Abstract
Laser induced fluorescence in submicrometer fluidic channels was used to characterize the synthesis of polymerase chain reaction (PCR) products from a model bacterial system in order to explore the advantages and limitations of on chip real time single molecule PCR analysis. Single oligonucleotide universal bacterial primers and PCR amplicons from the 16S rDNA of Thermobifida fusca (325 bp) were directly detected at all phases of the reaction with low sample consumption and without post-amplification purification or size screening. Primers were fluorescently labeled with single Alexa Fluor 488 or Alexa Fluor 594 fluorophores, resulting in double labeled, two color amplicons. PCR products were driven electrokinetically through a fused silica channel with a 250 nm by 500 nm rectangular cross section. Lasers with 488 nm and 568 nm wavelengths were focused and overlapped on the channel for fluorescence excitation. All molecules entering the channel were rapidly and uniformly analyzed. Photon burst analysis was used to detect and identify individual primers and amplicons, and fluorescence correlation and cross-correlation spectroscopy were used to account for analyte flow speed. Conventional gel and capillary electrophoresis were also used to characterize the PCR amplification, and the results of differences in detection sensitivity and analyte discrimination were examined. Limits were imposed by the purity and labeling efficiency of the PCR reagents, which must be improved in parallel with increases in detection sensitivity.
INTRODUCTION
The manipulation, detection and analysis of nucleic acids have become essential tasks in many biological applications ranging from genomic sequencing to microbial ecology. The analytical demands of these applications have stimulated the recent development of microfluidic and nanofluidic devices to increase detection sensitivity and analytical speed. In the last decade, a variety of these structures have been developed for biomolecular analysis, including solid state nanopores,1, 2, 3 nanometric slits,4 entropic traps,5, 6 zero mode waveguides,7, 8 and asymmetric diffusion arrays.9 Submicrometer and nanofluidic channels have also played a significant role in this field, having demonstrated their utility for a variety of DNA analyses,10, 11, 12, 13, 14, 15 single fluorophore detection16 and the multicolor detection of novel fluorescent labels for the enhanced analysis of single biomolecules.17, 18, 19 The presented work explores the use of submicrometer fluidic channels in conjunction with single molecule fluorescence spectroscopy to improve upon conventional methods of detecting and analyzing polymerase chain reaction (PCR) amplification products, and compares the results obtained from the various methods.
PCR is central to many methods of biomolecular analysis, as it enables the copying of an in vitro target gene to the extent that it can be detected and analyzed. Briefly, this process consists of the iterative 5′-3′ elongation of a single stranded DNA sequence and its compliment from double stranded regions resulting from the hybridization of the oligonucleotides. These PCR primers create an initiation site for the processive enzyme, and at each cycle an equal number of the two primers are incorporated into the newly synthesized copies. The number of synthesized copies at a given cycle can be expressed as Nn=N0⋅(1+E)n, where N0 is the initial quantity of the target sequence, E is the efficiency, which varies from one to zero over the course of the reaction, and n is the number of cycles. PCR kinetics can typically be divided into three phases as a function of the number of denaturation-hybridization-elongation cycles. In the first reaction phase, at low cycle numbers, DNA is amplified exponentially but there are not enough copies to be detected using widely accessible conventional methods. In the middle phase, between about 15 and 20 cycles, amplicon quantification becomes possible while the reaction is still exponential.20 During the final phase, the reaction reaches a plateau due to the low concentration of free remaining primers, but the final quantity of amplicons is no longer related to the initial quantity of the target gene as the reaction efficiency tends toward zero. This has motivated recent developments, including real time and quantitative PCR, to reduce the threshold level of amplicon detection to the middle phase of the reaction, allowing quantification when the number of amplicons is still related to the initial number of copies.21 These technologies have benefited from laser induced fluorescence and the increased signal-to-noise-ratios of quenched fluorophore oligonucleotides, such as molecular beacons or scorpion probes. The general trend in PCR quantification consists of decreasing DNA amplification and increasing DNA detection sensitivity. In the work presented, this trend is extended to the single molecule regime by the use of submicrometer fluidic channels to rapidly, directly and uniformly detect single PCR amplicons at all reaction phases, with low sample consumption and without post-amplification purification or size screening. This higher sensitivity and single molecule limit of detection is an important step toward the quantification of PCR products after a minimal number of amplification cycles, eliminating the hindering effects of varying reaction efficiency, and improving the speed and accuracy of measurements. As PCR and other biochemical reactions are implemented on chip more frequently, submicrometer fluidic channels become an increasingly attractive architecture for rapid, sensitive and economical biomolecular analysis.
The primary feature of the submicrometer fluidic channel is its subfemtoliter detection (or focal) volume, which isolates single molecules and reduces the amount of extraneous fluorescent material observed, such as buffer solution and associated impurities.16 This increases the signal to noise ratio of single molecule detection, which is an important consideration when target nucleic acids are labeled with only a single fluorophore, and increases the solution concentrations at which single molecule detection can be performed. While the channels used in this paper were fabricated in a fused silica substrate, suspended glass nanofluidic channels have also been utilized for single molecule detection, and have the potential for reduced background fluorescence.22 Because the submicrometer channel dimensions are on the order of, or less than, the wavelength of light used for fluorescence excitation, the focal volume can be defined as a region of approximately constant intensity in some region of the structure.18 This results in the uniform excitation and analysis of molecules entering the device, significantly improving the ability to analyze and quantify fluorescence emission. The channel architecture also enables precise control over fluid flow and particle motion. When compared to diffusion-limited single molecule spectroscopy, this results in more uniform analysis times, higher throughput and the detection of each molecule once and only once,10, 16 improving single molecule counting statistics. Moreover, when compared to conventional methods of genomic analysis, submicrometer fluidic channels consume less reagent,10 and because of their size and structure, are amenable to parallel on-chip integration for high throughput, multiplexed genomic analysis.
In the presented work, submicrometer fluidic channels were used to rapidly isolate individual PCR products from a model bacterial system, Thermobifida fusca (T. fusca), for enhanced detection with a confocal microscope. T. fusca is an extremophile bacteria of particular interest for its bioindustrial potential, as it produces stable and high activity cellulase enzymes for the conversion of cellulose into fermentable sugars. This system was used here to examine a variety of issues involved with single molecule PCR analysis, while at the industrial scale, new technologies are needed for the rapid and accurate analysis of biomolecular processes involving T. fusca for enzyme production and conversion processes. Two universal bacterial primers, labeled with either single Alexa Fluor 488 or Alexa Fluor 594 fluorophores, were used to amplify a fragment of the 16S rDNA (regions V6, V7 and V8) from T. fusca, resulting in double labeled PCR amplicons. PCR primers were identified by the detection of photon bursts of a single color, green or red, while amplicons were identified by the simultaneous detection of green and red photon bursts. Individual primers and amplicons were directly detected at all phases of the reaction, as opposed to an indirect determination of concentration via size screening and macroscopic intensity measurements, in order to characterize the PCR amplification. Fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS) were also used to determine the electrokinetic flow speed of the analytes. FCS is a technique for analyzing fluorescence fluctuations due to concentration changes in a microscopic detection volume. It was first implemented in the 1970s to study diffusion-driven concentration changes23, 24, 25 and later flowing systems.26 It has since been widely used to study a variety of biological systems, and in the last several years has been used in conjunction with nanofabricated structures for single molecule analysis.8, 16, 17, 18 In the late 1990s, dual-color FCCS was established as a viable experimental method,27 and was later used to quantify a PCR reaction in free solution.28 In order to determine analyte flow speed using FCS and FCCS, the fluorescence intensity of the two detection channels were both autocorrelated and cross-correlated. The resulting correlation functions were fit to analytical models to extract information pertaining to the flow speeds of the various analytes.
Gel and capillary gel electrophoresis were also used to analyze the PCR amplification ofT. fusca by indirectly determining PCR product concentration via size screening and macroscopic intensity measurements. These conventional methods were able to detect PCR amplicons after 15 to 20 reaction cycles and reproduced the trend observed with single molecule measurements, but with different amplitudes due to differences in the mechanism of analyte discrimination and detection sensitivity. A variety of issues related to biomolecular reagents were explored in order to fully characterize and compare these results. Limits were found to be imposed by the purity and labeling efficiency of the PCR constituents, which must be improved in parallel with detection sensitivity.
MATERIALS AND METHODS
Samples
Genomic DNA was extracted using a Master Pure™ Complete DNA Purification Kit from EPICENTRE Biotechnologies (Madison, WI) from a 24 h culture of T. fusca. The optical density of DNA at 260 nm was adjusted to 1 OD unit. A universal bacterial primer set, CoSe950f (5′-AACAGGATTAGATACCCTGGTAG-3′) 5′-labeled with Alexa Fluor 488 (495∕519) and CoSe1300rv (5′-CTCGTTGCGGGACTTAACC-3′) 5′-labeled with Alexa Fluor 594 (590∕617), was used to amplify a 325 bp fragment of the 16S rDNA. PAGE-purified oligonucleotides were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). The PCR mixture consisted of 50 mM buffer, 2 mM MgCl2, 2 mM dNTP, and 1.25 U taq polymerase (MasterAmp, Epicenter). Both primers were added at a final concentration of 0.2 μM. Genomic DNA (1 μl per reaction, approximately 50 ng of DNA) was added and 50 μl reactions were performed with an Icycler from Bio-Rad (Hercules, CA) using the following amplification program: 94 °C for 5 min for initial denaturation, then 94 °C for 30 s, 55 °C for 20 s, 72 °C for 20 s, and 72 °C for 5 min for the final extension. Tubes were sequentially removed and chilled on ice after 0, 4, 8, 12, 14, 16, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, and 40 cycles. PCR samples were diluted to approximately 100 pM in 5X TBE with 1% (wt∕vol) poly(vinylpyrrolidone) added to suppress electroosmotic flow29 and surface interactions for analysis in submicrometer fluidic channels.
Gel electrophoresis and capillary electrophoresis
PCR products and primers were quantified under UV light after electrophoresis in 2.5% (wt∕vol) agarose gel. Gels were imaged with a Syngene G:box HR gel documentation (Cambridge, U.K.) using either a short wavelength emission filter for Alexa Fluor 488 or a long wavelength emission filter for Alexa Fluor 594. Gels were then stained with Sybr Gold (Invitrogen) to verify the length of the PCR products by comparison with a 100 bp ladder. The intensities of each band, PCR products and remaining primers, were measured using Syngene GeneTools software for each fluorophore. Gel analyses were performed in triplicate using 5 μl of PCR sample per lane.
The quantification of PCR products and remaining primers by capillary electrophoresis was performed on a Spectrumedix Reveal™ Genetic Analysis System (State College, PA). The lasers and camera setup were optimized to detect Alexa Fluor 488. PCR samples were diluted (1∕100) in Spectrumedix 0.5X Reveal Dilution Buffer™. ILS600 ladder (0.5% vol∕vol) was loaded every 5 capillaries. Samples were injected for 30 s at 10 keV and electrophoresed in the Spectrumedix Reveal High Resolution Matrix™ for 4800 s, at 30 °C and 10 keV. Data collection started 5 min after injection. Electropherograms were analyzed with Spectrumedix Revelation™ software and the surface area of the primer and PCR product peaks. Each sample was injected 3 consecutive times, and the surface areas of primer and PCR product peaks were respectively integrated and summed for the three injections. PCR samples were analyzed in triplicate.
Submicrometer fluidic channels
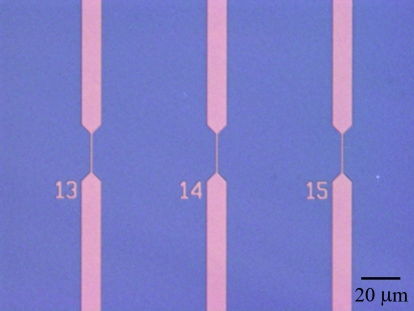
Fluidic channels with submicrometer dimensions were fabricated in fused silica. Positive channel and reservoir features were defined in a photoresist etch mask on a fused silica substrate. Several millimeters of microfluidic channel were used to connect each section of 500 nm wide channel to the sample reservoirs. A reactive ion etch was used to define a device depth of 250 nm. Inlet and outlet holes were powder-blasted through the substrate, which was cleaned using Piranha acid and RCA base baths and bonded and annealed to a 170 μm fused silica cover wafer. An optical micrograph of a parallel array of fluidic channels is shown in Fig. 1. A 100 V bias was applied across the two sample reservoirs using platinum electrodes, resulting in an electric field of approximately 1 kV∕cm along the length of submicrometer channel used for single molecule detection.
Figure 1.
Optical micrograph of a parallel array of submicrometer fluidic channels in fused silica. Single molecule detection was performed in the center region of the channel, with a width of 500 nm. The channel array was etched to a depth of 250 nm.
Optical setup
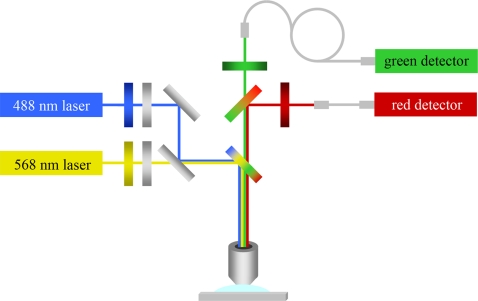
A confocal microscope was used in conjunction with the submicrometer fluidic channels for single molecule detection. A schematic of the optical setup is shown in Fig. 2. Unexpanded 488 nm and 568 nm laser beams with powers of 200 μW and 1000 μW, respectively, were overlapped with a beam splitter, directed toward a dual band dichroic mirror, and used to underfill a high numerical aperture objective. This resulted in focused but not diffraction-limited laser spots that illuminated the fluidic channel in an approximately uniform manner across its width and through its depth.18 A schematic of the submicrometer fluidic channel and excitation profile is shown in Fig. 3. The 250 nm depth and 500 nm width of the focal volume were defined by the dimensions of the fluidic channel, while the focal volume length was a function of the microscope objective and 50 μm optical fibers used as confocal pinhole apertures. The two laser spots were aligned to each other and the channel using a CCD camera. Fluorescence emission was collected with the objective, passed back through the dual band dichroic mirror and split into two color channels using a third dichroic mirror. The two color channel signals were band pass filtered and focused onto optical fibers connected to avalanche photodiodes for detection. Further details of the optical setup can be found elsewhere.18
Figure 2.
Schematic of the optical setup used for single molecule spectroscopy. 488 nm and 568 nm lasers were overlapped and focused on a submicrometer fluidic channel using a high numerical aperture microscope objective. Multicolor fluorescence emission was collected, split, filtered and focused on 50 mm optical fibers. Avalanche photodiodes were used for signal transduction and amplification.
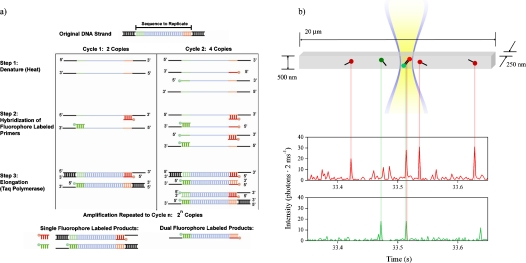
Figure 3.
(a) Schematic of PCR amplification process. Target DNA molecules were denatured by heating, and primers labeled with Alexa Fluor 488 and Alexa Fluor 594 were hybridized to the target strands. Taq polymerase was used for elongation. After several cycles, double labeled two color products were the majority product. (b) Schematic of the submicrometer fluidic channel, laser excitation profile and PCR primer and amplicon detection. The channel was illuminated in an approximately uniform manner through its width and depth, resulting in a focal volume of 180 aL. As single labeled primers and double labeled amplicons were driven electrophoretically through the focal volume, bursts of fluorescence were detected in the two color channels. Primers were detected as single color red or green bursts. Amplicons were detected as simultaneous bursts of fluorescence in both color channels.
RESULTS AND DISCUSSION
A combination of fluorescence correlation spectroscopy and photon burst analysis was used to characterize the PCR amplification kinetics of the genomic rDNA of T. fusca. FCS and FCCS were used to determine and account for the average electrophoretic flow speeds of the primers and products, and single photon bursts were detected and evaluated to identify individual PCR analytes.
Concentration
PCR samples were diluted to 100 pM to ensure with a high degree of confidence that the simultaneous detection of bursts of fluorescence in both color channels was the result of the detection of individual double labeled PCR amplicons. Based on Poisson statistics, the probability of red and green PCR primers randomly occupying the focal volume simultaneously at this concentration was reduced to 3×10−5. Because the presented method achieves single molecule detection with high sensitivity,17 PCR samples could be further diluted to lower concentrations at the cost of increased analysis time and decreased statistical validity. The concentration limit of detection with this method (and single molecule methods in general) is therefore a function of analysis time and statistical validity, and the relative importance of these various parameters must be balanced for a given application. Accordingly, PCR samples could also be analyzed at higher concentrations for increased throughput, but with an increased probability of false amplicon detection. 100 pM was selected as a reasonable compromise between rapid analysis and high statistical confidence.
FCS and FCCS
FCS and FCCS are methods for analyzing temporal fluctuations in fluorescence due to concentration changes in a microscopic detection volume. The normalized correlation function is generally defined as:
| (1) |
In this equation, G is the correlation function, δF is a fluctuation in fluorescence intensity, and i and j represent the green and red detection channels, respectively. The autocorrelation function for either detection channel results from i=j, while setting i≠j gives the cross-correlation function.
Fluctuations in fluorescence intensity resulted from red and green molecules entering and leaving the focal volume. Autocorrelating the fluorescence intensity in one of the detection channels provided time averaged information on the duration of fluorescence fluctuations of that color, as well as the concentration of all molecules of that color. Cross-correlating the fluorescence intensity in the two detection channels gave similar information for simultaneous fluctuations in fluorescence in both detection channels. Spectral cross-talk from the red to green detection channels was not observed, and cross-talk from the green to red detection channels was found to be negligible, and subsequently disregarded. A full treatment can be found elsewhere.27 Neglecting spectral cross-talk and differences in focal volume size, the cross-correlation function at τ=0 can be expressed as:
| (2) |
In this equation, NGR is the number of molecules per unit focal volume that are simultaneously green and red, e.g., double labeled PCR amplicons. NG and NR are the numbers of single labeled green and red molecules per unit focal volume, respectively. Because the autocorrelation functions include information on all fluorescent fluctuations of one color, including both single color molecules and amplicons, the denominator terms are equal to the zero-intercepts of the measured autocorrelation functions. In order to extract physical parameters of interest, the auto- and cross-correlation functions were fit to an analytical model accounting for decay of fluorescence fluctuations due to one dimensional diffusion24 and flow26:
| (3) |
In this equation, G0 is the intercept of the correlation function at τ=0, τd is the characteristic decay time due to diffusion and τf is the characteristic decay time due to electrophoretic flow. Using the value of τf and the size of the focal volumes, the electrophoretic speed of single labeled molecules and double labeled amplicons was determined. The results are shown in Table 1.
Table 1.
Characteristic flow time and molecular speed.
| Green molecules | Red molecules | Two color amplicons | |
|---|---|---|---|
| τf(μs) | 324±12 μs | 259±17 μs | 353±9 μs |
| Speed (mm∕s) | 1.29±0.05 mm∕s | 1.61±0.10 mm∕s | 1.18±0.03 mm∕s |
Because single color molecules had higher electrophoretic mobilities than the double labeled amplicons, a proportionally higher number of single color molecules than amplicons were detected. This introduces a multiplicative correction factor
equal to the ratio of the single color analyte to amplicon flow speeds, which is needed to determine the actual percent of primers reacted.
Photon burst analysis and single molecule detection
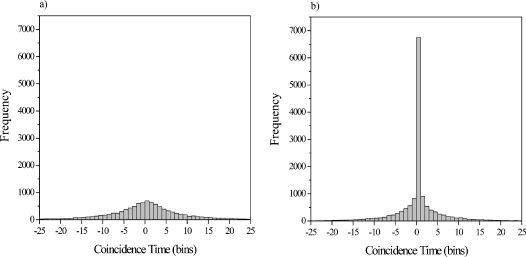
Based on analysis of the autocorrelation and cross-correlation function decay rates, a 2 ms bin time was selected to confine each detection event to a single bin. A burst height histogram was then generated for both color channels and the background noise peaks were fit to Poisson distributions. Single molecule detection thresholds of four standard deviations above the mean background noise were defined to exclude background noise from inadvertent analysis. Photon bursts meeting or exceeding the threshold in each color channel were identified and located. To identify coincident photon bursts, the color channel with fewer bursts was selected, and for each photon burst above the threshold in that color channel, the nearest photon burst in the other color channel was located and the time between the two bursts was determined. Bursts in both color channels that were detected in the same bin were considered coincident. Double labeled PCR amplicons were identified in this manner for each PCR cycle investigated. A histogram of the time between red and green fluorescent bursts at PCR cycle 0 is shown in Fig. 4a, corresponding to the detection of uncorrelated single color molecules. The same histogram for PCR cycle 40 is shown in Fig. 4b. A large central peak is observed, resulting from the coincident detection of red and green fluorescent bursts, due to the large number of PCR amplicons incorporating both primers. Histograms for each PCR cycle analyzed in addition to cycles 0 and 40 (cycles 4, 8, 12, 16, 20, 24, 28, 32, results not shown) display an increasing number of amplicons detected as they are synthesized over the course of the reaction. Around 15,000 photon bursts of each color were detected in 15 sequential one minute runs for each cycle number analyzed. The number of double labeled PCR amplicons detected varied from approximately zero at low cycle numbers to almost 7,000—about 2 attograms of DNA—at the end of the reaction. Approximately 200 picoliters of sample was consumed on-chip by this particular analysis, but like concentration, consumption can be varied while remaining balanced against other experimental parameters to meet the goals of a particular application.
Figure 4.
(a) Distribution of the time between red and green fluorescent bursts at PCR cycle 0, corresponding to the detection of uncorrelated PCR primers. (b) Distribution of the time between red and green fluorescent bursts at PCR cycle 40. A large central peak is observed, corresponding to the coincident detection of red and green fluorescent bursts, resulting from PCR amplicons incorporating both primers. The bin time is 2 ms.
Single molecule PCR analysis
The numbers of single color molecules and two color amplicons detected at each PCR cycle analyzed were used to determine the percent of primers reacted and characterize the amplification process. The percent of reacted single color molecules of type i was defined as the number of PCR amplicons divided by the sum of the amplicons and type i molecules, and multiplied by 100 as well as the correction factor for analyte flow speed:
| (4) |
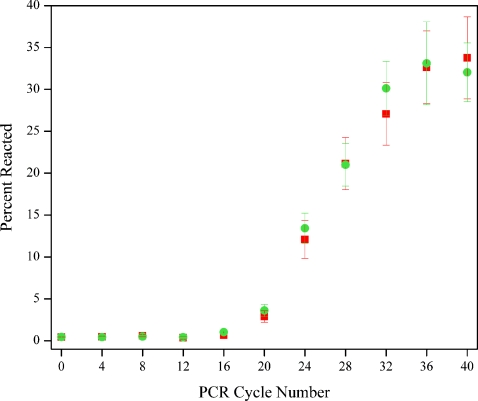
The percent of primers reacted as a function of PCR cycle number for the two color channels is shown in Fig. 5. The error bars represent the standard deviation of 15 sequential equilibrated measurements. The curve in Fig. 5 displays the characteristic sigmoidal shape representative of PCR amplification, with the percent reacted at each point reflecting the increasing number of PCR amplicons detected. The maximum plateau value of the percent reacted was found to be approximately 33%.
Figure 5.
Percent of primers reacted as a function of PCR cycle number, measured using submicrometer fluidic channels to isolate PCR primers and amplicons for detect detection at all reaction phases. Green circles and red squares represent the percent of reacted primers labeled with Alexa Fluor 488 and Alexa Fluor 596, respectively. Data is shown corrected for analyte velocity, and error bars correspond to one standard deviation.
Conventional PCR analysis
Conventional methods of genomic analysis were also applied to characterize the PCR amplification. Gel and capillary electrophoresis were used to separate the primers and product based on their electrophoretic mobility, after which the bands were quantified via macroscopic fluorescence spectroscopy. No primer dimers or PCR side products were observed. The percent of reacted primers was calculated by dividing the intensity of the PCR product band or peak, for gel or capillary electrophoresis, respectively, by the sum of the intensities of the PCR product and one of the two primer bands and multiplying by 100:
| (5) |
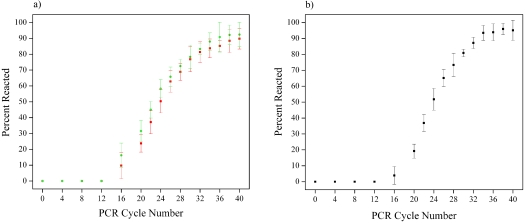
The percent of reacted primers as a function of cycle number as measured by these methods is shown in Fig. 6. Identical amplification curves were measured for both primer bands and peaks. While these curves exhibited trends similar to the one measured by single molecule spectroscopy in submicrometer fluidic channels, the percent of reacted primers reached plateau values of approximately 95% and 90%, when measured by capillary gel electrophoresis and gel electrophoresis, respectively.
Figure 6.
(a) Percent of primers reacted as a function of PCR cycle number measured using gel electrophoresis. (b) Percent of primers reacted as a function of PCR cycle number measured using capillary gel electrophoresis.
Comparison of single molecule and conventional measurements
In order to fully characterize and compare the results obtained from single molecule and conventional methods of PCR analysis, two issues in particular must be considered—the incidence of free dye molecules and imperfect labeling efficiency. Because of the sensitivity of single molecule measurements, these issues result in the presence and detection of contaminants in addition to single labeled primers and double labeled amplicons, including free dye molecules and single labeled PCR amplicons resulting from unlabeled PCR primers. These problems are encountered frequently and variably when samples are obtained from commercial vendors, can be difficult to characterize, significantly impact the results obtained from single molecule measurements, and are an impediment to quantitative analysis. They are therefore given a careful treatment here. These two issues do not, however, have the same effect on less sensitive conventional methods of PCR characterization. While free Alexa Fluor dye molecules are electrophoresed along with the PCR products, they are poorly resolved by the gel matrix, and only the sharp bands corresponding to the primers and amplicons are considered for analysis. The free dye concentration is therefore not taken into account in the final quantification of the PCR reaction. Moreover, gel-based electrophoresis methods do not provide information on the coincidence of the two fluorophores, as the quantification of the ratio of PCR products to primers is performed individually for each color channel. As the labeling efficiency is a constant parameter of the PCR amplification, and assuming that unlabelled primers are not preferentially incorporated, the ratio of PCR product over the total fluorescence is not influenced by the degree of labeling of the primers.
To determine the amount of free dye molecules, a fluorospectrometer was used to measure the difference between DNA absorbance at 260 nm and fluorophore absorbance at the given excitation wavelength. This provided a measure of the number of fluorophores per base, and because the number of bases per oligonucleotide was known, the average number of fluorophores per PCR primer could be calculated. Using this technique, 1.81±0.11 Alexa Fluor 488 molecules and 1.44±0.04 Alexa Fluor 594 molecules were observed for each associated primer. Both negatively charged dye molecules were expected to flow in a manner comparable to that of the labeled primers, which corresponded with autocorrelation curves for both color channels that were well-described by a single species flow model (not shown). Because the free dye molecules were detected and analyzed in a manner indistinguishable from that of labeled primers, there appeared to be an excess of unreacted primers, reducing the measured value of the percent of primers reacted. The ratio of the number of free dye molecules to the number of oligomers introduces another correction factor,
needed to account for the actual percent of primers reacted.
Another result from the measured ratios of dye molecules to oligonucleotides is that a direct measurement of the labeling efficiency of the primers is obscured. Labeling efficiency is an important variable to consider when determining the percent of primers reacted. Unlabeled primers would result in a distribution of single labeled PCR amplicons which are then detected as primers of the other color, reducing the measured amplitude value of the amplification curve. This would introduce a third correction factor,
where η is the fraction of labeled oligomers, needed to account for the actual percent of primers reacted. Prior to the presented experiments, several PCR primers from different vendors were obtained and analyzed. Measurements of the dye to oligonucleotide ratio demonstrated that the values of free dye concentration and labeling efficiency were variable and deviated significantly from the ideal value of one dye molecule per oligonucleotide primer, even when the highest available standards of purification were specified, including dual-HPLC and PAGE. Assuming zero free dye molecules for these previous measurements, labeling efficiencies were found to be between 70% and 90%. The likely presence of unaccounted for free dye in these measurements would further reduce these values.
The possible effects of heat cycling, including loss and degradation of fluorophores from PCR primers, were also considered but for the most part discounted. In the event that heat cycling resulted in fluorophore degradation and unlabeled primers, a population of single labeled amplicons would be generated and detected as primers of the other color. Loss of fluorophores from labeled primers would also result in unlabeled primers in addition to additional free dye molecules. Macroscopic fluorescence measurements (not shown) demonstrated no decrease in fluorescence after 40 PCR cycles, indicating that the two fluorophores were unaffected by the heat cycling process. Because of the covalent linkage mechanism between the oligonucleotides and fluorescent labels, separation of the two due to heat cycling was considered unlikely, although still a possibility. A variety of optical issues were also considered and largely discounted, including FRET and photobleaching of individual dye molecules to the extent that they were not detected as they passed through the focal volume.
Considering these various factors, the difference between the amplification curve amplitudes is best addressed by the correction factors CFFD and CFLE. In the unlikely event of perfect labeling efficiency for both primers, CFLE=1, and to account for free dye molecules the single molecule amplification curve would be multiplied by CFFDi=1.81 for the green primers and CFFDj=1.44 for the red primers. Labeling efficiencies of 70% to 90%, consistent with that of previous measurements, would increase CFLE to the extent that the amplitudes of the amplification curves measured using single molecule and conventional techniques would agree. Any remaining uncertainty about these results must be addressed by the development and application of more stringent methods of filtration and recovery than are commercially available to increase the accuracy of reagent characterization.
CONCLUSION
The PCR amplification of a fragment of 16S rDNA from T. fusca was analyzed using single molecule fluorescence spectroscopy in a submicrometer fluidic channel, a potential structure for integrated on chip single molecule analysis. This technique resulted in the ability to directly, rapidly and uniformly detect individual PCR primers and amplicons at all phases of the amplification process, with low sample consumption and without post-amplification purification or size screening. Gel and capillary gel electrophoresis were also used for PCR analysis, and provided an indirect determination of PCR product concentration via size screening and macroscopic intensity measurements. Amplicons were detected after 15 to 20 cycles with these methods, which were found to measure an amplification trend similar to that observed using single molecule methods but with different plateau amplitudes due to differences in the mechanism of analyte discrimination and detection sensitivity. Biomolecular parameters and reagents significantly influenced the analysis of the target products. Labeling efficiency and free dye molecules were primary factors in explaining the differing amplitude of the amplification curves. Because each fluorescent molecule in solution was detected, great care was taken to eliminate contamination from buffers and additives, but limits were imposed by the purity and labeling efficiency of the PCR constituents. Single molecule methods place a premium on control over sample purity and labeling, and as the sensitivity of biomolecular detection and quantification increases, control over the purity of biomolecular reagents such as primers and probes should be improved in parallel.
The future use of smaller fluidic channels will further improve detection sensitivity and uniformity and increase the concentration range at which single molecule analysis can be performed, and the on-chip implementation of parallel fluidic channels and detectors will significantly increase throughput. The complexity and utility of biomolecular reactions for the detection and quantification of microorganisms can also be greatly increased by the use of more than two fluorescent labels with a combination of universal or specific PCR primers. With these advances, the presented method will be ideal for the rapid detection and analysis of PCR products after a minimal number of amplification cycles, eliminating the limiting effects of varying reaction efficiency on quantitative PCR analysis, improving the speed and accuracy of measurements, and providing a multiparametric analysis as each double labeled product corresponds to a given DNA sequence.
An important consideration is that submicrometer fluidic channels would be most useful when integrated as a component of a micro-total-analysis-system. Such a system could perform PCR amplification on-chip, and manipulate the microscopic sample volumes encountered at this scale. Without the benefits of this technology, the amplification was performed conventionally and a diluted fraction of this sample was analyzed on-chip. The concentration and volume of off-chip PCR products were not considered to be of particular importance in the presented work as the intent was to demonstrate a single molecule analysis of PCR products. It is evident, however, that submicrometer fluidic channels stand to benefit from increased integration, and future work must continue to address this issue.
ACKNOWLEDGMENTS
This work was supported by the Nanobiotechnology Center (NBTC), an STC Program of the National Science Foundation under Agreement No. ECS-9876771, and the New York State Office of Science, Technology and Academic Research (NYSTAR). The authors would like to thank Joshua Edel, Kevan Samiee, Christian Reccius, and Elizabeth Strychalski for helpful discussions.
References
- Li J., Stein D., McMullan C., Branton D., Aziz M. J., and Golovchenko J. A., Nature (London) 10.1038/35084037 412, 166 (2001). [DOI] [PubMed] [Google Scholar]
- Storm A. J., Chen J. H., Zandbergen H. W., and Dekker C., Phys. Rev. E 10.1103/PhysRevE.71.051903 71, 051903 (2005). [DOI] [PubMed] [Google Scholar]
- Storm A. J., Storm C., Chen J. H., Zandbergen H., Joanny J. F., and Dekker C., Nano Lett. 10.1021/nl048030d 5, 1193 (2005). [DOI] [PubMed] [Google Scholar]
- Wei C. H., Tsao P. H., Farm W., Wei P. K., Tegenfeldt J. O., and Austin R. H., J. Opt. Soc. Am. B 10.1364/JOSAB.21.001005 21, 1005 (2004). [DOI] [Google Scholar]
- Han J. and Craighead H. G., Science 10.1126/science.288.5468.1026 288, 1026 (2000). [DOI] [PubMed] [Google Scholar]
- Turner S. W. P., Cabodi M., and Craighead H. G., Phys. Rev. Lett. 10.1103/PhysRevLett.88.128103 88, 128103 (2002). [DOI] [PubMed] [Google Scholar]
- Levene M. J., Korlach J., Turner S. W., Foquet M., Craighead H. G., and Webb W. W., Science 10.1126/science.1079700 299, 682 (2003). [DOI] [PubMed] [Google Scholar]
- Samiee K. T., Foquet M., Guo L., Cox E. C., and Craighead H. G., Biophys. J. 10.1529/biophysj.104.052795 88, 2145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabodi M., Chen Y. F., Turner S. W. P., Craighead H. G., and Austin R. H., Electrophoresis 23, 3496 (2002). [DOI] [PubMed] [Google Scholar]
- Foquet M., Korlach J., Zipfel W., Webb W. W., and Craighead H. G., Anal. Chem. 10.1021/ac011076w 74, 1415 (2002). [DOI] [PubMed] [Google Scholar]
- Chou H. P., Spence C., Scherer A., and Quake S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.96.1.11 96, 11 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehn R., Lu M. C., Wang Y. M., Lim S. F., Cox E. C., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0503809102 102, 10012 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner W., Morton K. J., Riehn R., Wang Y. M., Yu Z. N., Rosen M., Sturm J. C., Chou S. Y., Frey E., and Austin R. H., Phys. Rev. Lett. 10.1103/PhysRevLett.94.196101 94, 196101 (2005). [DOI] [PubMed] [Google Scholar]
- Tegenfeldt J. O., Cao H., Reisner W. W., Prinz C., Austin R. H., Chou S. Y., Cox E. C., and Sturm J. C., Biophys. J. 10.1529/biophysj.103.039040 86, 596A (2004). [DOI] [Google Scholar]
- Tegenfeldt J. O., Prinz C., Cao H., Chou S., Reisner W. W., Riehn R., Wang Y. M., Cox E. C., Sturm J. C., Silberzan P., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0403849101 101, 10979 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foquet M., Korlach J., Zipfel W. R., Webb W. W., and Craighead H. G., Anal. Chem. 10.1021/ac035088o 76, 1618 (2004). [DOI] [PubMed] [Google Scholar]
- Stavis S. M., Edel J. B., Samiee K. T., and Craighead H. G., Lab Chip 10.1039/b416161k 5, 337 (2005). [DOI] [PubMed] [Google Scholar]
- Stavis S. M., Edel J. B., Li Y. G., Samiee K. T., Luo D., and Craighead H. G., Nanotechnology 10.1088/0957-4484/16/7/002 16, S314 (2005). [DOI] [PubMed] [Google Scholar]
- Stavis S. M., Edel J. B., Li Y. G., Samiee K. T., Luo D., and Craighead H. G., J. Appl. Phys. 10.1063/1.2005371 98, 044903 (2005). [DOI] [Google Scholar]
- Saunders N. A., Real-Time PCR: An Essential Guide, edited by Edwards K., Logan J., and Saunders N. (Horizon Bioscience, Wymondham, Norfolk, UK, 2004), Chap. 6, pp. 103–123. [Google Scholar]
- Rutledge R. G. and Cote C., Nucleic Acids Res. 10.1093/nar/gng093 31, 93 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbridge S. S., Edel J. B., Stavis S. M., Moran-Mirabal J. M., Allen S. D., Coates G., and Craighead H. G., J. Appl. Phys. 10.1063/1.1924888 97, 124317 (2005). [DOI] [Google Scholar]
- Magde D., Webb W. W., and Elson E., Phys. Rev. Lett. 10.1103/PhysRevLett.29.705 29, 705 (1972). [DOI] [Google Scholar]
- Magde D., Elson E. L., and Webb W. W., Biopolymers 10.1002/bip.1974.360130103 13, 29 (1974). [DOI] [PubMed] [Google Scholar]
- Elson E. L. and Magde D., Biopolymers 10.1002/bip.1974.360130102 13, 1 (1974). [DOI] [PubMed] [Google Scholar]
- Magde D. and Elson E. L., Biopolymers 10.1002/bip.1978.360170208 17, 361 (1978). [DOI] [PubMed] [Google Scholar]
- Schwille P., MeyerAlmes F. J., and Rigler R., Biophys. J. 72, 1878 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigler R., Foldes-Papp Z., Meyer-Alme F. J., Sammet C., Volcker M., and Schnetz A., J. Biotechnol. 10.1016/S0168-1656(98)00079-0 63, 97 (1998). [DOI] [PubMed] [Google Scholar]
- Gao Q. F. and Yeung E. S., Anal. Chem. 10.1021/ac970999h 70, 1382 (1998). [DOI] [PubMed] [Google Scholar]