation of NADPH to mitochondrial electron transport, and serve as a mechanism for energy production (28). The conversion of P5C to proline ultimately leads to stimulation of the oxidation of glucose through the pentose phosphate pathway, resulting in the production of ribose 5-phosphate. Increases in ribose 5-phosphate lead to increases in phosphoribosylpyrophosphate, a critical intermediate for nucleotide synthesis (28). An increase in the concentrations of nucleotide pools is thus implicated as one outcome of the induction of the proline/P5C pathway by p53, and this increase might influence gene expression and activities of caspases. For instance, nucleotide pools have been shown to be important in influencing gene expression in HL60 promyelocytic leukemia cells (32), and a p53-inducible ribonucleotide reductase is required to mediate efficient repair of damaged DNA (21). Moreover, cellular nucleotide pools have been implicated in a reversible p53-dependent G0/G1 cell cycle arrest (33) and in the role of dATP in the activation of caspase-induced apoptosis (34). Finally, ATP appears to be required for downstream events in apoptosis (35).
Mitochondria participate in apoptosis by activating caspases through the release of cytochrome c into the cytosol, where it binds to Apaf-1 and activates caspase pathways (34, 36). It is thus intriguing that ferricytochrome c is a required cofactor in the conversion of proline to P5C by proline oxidase in vitro (37). Moreover, caspases are cysteine-dependent enzymes and appear to be redox sensitive (38). A truncated recombinant proline oxidase protein was still capable of localizing to mitochondria and inducing apoptosis in H1299 cells. The shuttling of redox potential from the mitochondria to the cytoplasm through the proline/P5C pathway might influence caspase activity. However, we acknowledge that proline oxidase may mediate apoptosis through pathways other than the P5C metabolite.
The differential sensitivity of ECV-304 cells and DECV cells to p53 provides a model system for identifying molecular events important in the p53-mediated apoptosis of tumor cells. Using this system, we have identified several potential targets of p53 that may play roles in growth suppression or apoptosis or both. All but one (protocadherin-42) of the genes identified to be differentially expressed between p53-induced ECV-304 and DECV cells were found to be regulated by p53, indicating that this system will allow for the identification of other genes regulated by p53. The complexity of differential gene expression regulated by p53 identified by DNA microarray in our system indicates that a diverse array of genes may potentially play synergistic or additive roles in p53-mediated apoptosis in vitro and in vivo.
Acknowledgments
This work was supported by a grant (1-RO1-HL59373) from the National Heart, Lung, and Blood Institute.
Abbreviations
- RT-PCR
reverse transcription–PCR
- P5C
pyrroline-5-carboxylate
- DECV
differentiated ECV, a derivative line of ECV-304 cells that is resistant to p53-mediated apoptosis
- GFP
green fluorescent protein
- FACS
fluorescence-activated cell sorter
- DNP
2,4-dinitrophenylhydrazine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230445997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230445997
References
- 1.Martinez J, Georgoff I, Martinez J, Levine A J. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 2.Diller I, Kassel J, Nelson C E, Gryka M A, Litwak G, Gebhart M, Bressac B, Ozturk M, Baker S J, Vogelstein B, et al. Mol Cell Biol. 1990;11:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin D, Shields M T, Ullrich S J, Apella E, Mercer W E. Proc Natl Acad Sci USA. 1992;89:9210–9214. doi: 10.1073/pnas.89.19.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonigrinstein R, Zanbar I, Alboum I, Goldfinger N, Rotter V. Oncogene. 1993;8:3297–3305. [PubMed] [Google Scholar]
- 5.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 6.Livingstone S R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 7.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 8.Stromblad S, Becker J C, Yebra M, Brooks P C, Cheresh D A. J Clin Invest. 1996;98:426–433. doi: 10.1172/JCI118808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yonish-Rouach E, Grunwald D, Wilder S, Kimchi A, May E, Lawrence J J, May P, Oren M. Mol Cell Biol. 1993;13:1415–1423. doi: 10.1128/mcb.13.3.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debbas M, White E. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 11.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hoope M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 12.Gottlieb E, Haffner R, Von Ruden T, Wagner E F, Oren M. EMBO J. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt A J, Potten C S, Kemp C J, Hickman J A, Balmain A, Lane D P, Hall P A. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- 14.Lane D P. Nature (London) 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 15.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 16.Zhan Z, Carrier F, Fornace A J. Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollstein M, Sidransky D, Vogelstein B, Harris C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 18.Bennett W P, Hussain S P, Vahakangas K H, Khan M A, Shields P G, Harris C C. J Pathol. 1999;187:8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 20.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukada S, Matsul K, Takel Y, Nakamura Y. Nature (London) 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell S A, Davis G E. Apoptosis. 2000;5:277–288. doi: 10.1023/a:1009660714216. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W-W, Fang X, Branch C D, Mazur W, French B A, Roth J A. BioTechniques. 1993;15:868–872. [PubMed] [Google Scholar]
- 24.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unger T, Sionov R V, Moallem E, Yee C L, Howley P M, Oren M, Haupt Y. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 26.Moreno M, Puigserver P, Llull J, Gianotti M, Lanni A, Goglia F, Palou A. Biochem J. 1994;300:463–468. doi: 10.1042/bj3000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell S A, Acosta S A, Tombusch K, Davis G E. Apoptosis. 1997;2:442–454. doi: 10.1023/a:1026418010549. [DOI] [PubMed] [Google Scholar]
- 28.Phang J M. Curr Top Cell Regul. 1985;25:91–132. doi: 10.1016/b978-0-12-152825-6.50008-4. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 30.Basch J J, Wickham E D, Farell H M., Jr J Dairy Sci. 1997;80:3241–3248. doi: 10.3168/jds.S0022-0302(97)76298-2. [DOI] [PubMed] [Google Scholar]
- 31.Meyer J. Arch Biochem Biophys. 1977;178:387–395. doi: 10.1016/0003-9861(77)90208-9. [DOI] [PubMed] [Google Scholar]
- 32.Lucas D L, Webster H K, Wright D G. J Clin Invest. 1983;72:1889–1900. doi: 10.1172/JCI111152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linke S P, Clarkin K C, DiLeonardo A, Tsou A, Wahl G M. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Want X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 35.Eguchi Y, Shimizu S, Tsujimoto Y. Cancer Res. 1997;57:1835–1840. [PubMed] [Google Scholar]
- 36.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 37.Herzfeld A, Mezl V A, Knox W E. Biochem J. 1977;166:95–103. doi: 10.1042/bj1660095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampton M B, Fadeel B, Orrenius S. Ann NY Acad Sci. 1998;854:328–335. doi: 10.1111/j.1749-6632.1998.tb09913.x. [DOI] [PubMed] [Google Scholar]
- 39.Yang R, Muller C, Huynh V, Fung Y K, Yee A S, Koeffler H P. Mol Cell Biol. 1999;19:2400–2407. doi: 10.1128/mcb.19.3.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, MacLachlan T K, De Luca A, Claudio P P, Condorelli G, Giordano A. Cancer Res. 1995;55:3992–3995. [PubMed] [Google Scholar]
- 41.Deed R W, Bianchi S M, Atherton G T, Johnston D, Santibanez-Koref M, Murphy J J, Norton J D. Oncogene. 1993;8:599–607. [PubMed] [Google Scholar]
- 42.Obata S, Sago H, Mori N, Rochelle J M, Seldin M F, Davidson M, St. John T, Taketani S, Suzuki S T. J Cell Sci. 1995;108:3765–3773. doi: 10.1242/jcs.108.12.3765. [DOI] [PubMed] [Google Scholar]
- 43.Slee E A, Harte M T, Kluck R M, Wolf B B, Casiano C A, Newmeyer D D, Wang H G, Reed J C, Nicholson D W, Alnemri E S, et al. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Winter J P, Waisfisz Q, Rooimans M A, van Berkel C G, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, et al. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 45.Rieber M, Strasberg-Rieber M. Int J Cancer. 1998;76:757–760. doi: 10.1002/(sici)1097-0215(19980529)76:5<757::aid-ijc22>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 46.Rouault J P, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, et al. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]