Abstract
Purpose
MDM2 regulates p53, which controls cell cycle arrest and apoptosis. Both proteins, along with Ki-67, which is an established strong determinant of metastasis, have shown promise in predicting the outcome of men treated with radiation therapy (RT) with or without short-term androgen deprivation (STAD). This report compares the utility of abnormal expression of these biomarkers in estimating progression in a cohort of men treated on RTOG 92-02.
Patients and Methods
Adequate tissue for immunohistochemistry was available for p53, Ki-67, and MDM2 analyses in 478 patient cases. The percentage of tumor nuclei staining positive (PSP) was quantified manually or by image analysis, and the per-sample mean intensity score (MIS) was quantified by image analysis. Cox regression models were used to estimate overall mortality (OM), and Fine and Gray's regressions were applied to the end points of distant metastasis (DM) and cause-specific mortality (CSM).
Results
In multivariate analyses that adjusted for all markers and treatment covariates, MDM2 overexpression was significantly related to DM (P = .02) and OM (P = .003), and Ki-67 overexpression was significantly related to DM (P < .0001), CSM (P = .0007), and OM (P = .01). P53 overexpression was significantly related to OM (P = .02). When considered in combination, the overexpression of both Ki-67 and MDM2 at high levels was associated with significantly increased failure rates for all end points (P < .001 for DM, CSM, and OM).
Conclusion
Combined MDM2 and Ki-67 expression levels were independently related to distant metastasis and mortality and, if validated, could be considered for risk stratification of patients with prostate cancer in clinical trials.
INTRODUCTION
The MDM2 oncoprotein is an established regulator of p53 via effects on p53 degradation and negative feedback inhibition. Downregulation of p53 results in the prevention of p53-mediated apoptosis and cell cycle arrest.1–3 In addition, MDM2 interacts with other regulatory proteins, such as pRB4 and E2F-1,5 independent of p53. In prostate cancer, MDM2 knockdown increases the sensitivity of the tumor cells to androgen deprivation and radiation both in vitro and in vivo6–8 and enhances tumor growth inhibition in androgen-insensitive cells.9
In an earlier report that evaluated the association between MDM2 overexpression and outcome of patients with prostate cancer, we observed a relationship to Gleason score and a trend of an association with distant metastases (DM) in men treated with radiation therapy (RT) with or without short-term androgen deprivation (STAD) in Radiation Therapy Oncology Group (RTOG) protocol 86-10.10 A relatively small sample size was available for this analysis, and prostate-specific antigen (PSA) information was limited.
RTOG protocol 92-02 is a much larger, multi-institutional, phase III, randomized trial that compared RT + STAD to RT + long-term androgen deprivation (LTAD).11–14 We have previously published that Ki-6712 and p53,11 when tested individually, are predictive of DM and cause-specific mortality (CSM) in men treated on RTOG 92-02. MDM2 is a key regulator of p53 and, hence, proliferation, and it is a potential therapeutic target; thus, this investigation explores the relationships between MDM2, Ki-67, and p53 expression with patient outcome for men treated with RT + STAD and RT + LTAD on RTOG 92-02.
PATIENTS AND METHODS
Patients Characteristics
The study details of RTOG protocol 92-02 have been described elsewhere.12,13 To summarize, RTOG 92-02 was a randomized trial that compared LTAD (28 months) with STAD (4 months) in addition to RT in patients with locally advanced prostate cancer. During a median follow-up of 10 years, RT + LTAD treatment was associated with reduced biochemical failure, local progression, disease progression, DM, and disease-specific mortality; however, a reduction in overall mortality was not observed. An effect of RT + LTAD on overall mortality was seen in men who had cancer with Gleason scores of 8 to 10.14 There were 1,521 analyzable patients, of whom 478 (31.4%) had pretreatment diagnostic tumor tissue (which consisted of 428 needle-core biopsies and 49 transurethral resection specimens) adequately stained for all three biomarkers in this report. All data were de-identified for analysis, and institutional review board approval for this study was given by the RTOG and by the Fox Chase Cancer Center, Philadelphia, PA.
Immunohistochemical Analysis
The immunohistochemical staining protocol has been detailed in prior reports.11,12 Briefly, pretreatment formalin-fixed, paraffin-embedded tissue specimens were cut onto poly-l-lysine slides, were deparaffinized in xylene, were rehydrated, and were washed. A pressure cooker was used for antigen retrieval in citrate buffer. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. The primary antibodies used were MDM2 (No. M7146, Clone SMP14, 1:100 dilution; Dako Corp, Carpinteria, CA), MIB-1 (No. M7240, 1:100 dilution; Dako Corp), and p53 (No. M7001, Clone DO7, 1:100 dilution; Dako Corp). The current MDM2 antibody was selected, as validation studies in our laboratory demonstrated clearer nuclear staining than seen in our previous report (Zymed Laboratories, Inc, South San Francisco, CA).10 Although both antibodies stained in the nuclear and cytoplasmic compartments, the current antibody showed better compartmentalization and lower background. Immune complexes were detected by the labeled streptavidin-biotin method (Dako LSAB 2 Kit; Dako Corp) for MDM2 and Ki-67 and by the ABC method, using 3-amino-9-ethylcarbazole as the chromogen for p53. A hematoxylin counterstain was used (Dako Corp). The positive controls were human prostate carcinoma (MDM2), a colon carcinoma with a known p53 mutation (p53), and normal tonsil sections (Ki-67). Negative controls were produced by omitting the primary antibody.
Image Analysis
Semi-automated image analysis was performed, as previously described.10 All the slides were scanned and analyzed (by L.-Y.K.). These patient cases were reviewed for tumor tissue together with an oncologic pathologist (T.A-S.) in a previous study.12 The slides were scanned at 10× magnification according to the scanning protocol of the imaging system (ACIS II; Clarient Chromavision Inc, San Juan Capistrano, CA). Digital images were captured by a three-chip CCD camera (Sony, New York, NY). Color thresholds then were set for each slide to eliminate background. Tumor regions were outlined either with a 40× circular field or by freehand around the glands of interest, which thereby excluded stromal tissue and unwanted artifacts. At least six regions of interest were chosen for every patient case with sufficient tumor. Pixel-based scores that represented the percentage of tumor nuclei staining positive (PSP) and the mean intensity score (MIS) were recorded.15–17
Definition of Disease End Points
Disease end points have been described previously.11–13,18 Briefly, DM was defined as clinical evidence of distant disease by any method. CSM was death certified as a result of prostate cancer, death as a result of treatment complications, death as a result of unknown causes with active malignancy (clinical disease relapse), or death as a result of another cancer with documented bone metastases attributed to prostate cancer before the appearance of the second independent cancer. Finally, overall mortality (OM) was defined as death as a result of any cause. Time to DM, CSM, and OM were measured from the date of random assignment to the date of death or last follow-up.
Statistical Analysis
χ2 test statistics were used to compare pretreatment characteristics of patient cases. Pretreatment characteristics were categorized as follows: median age in the entire cohort (< 70 v ≥ 70 years); Gleason score (2 to 6 v 7 v 8-10), initial pretreatment PSA (iPSA; ≤ 30 v > 30 ng/mL), and T stage (2 v 3 or 4) were categorized by their stratification groupings in RTOG 92-02. The Kaplan-Meier method was used to estimate the survival rate for OM. Log-rank test statistics were used to compare the estimated OM rates. The cumulative incidence method was used to estimate rates of DM and CSM, because it considers other competing causes of mortality. Gray's test statistics were used to compare cumulative incidence rates. To analyze whether each biomarker was independently associated with the end points while analysis was adjusted for other covariates, a Cox proportional hazards regression model was used for OM, and Fine and Gray's regression models19 were used for DM and CSM. Unadjusted and adjusted hazard ratios (HRs) were calculated by using either the Cox proportional hazards model or Fine and Gray's regression model with associated 95% CIs and P values. All statistical tests were two-sided, and P less than .05 was considered statistically significant. Statistical Analysis System (SAS Institute, Cary, NC) and R software (http://www.r-project.org) were used for all statistical analyses.
RESULTS
Adequate tissue was available for immunohistochemical staining of p53 in 780 (51.3%) patient cases, of Ki-67 in 637 (41.9%) patient cases, and of MDM2 in 589 (38.7%) patient cases, of the 1,521 total patient cases in the parent cohort. There were 478 patient cases (31.4%) with complete biomarker data for all three markers. The pretreatment characteristics of the groups with individual or complete biomarker data versus those without biomarker data were similar, except for Gleason Score, in which a higher proportion of the study cohort had Gleason scores of 8 to 10 (Ki-67, P = .04; MDM2, P = .007; all markers, P = .006) and had assigned treatment (MDM2, P = .0006; all markers, P = .02; Table 1). Although these study cohorts may not represent a random sample of the parent cohort, there were no statistically significant differences between the groups with marker data (whether individual or complete) and those without for the end points DM, CSM, and OM (Appendix Table 1, online only).
Table 1.
Distribution of Patients by Missing v Determined Biomarkers
| Patient Demographics and Clinical Characteristics | Biomarker |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p53 |
Ki-67 |
MDM2 |
All |
|||||||||||||||||
| Missing |
Determined |
P* | Missing |
Determined |
P* | Missing |
Determined |
P* | Missing |
Determined |
P* | |||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |||||
| No. of eligible patients | 905 | 60 | 616 | 41 | — | 884 | 58 | 637 | 42 | 932 | 61 | 589 | 39 | 1043 | 69 | 478 | 31 | — | ||
| Age, years | ||||||||||||||||||||
| < 70 | 403 | 45 | 278 | 45 | .82 | 394 | 45 | 287 | 45 | .85 | 415 | 45 | 226 | 45 | .81 | 462 | 44 | 219 | 46 | .58 |
| ≥ 70 | 502 | 55 | 338 | 55 | 490 | 55 | 350 | 55 | 517 | 55 | 523 | 55 | 581 | 56 | 259 | 54 | ||||
| Median | 70 | 70 | 70 | 70 | 70 | 70 | 70 | 70 | ||||||||||||
| Range | 43-88 | 44-88 | 43-88 | 44-88 | 43-87 | 43-88 | 43-88 | 44-88 | ||||||||||||
| Gleason score* | ||||||||||||||||||||
| 2-6 | 361 | 43 | 222 | 39 | .055 | 356 | 43 | 227 | 38 | .04 | 381 | 44 | 202 | 37 | .007 | 424 | 43 | 159 | 36 | .006 |
| 7 | 290 | 34 | 187 | 33 | 281 | 34 | 196 | 33 | 291 | 33 | 186 | 34 | 324 | 33 | 153 | 34 | ||||
| 8-10 | 196 | 23 | 165 | 29 | 190 | 23 | 171 | 29 | 199 | 23 | 162 | 29 | 227 | 23 | 134 | 30 | ||||
| Unknown | 58 | 42 | 57 | 43 | 61 | 39 | 68 | 32 | ||||||||||||
| iPSA, ng/mL | ||||||||||||||||||||
| ≤ 30 | 615 | 68 | 406 | 66 | .40 | 600 | 68 | 421 | 66 | .47 | 627 | 67 | 394 | 67 | .88 | 706 | 68 | 315 | 66 | .49 |
| > 30 | 290 | 32 | 210 | 34 | 284 | 32 | 216 | 34 | 305 | 33 | 195 | 33 | 337 | 32 | 163 | 34 | ||||
| Median | 19.2 | 21.4 | 194 | 21.0 | 19.5 | 20.8 | 19.2 | 21.5 | ||||||||||||
| Range | 0.8-250 | 0.1-219.7 | 0.8-250 | 0.1-219.7 | 0.4-250 | 0.1-219.7 | 0.4-250 | 0.1-219.7 | ||||||||||||
| T stage | ||||||||||||||||||||
| 2 | 409 | 45 | 282 | 46 | .82 | 397 | 45 | 294 | 46 | .63 | 423 | 45 | 268 | 46 | .97 | 475 | 46 | 216 | 45 | .90 |
| 3-4 | 496 | 55 | 334 | 54 | 487 | 55 | 343 | 54 | 509 | 55 | 321 | 55 | 568 | 54 | 262 | 55 | ||||
| Assigned treatment | ||||||||||||||||||||
| LTAD + RT | 440 | 49 | 318 | 52 | .25 | 426 | 48 | 332 | 52 | .13 | 432 | 46 | 326 | 55 | .0006 | 498 | 48 | 260 | 54 | .02 |
| STAD + RT | 465 | 51 | 298 | 48 | 458 | 52 | 305 | 48 | 500 | 54 | 263 | 45 | 545 | 52 | 218 | 46 | ||||
Abbreviations: iPSA, initial pretreatment prostate-specific antigen; LTAD, long-term androgen deprivation; RT, radiotherapy; STAD, short-term androgen deprivation.
χ2 test statistics.
In terms of pretreatment characteristics, there were no significant differences in the distribution of patients by assigned treatment in each of these data sets. The median age of the patients with all three biomarkers was 70 years (range, 44 to 88 years), and the median iPSA was 21.5 ng/mL (range, 0.1 to 219.7 ng/mL). One hundred fifty-three patients (34%) and 134 patients (30%) had Gleason scores of 7 and of 8 to 10, respectively, and 262 (55%) patient cases had T3-4 disease. Two hundred sixty patients (54%) and 218 patients (46%) were assigned to the RT + LTAD and RT + STAD treatment arms, respectively. Median follow-up for all patients still alive with complete biomarker data was 9.3 years (0.04 to 13.9 years), and median follow-up was 11.4 years (range, 1.6 to 13.9 years).
We have observed that the median cut point is often not the most optimal for predicting outcome. Rather than undergo a random search for an optimal dichotomous cut point for each biomarker, we systematically investigated the relationships to OM, CSM, and DM for cut points at the 25th (Q1), 50th (median), and 75th percentiles (Q3). Also considered were the relative differences in these associations by using the manual and image-analysis PSP and image-analysis MIS parameters as dichotomous variables. Manual scoring of the MDM2 patient cases was not performed, as image analysis was found to be more reliable in our previous report.10 The optimal cut point for each marker was determined by the results of multivariate analyses that were adjusted for age, iPSA, Gleason score, clinical stage, and assigned treatment (Table 2). The manual PSP median (0%) for p53 (of note, any staining of ≤ 5% was counted as 0%), manual PSP Q3 (11.3%) for Ki-67, and image-analysis MIS Q3 (184 arbitrary units) for MDM2 were chosen on the basis of being associated with the highest HRs for all significant end points.
Table 2.
Multivariate Analyses of Markers as Dichotomous Variables: Individual Biomarker Models
| End Point by Marker and Evaluation Type | Multivariate Analyses |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median |
Q1 |
Q3 |
||||||||||
| Cut Point | HR | 95% CI | P* | Cut Point | HR | 95% CI | P* | Cut Point | HR | 95% CI | P* | |
| Manual PSP | ||||||||||||
| p53 | 0 | NA | 10 | |||||||||
| DM | 1.63 | 1.15 to 2.31 | .007 | 1.48 | 1.01 to 2.17 | .045 | ||||||
| CSM | 1.25 | 0.84 to 1.84 | .27 | 1.15 | 0.74 to 1.77 | .54 | ||||||
| OM | 0.94 | 0.76 to 1.16 | .54 | 0.93 | 0.73 to 1.18 | .55 | ||||||
| Ki-67 | 6.3 | 3.7 | 11.3 | |||||||||
| DM | 1.67 | 1.13 to 2.46 | .01 | 1.78 | 1.06 to 3.00 | .03 | 2.63 | 1.78 to 3.90 | < .0001 | |||
| CSM | 1.62 | 1.06 to 2.47 | .026 | 1.55 | 0.91 to 2.66 | .11 | 2.22 | 1.44 to 3.43 | .0003 | |||
| OM | 1.31 | 1.05 to 1.64 | .02 | 1.11 | 0.86 to 1.44 | .43 | 1.40 | 1.09 to 1.80 | .008 | |||
| Image-analysis PSP | ||||||||||||
| p53 | 6 | 1 | 27 | |||||||||
| DM | 1.30 | 0.92 to 1.85 | .14 | 1.21 | 0.81 to 1.80 | .36 | 1.13 | 0.76 to 1.67 | .56 | |||
| CSM | 1.17 | 0.80 to 1.72 | .41 | 1.02 | 0.67 to 1.55 | .94 | 1.30 | 0.86 to 1.98 | .22 | |||
| OM | 0.97 | 0.80 to 1.19 | .78 | 1.01 | 0.81 to 1.26 | .94 | 0.85 | 0.67 to 1.08 | .19 | |||
| Ki-67 | 7.7 | 4.5 | 12.8 | |||||||||
| DM | 1.99 | 1.34 to 2.96 | .0007 | 1.42 | 0.88 to 2.28 | .15 | 2.39 | 1.59 to 3.59 | < .0001 | |||
| CSM | 1.68 | 1.10 to 2.59 | .018 | 1.37 | 0.81 to 2.32 | .23 | 2.05 | 1.31 to 3.19 | .0016 | |||
| OM | 1.34 | 1.07 to 1.68 | .01 | 1.23 | 0.94 to 1.60 | .13 | 1.16 | 0.90 to 1.50 | .26 | |||
| MDM2 | 41 | 16 | 61 | |||||||||
| DM | 1.16 | 0.79 to 1.71 | .46 | 1.20 | 0.74 to 1.95 | .45 | 1.19 | 0.77 to 1.82 | .43 | |||
| CSM | 1.22 | 0.79 to 1.86 | .37 | 1.36 | 0.80 to 2.30 | .26 | 1.18 | 0.74 to 1.89 | .48 | |||
| OM | 1.14 | 0.91 to 1.44 | .25 | 1.08 | 0.83 to 1.41 | .55 | 1.14 | 0.88 to 1.47 | .32 | |||
| Image-analysis MIS | ||||||||||||
| p53 | 130 | 91 | 151 | |||||||||
| DM | 1.04 | 0.73 to 1.48 | .83 | 1.21 | 0.80 to 1.82 | .37 | 1.47 | 1.01 to 2.15 | .045 | |||
| CSM | 0.85 | 0.58 to 1.24 | .39 | 0.97 | 0.63 to 1.48 | .87 | 1.15 | 0.75 to 1.75 | .53 | |||
| OM | 1.00 | 0.82 to 1.22 | .96 | 1.01 | 0.81 to 1.27 | .91 | 0.94 | 0.74 to 1.19 | .61 | |||
| Ki-67 | 182.95 | 167.1 | 195.25 | |||||||||
| DM | 1.52 | 1.02 to 2.27 | .04 | 1.43 | 0.91 to 2.26 | .12 | 0.97 | 0.61 to 1.54 | .89 | |||
| CSM | 1.14 | 0.75 to 1.75 | .54 | 1.43 | 0.88 to 2.32 | .15 | 0.79 | 0.47 to 1.34 | .38 | |||
| OM | 1.17 | 0.93 to 1.46 | .18 | 1.32 | 1.01 to 1.72 | .04 | 0.95 | 0.73 to 1.24 | .72 | |||
| MDM2 | 167 | 147 | 184 | |||||||||
| DM | 1.73 | 1.17 to 2.56 | .006 | 1.47 | 0.92 to 2.36 | .11 | 1.84 | 1.21 to 2.78 | .004 | |||
| CSM | 1.74 | 1.12 to 2.69 | .013 | 1.35 | 0.81 to 2.26 | .25 | 1.65 | 1.04 to 2.61 | .032 | |||
| OM | 1.17 | 0.93 to 1.47 | .18 | 1.29 | 0.99 to 1.69 | .06 | 1.42 | 1.10 to 1.83 | .007 | |||
NOTE. Analyses were adjusted for age (< 70 v ≥ 70 years), iPSA (≤ 30 v > 30 ng/mL), Gleason score (2-6 v 7 v 8-10), clinical stage (T2 v T3-4), and assigned treatment. Each biomarker is considered without the other biomarkers included in the models.
Abbreviations: Q1, lowest quartile; Q3, upper quartile; HR, hazard ratio; PSP, percent staining positive; NA, not applicable; DM, distant metastasis; CSM, cause-specific mortality; OM, overall mortality; MIS, mean intensity score.
P values are from Fine and Gray's regression models for CSM and DM and from Cox regression models for OM.
Table 3 lists the crude numbers of failures by each biomarker-dichotomous cut point for each end point. Table 3 also displays the HR results of the multivariate analyses by each end point, after adjustment for age, iPSA, Gleason score, clinical stage, and assigned treatment and for all the covariates above plus the other biomarkers. After analysis was adjusted for the covariates, including the other markers, p53 lost its significance for DM (HR, 1.04; 95% CI, 0.68 to 1.61; P = .85), and MDM2 was no longer significant for CSM (HR, 1.55; 95% CI, 0.94 to 2.55; P = .08). The other previously significant relationships defined in individual biomarker multivariate analyses in Table 2 remained. We also performed these analyses with iPSA as a continuous covariate (Appendix Table 2, online only), and the significant biomarker relationships to DM, CSM, and OM were not substantively different.
Table 3.
Summary of Failures and Adjusted MVA Results
| End Point and Cut Point by Marker and Evaluation Type | Failure Summary |
MVA Results |
|||
|---|---|---|---|---|---|
| Failures | Total | HR | 95% CI | P* | |
| p53 by manual PSP (n = 780) | |||||
| DM | |||||
| ≤ 0 | 77 | 523 | 1.04 | 0.68 to 1.61 | .85 |
| > 0 | 70 | 257 | |||
| CSM | |||||
| ≤ 0 | 69 | 523 | 0.86 | 0.54 to 1.38 | .54 |
| > 0 | 49 | 257 | |||
| OM | |||||
| ≤ 0 | 293 | 523 | 0.72 | 0.55 to 0.95 | .02 |
| > 0 | 143 | 257 | |||
| Ki-67 by manual PSP, % (n = 616) | |||||
| DM | |||||
| ≤ 11.3 | 69 | 478 | 2.95 | 1.89 to 4.60 | < .0001 |
| > 11.3 | 54 | 159 | |||
| CSM | |||||
| ≤ 11.3 | 57 | 478 | 2.35 | 1.43 to 3.85 | .0007 |
| > 11.3 | 42 | 159 | |||
| OM | |||||
| ≤ 11.3 | 244 | 478 | 1.44 | 1.09 to 1.90 | .01 |
| > 11.3 | 100 | 159 | |||
| MDM2 by image-analysis MIS (n = 589) | |||||
| DM | |||||
| ≤ 184 | 81 | 445 | 1.72 | 1.09 to 2.69 | .02 |
| > 184 | 39 | 144 | |||
| CSM | |||||
| ≤ 184 | 65 | 445 | 1.55 | 0.94 to 2.55 | .08 |
| > 184 | 30 | 144 | |||
| OM | |||||
| ≤ 184 | 236 | 445 | 1.54 | 1.16 to 2.05 | .003 |
| > 184 | 88 | 144 | |||
NOTE. Analyses were adjusted for age (< 70 v ≥ 70 years), initial pretreatment prostate-specific antigen (≤ 30 v > 30 ng/mL), Gleason score (2-6 v 7 v 8-10), clinical stage (T2 v T3-4), assigned treatment, and the other biomarkers.
Abbreviations: MVA, multivariate analysis; HR, hazard ratio; PSP, percent staining positive; DM, distant metastasis; CSM, cause-specific mortality; OM, overall mortality; MIS, mean intensity score.
P values are from Fine and Gray's regression model for CSM and DM and from the Cox regression model for OM.
The biomarker data were also considered as continuous variables for all end points (data not shown). In these multivariate analyses, only Ki-67 was significantly related to all the end points, after adjustment for all covariates, including the other markers (DM, P < .0001; CSM, P < .0001; OM, P = .001).
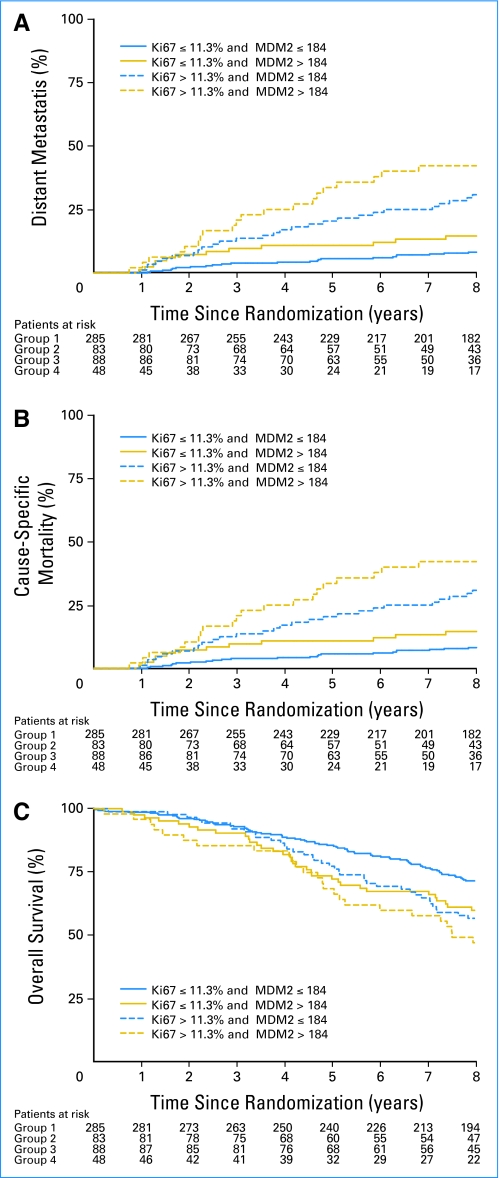
As dichotomous covariates, both Ki-67 and MDM2 were independent predictors of DM. As such, the combination of the Ki-67 and MDM2 data were considered together. Of note, when using either the Ki-67 manual or image-analysis data, the results were comparable with the four groups statistically different with respect to DM, CSM, and OM. The manual Ki-67 data were used in Table 3, because the results were stronger than when the image-analysis data were used; therefore, the manual Ki-67 results were combined with the image analysis results for MDM2. The group with high Ki-67 and MDM2 expression was more likely to experience failure events for all three end points than those groups with low Ki-67 and/or MDM2 expression (Fig 1). Figures 1A and 1B display the cumulative incidence curves (unadjusted) for DM and CSM, and Figure 1C displays the Kaplan-Meier OM curves (unadjusted), subgrouped by the combination of Ki-67 and MDM2 expression data. The curves demonstrate that patients with high Ki-67 PSP and intensity of MDM2 had much greater risks of DM and death. These relationships remained after analysis was adjusted for age, iPSA, Gleason score, clinical stage, assigned treatment, and p53 (Table 4). As Table 4 lists, high Ki-67 (manual PSP > 11.3%) and high MDM2 (image-analysis MIS > 184) were predictive of DM (P < .0001), CSM (P < .0001), and OM (P = .0002) independent of the other covariates. Of note, these findings were not changed substantively when iPSA was included as a continuous covariate (Appendix Table A3, online only).
Fig 1.
Cumulative incidence failure curves of (A) distant metastasis and (B) cause-specific mortality. (C) Kaplan-Meier survival curves of overall mortality subdivided by the combination of Ki-67 percentage of tumor nuclei staining positive and MDM2 mean intensity score. Unadjusted rates are shown.
Table 4.
Multivariate Analysis of Combined MDM2 and Ki-67 Marker Data
| Covariate by End Point | Multivariate Analysis |
||
|---|---|---|---|
| HR | 95% CI | P * | |
| DM | |||
| Age ≥ 70 years | 0.57 | 0.37 to 0.90 | .02 |
| GLSC | |||
| 7 | 1.21 | 0.60 to 2.36 | .57 |
| 8-10 | 3.85 | 2.14 to 6.93 | < .0001 |
| iPSA > 30 ng/mL | 1.34 | 0.88 to 2.05 | .17 |
| Clinical stage T3-4 | 1.77 | 1.12 to 2.79 | .01 |
| Assigned treatment of STAD + RT | 2.13 | 1.37 to 3.32 | .0009 |
| p53 manual PSP > 0 | 1.05 | 0.68 to 1.63 | .82 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.54 | 0.79 to 2.99 | .20 |
| > 11.3% and ≤ 184 | 2.75 | 1.57 to 4.82 | .0004 |
| > 11.3% and > 184 | 5.12 | 2.93 to 8.97 | < .0001 |
| CSM | |||
| Age ≥ 70 years | 0.62 | 0.38 to 1.00 | .052 |
| GLSC | |||
| 7 | 1.28 | 0.64 to 2.57 | .49 |
| 8-10 | 3.65 | 1.95 to 6.84 | < .0001 |
| iPSA > 30 ng/mL | 1.12 | 0.70 to 1.78 | .64 |
| Clinical stage T3-4 | 2.50 | 1.50 to 4.17 | .0004 |
| Assigned treatment of STAD + RT | 2.07 | 1.27 to 3.36 | .003 |
| p53 manual PSP > 0 | 0.87 | 0.54 to 1.39 | .55 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.49 | 0.74 to 2.97 | .26 |
| > 11.3% and ≤ 184 | 2.27 | 1.21 to 4.26 | .01 |
| > 11.3% and > 184 | 3.68 | 1.94 to 6.95 | < .0001 |
| ΟΜ | |||
| Age ≥ 70 years | 1.59 | 1.22 to 2.06 | .0005 |
| GLSC | |||
| 7 | 1.58 | 1.15 to 2.18 | .005 |
| 8-10 | 1.15 | 0.88 to 1.50 | .31 |
| iPSA > 30 ng/mL | 1.12 | 0.81 to 1.50 | .50 |
| Clinical stage T3-4 | 1.26 | 0.97 to 1.64 | .08 |
| Assigned treatment of STAD + RT | 1.26 | 0.98 to 1.63 | .08 |
| p53 Manual PSP > 0 | 0.72 | 0.55 to 0.95 | .02 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.62 | 1.13 to 2.31 | .009 |
| > 11.3% and ≤ 184 | 1.50 | 1.06 to 2.12 | .02 |
| > 11.3% and > 184 | 2.14 | 1.43 to 3.20 | .0002 |
NOTE. Multivariate analyses were performed on 446 patient cases.
Abbreviations: HR, hazard ratio; DM, distant metastasis; GLSC, Gleason score; STAD, short-term androgen deprivation; RT, radiation therapy; PSP, percent staining positive; MIS, mean intensity score; CSM, cause-specific mortality; OM, overall mortality.
P values are from Fine and Gray's regression model for CSM and DM and from the Cox regression model for OM.
The outcome of the patients in RTOG protocol 92-02 was dependent on the length of androgen deprivation received. To study if this was associated with the combination of Ki-67 and MDM2 expression, tests for an interaction between the assigned treatment arm and each combined Ki-67/MDM2 group were performed (Table 5); there were no statistically significant differences for all end points tested. These results indicate that assigned treatment as a covariate was valid for interpretation, and they substantiated treatment association with DM and CSM (Table 4). We then questioned whether any of the combined Ki-67 and MDM2 subgroups (ie, ≤ 11.3% and ≤ 184 v ≤ 11.3% and > 184 v > 11.3% and ≤ 184 v > 11.3% and > 184) had reduced DM by RT + LTAD treatment. For example, if the high Ki-67 and high MDM2 (ie, > 11.3% and > 184) subgroup had reduced DM with RT + LTAD compared with RT + STAD, then we would have a better idea of which patients require RT + LTAD. Although a distinct trend was not observed, the small numbers of events in three of the four subgroups precluded meaningful statistical analyses (data not shown).
Table 5.
Test for an Interaction Between the Ki-67 and MDM2 Combination and Treatment Arm
| Covariate by End Point | Analysis |
||
|---|---|---|---|
| HR | 95% CI | P* | |
| DM | |||
| Ki-67 and MDM2 | |||
| ≤ 11.3% and ≤ 184 | RL | — | — |
| ≤ 11.3% and > 184 | 0.79 | 0.22 to 2.76 | .71 |
| > 11.3% and ≤ 184 | 4.66 | 2.23 to 9.75 | < .0001 |
| > 11.3% and > 184 | 5.34 | 2.25 to 12.69 | .0002 |
| Treatment arm | |||
| STAD + RT | 2.63 | 1.34 to 5.16 | .005 |
| Group 1 and treatment interaction | 2.44 | 0.57 to 10.41 | .23 |
| Group 2 and treatment interaction | 0.43 | 0.16 to 1.20 | 0.11 |
| Group 3 and treatment interaction | 0.90 | 0.31 to 2.66 | 0.85 |
| CSM | |||
| Ki-67 and MDM2 | |||
| ≤ 11.3% and ≤ 184 | RL | — | — |
| ≤ 11.3% and > 184 | 0.45 | 0.10 to 1.93 | .28 |
| > 11.3% and ≤ 184 | 2.38 | 1.06 to 5.37 | .04 |
| > 11.3% and > 184 | 3.92 | 1.67 to 9.20 | .002 |
| Treatment arm | |||
| STAD + RT | 1.62 | 0.82 to 3.18 | .16 |
| Group 1 and treatment interaction | 4.57 | 0.87 to 24.01 | .07 |
| Group 2 and treatment interaction | 1.10 | 0.36 to 3.31 | .87 |
| Group 3 and treatment interaction | 1.00 | 0.32 to 3.15 | 1.00 |
| OM | |||
| Ki-67 and MDM2 | |||
| ≤ 11.3% and ≤ 184 | RL | — | — |
| ≤ 11.3% and > 184 | 1.35 | 0.85 to 2.16 | .21 |
| > 11.3% and ≤ 184 | 1.52 | 1.02 to 2.25 | .04 |
| > 11.3% and > 184 | 2.41 | 1.45 to 4.01 | .0007 |
| Treatment arm | |||
| STAD + RT | 1.18 | 0.85 to 1.63 | .32 |
| Group 1 and treatment interaction | 1.05 | 0.53 to 2.07 | .90 |
| Group 2 and treatment interaction | 0.96 | 0.50 to 1.85 | .91 |
| Group 3 and treatment interaction | 0.66 | 0.31 to 1.41 | .28 |
P value is from χ2 test with the Cox proportional hazards model.
Abbreviations: HR, hazard ratio; DM, distant metastasis; RL, reference; STAD, short-term androgen deprivation; RT, radiotherapy; CSM, cause-specific mortality; OM, overall mortality.
DISCUSSION
MDM2 is a regulator of p53-mediated cell cycle arrest and apoptosis; it also reduces androgen-receptor signaling in prostate cancer via p53 inhibition20 and is implicated in the ubiquitination and degradation of androgen receptors via the Akt pathway.21,22 In our laboratory, MDM2 knockdown by antisense oligonucleotide-sensitized, androgen-sensitive, and selected androgen-insensitive, prostate cancer cells to androgen deprivation and RT.6–9 Because MDM2 is overexpressed in some prostate cancers and because overexpression affects response to RT and androgen deprivation, this biomarker has the potential for identification of patients who would benefit from more aggressive therapy, including agents that knock down or interfere with MDM2 actions.
There are three major MDM2 isoforms (90, 75, and 60 kDa) reported in the literature. Of these, the 90 and 60 kDa isoforms appear to be regulated by p53, whereas regulation is unclear for the p75 isoform. The p60 isoform is a caspase 3–mediated, degraded product of MDM2. Both the p90 and p60 forms interact with p53.23 The antibody used in this article (M7146) recognizes the 154-to-167 epitope, which is present in all three major isoforms. There are no MDM2 gene polymorphisms known; however, there is a 309 T/g polymorphism in the promoter; therefore, this polymorphism would not affect antibody reactivity. In addition, the polymorphic site on the promoter does not alter any transcription factor binding site and, hence, does not affect the protein product of MDM2.
The examination of MDM2 expression in patients enrolled on RTOG 92-02 makes this, to our knowledge, the largest investigation of this biomarker in men with prostate cancer. Abnormally high levels of p53 and a high Ki-67 staining index have been significant predictors of DM in previous reports on patient cases from RTOG 92-02.11,12 Our prior analysis of MDM2 expression in patients from RTOG 86-10,10 a smaller study, suggested that this biomarker has promise. The testing of MDM2 with p53 and Ki-67 in patient cases from RTOG 92-02 allows for an assessment of the relative importance of two proteins (ie, p53 and MDM2) that feedback-regulate each other.
The decreasing number of available patient cases for each biomarker is a common problem of immunohistochemical studies of multiple markers and is due to limited tissue in the pretreatment diagnostic needle biopsy specimens. The p53 analysis was the earliest performed and yielded data from 780 patient cases, which was followed by 637 for Ki-67 and finally 589 for MDM2. The final study cohort with all marker data comprised 478 cases. The reduction in available cases is a potential source of bias, which was reflected in differences in the distribution of patients by Gleason score and assigned treatment compared with those who did not have biomarkers available. However, there was no difference in any of the end points tested between those that had biomarkers available and those that did not. Moreover, this is the largest multiple biomarker study in men treated with radiotherapy and sets the stage for validation studies in another patient cohort.
The previous MDM2 study on RTOG protocol 86-10,10 as well as this study, employed automated image analysis as a more reproducible method of evaluating the marker. For uniformity and to test for its usefulness, the p53 and Ki-67 patient cases were also analyzed by image analysis (Table 2). In the multivariate analysis that controlled for clinical and treatment covariates, the image-analysis data for both Ki-67 and p53 were weaker than the manual data, and p53 lost its significance entirely. The p53 manual analysis performed by Che et al11 took into account moderately to strongly-stained nuclei as positive for p53 overexpression. In this study, image-analysis quantification considered all positive nuclei of any intensity. Che et al11 also reported a significant cut point of 20% for p53 overexpression by manual analysis on the basis of prior reports.24–26 When tested on the current data, the 20% cut point was not optimal. For the sake of simplicity, all data were dichotomized at the quartiles.
MDM2 was found to be most predictive of patient outcome as a categoric, rather than a continuous, covariate. These data suggest that, beyond a certain expression threshold level, there is an increased risk of DM. The Ki-67 PSP score was the strongest predictor of DM of all the markers, whereas p53 was weakest in the multivariate analyses. Agus et al27 demonstrated in tumor models that response to androgen deprivation involves a short-term p53 increase that causes cell cycle arrest and then a reduction in the proliferative index by p16 and p27, the cyclin-dependent kinase inhibitors. MDM2 and cyclin D1 are subsequently overexpressed in conjunction with the development of androgen resistance, as a result of a release from cell cycle arrest. Also, in studies of hormone-resistant prostate cancer cell lines, Wu et al28 demonstrated greater in vivo tumor growth rate and increased radioresistance in xenografts in which MDM2 was overexpressed. These results support the findings herein, which suggest that Ki-67 and MDM2 are important in determining the response of patients to androgen deprivation.
In summary, this is the largest quantitative expression study of MDM2, to our knowledge, in a contemporary cohort of men with high risk prostate cancer. The combination of Ki-67 and MDM2 overexpression appear to significantly predispose to DM and OM. If validated in a separate patient population, such as RTOG 94-13, the parameters defined could be applied to predict patient outcome and could prove valuable for risk stratification in clinical trials.
Appendix
Table A1.
Univariate Cox Proportional Hazards Model of Missing v Determined Marker Data
| Marker Data by End Point | Patients |
Analysis |
|||
|---|---|---|---|---|---|
| No. of Patients | No. Who Experienced Failure | HR* | 95% CI | P† | |
| Overall mortality | |||||
| Mssing | 1,043 | 565 | 0.98 | 0.85 to 1.14 | .82 |
| Determined | 478 | 261 | |||
| Cause-specific mortality | |||||
| Missing | 1,043 | 146 | 1.21 | 0.92 to 1.58 | .17 |
| Determined | 478 | 82 | |||
| Distant metastasis | |||||
| Missing | 1,043 | 189 | 1.12 | 0.92 to 1.49 | .20 |
| Determined | 478 | 102 | |||
Abbreviation: HR, hazard ratio.
A hazard ratio of 1 indicated no difference between the two subgroups. The marker data indicator was coded as follows: 0, missing; 1, determined.
P value from χ2 test by using the Cox proportional hazards model.
Table A2.
Summary of Multivariate Analyses of Biomarkers With iPSA As a Continuous Covariate
| Endpoint by Marker | Cut Point | Analysis |
|||||
|---|---|---|---|---|---|---|---|
| Adjusted for Clinical and Treatment Covariates* |
Adjusted for All Biomarkers and Other Covariates† |
||||||
| HR | 95% CI | P‡ | HR | 95% CI | P‡ | ||
| p53 manual PSP | 0 | ||||||
| DM | 1.68 | 1.18 to 2.39 | .004 | 1.08 | 0.70 to 1.67 | .74 | |
| CSM | 1.28 | 0.86 to 1.91 | .23 | 0.86 | 0.54 to 1.39 | .55 | |
| OM | 0.94 | 0.76 to 1.16 | .55 | 0.72 | 0.55 to 0.95 | .02 | |
| Ki-67 manual PSP | 11.3 | ||||||
| DM | 2.57 | 1.73 to 3.82 | < .0001 | 2.87 | 1.84 to 4.47 | < .0001 | |
| CSM | 2.17 | 1.39 to 3.38 | .0006 | 2.33 | 1.41 to 3.85 | .001 | |
| OM | 1.39 | 1.09 to 1.78 | .008 | 1.42 | 1.08 to 1.88 | .01 | |
| MDM2 image-analysis MIS | 184 | ||||||
| DM | 1.83 | 1.21 to 2.77 | .004 | 1.78 | 1.13 to 2.79 | .01 | |
| CSM | 1.63 | 1.03 to 2.58 | .04 | 1.57 | 0.97 to 2.56 | .07 | |
| OM | 1.42 | 1.10 to 1.83 | .007 | 1.55 | 1.17 to 2.07 | .002 | |
Abbreviations: iPSA, initial pretreatment prostate-specific antigen; HR, hazard ratio; PSP, percent staining positive; DM, distant metastasis; CSM, cause-specific mortality; OM, overall mortality; MIS, mean intensity score.
Analyses were adjusted for age (< 70 v ≥ 70 years), iPSA (continuous), Gleason score (2-6 v 7 8-10), clinical stage (T2 v T3-4) and assigned treatment.
Adjusted for all biomarker and the following covariates: age, iPSA, Gleason score, clinical stage, and assigned treatment.
P values are from Fine and Gray's regression model for CSM and DM and from the Cox regression model for OM.
Table A3.
Multivariate Analyses of Combined MDM2 and Ki-67 With iPSA As a Continuous Covariate
| Covariate by End Point | Analysis |
||
|---|---|---|---|
| HR | 95% CI | P* | |
| DM | |||
| Age ≥ 70 years | 0.56 | 0.36 to 0.88 | .01 |
| GLSC | |||
| 7 | 1.16 | 0.59 to 2.27 | .67 |
| 8-10 | 3.77 | 2.10 to 6.77 | < .0001 |
| Baseline PSA, continuous | 1.05 | 0.999 to 1.01 | .08 |
| Clinical stage T3-4 | 1.71 | 1.09 to 2.69 | .02 |
| Assigned treatment of STAD + RT | 2.08 | 1.32 to 3.25 | .002 |
| p53 manual PSP > 0 | 1.09 | 0.70 to 1.69 | .71 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.54 | 0.79 to 3.01 | .20 |
| > 11.3% and ≤ 184 | 2.71 | 1.56 to 4.72 | .0004 |
| > 11.3% and > 184 | 4.87 | 2.75 to 8.60 | < .0001 |
| CSM | |||
| Age ≥ 70 years | 0.61 | 0.38 to 0.995 | .048 |
| GLSC | |||
| 7 | 1.28 | 0.63 to 2.57 | .49 |
| 8-10 | 3.65 | 1.95 to 6.82 | < .0001 |
| iPSA, continuous | 1.00 | 0.99 to 1.01 | .82 |
| Clinical stage T3-4 | 2.49 | 1.49 to 4.14 | .0005 |
| Assigned treatment of STAD + RT | 2.06 | 1.26 to 3.38 | .004 |
| p53 manual PSP > 0 | 0.87 | 0.54 to 1.40 | .56 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.48 | 0.74 to 2.97 | .27 |
| > 11.3% and ≤ 184 | 2.25 | 1.20 to 4.23 | .01 |
| > 11.3% and > 184 | 3.65 | 1.89 to 7.04 | .0001 |
| OM | |||
| Age ≥ 70 years | 1.57 | 1.21 to 2.04 | .0007 |
| GLSC | |||
| 7 | 1.14 | 0.83 to 1.58 | .42 |
| 8-10 | 1.59 | 1.16 to 2.19 | .004 |
| iPSA, continuous | 1.00 | 0.996 to 1.003 | .86 |
| Clinical stage T3-4 | 1.27 | 0.98 to 1.65 | .08 |
| Assigned treatment of STAD + RT | 1.27 | 0.98 to 1.65 | .07 |
| p53 manual PSP > 0 | 0.72 | 0.55 to 0.95 | .02 |
| Ki-67 PSP and MDM2 MIS | |||
| ≤ 11.3% and > 184 | 1.60 | 1.12 to 2.30 | .01 |
| > 11.3% and ≤ 184 | 1.46 | 1.04 to 2.06 | .03 |
| > 11.3% and > 184 | 2.16 | 1.44 to 3.24 | .0002 |
NOTE. Multivariate analyses were performed on 446 patient cases.
P values are from Fine and Gray's regression model for CSM and DM and from the Cox regression model for OM.
Abbreviations: iPSA, initial pretreatment prostate-specific antigen; HR, hazard ratio; DM, distant metastasis; GLSC, Gleason Score; STAD, short term androgen deprivation; RT, radiation therapy; PSP, percent staining positive; MIS, mean intensity score; CSM, cause-specific mortality; OM, overall mortality.
Footnotes
Supported in part by Grants No. CA-006927, CA-101984-01, CA-21661, and CA-32115 from the National Cancer Institute and by grants from Varian Medical Systems, Palo Alto, CA and the Pennsylvania Department of Health. The contents are solely the responsibility of the authors and do not necessarily represent the official views of these organizations.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Alan Pollack, Varian Medical Systems Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Li-Yan Khor, Kyounghwa Bae, M. Elizabeth Hammond, Alan Pollack
Collection and assembly of data: Li-Yan Khor, Tahseen Al-Saleem, M. Elizabeth Hammond, David J. Grignon, Mingxin Che
Data analysis and interpretation: Li-Yan Khor, Kyounghwa Bae, Gerald E. Hanks, Howard M. Sandler
Manuscript writing: Li-Yan Khor, Kyounghwa Bae, Rebecca Paulus, Tahseen Al-Saleem, M. Elizabeth Hammond, David J. Grignon, Mingxin Che, Varagur Venkatesan, Roger W. Byhardt, Marvin Rotman, Gerald E. Hanks, Howard M. Sandler, Alan Pollack
Final approval of manuscript: Li-Yan Khor, Kyounghwa Bae, Rebecca Paulus, Tahseen Al-Saleem, M. Elizabeth Hammond, David J. Grignon, Mingxin Che, Varagur Venkatesan, Roger W. Byhardt, Marvin Rotman, Gerald E. Hanks, Howard M. Sandler, Alan Pollack
REFERENCES
- 1.Momand J, Zambetti GP, Olson DC, et al. The MDM2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 2.Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Marechal V, Levine AJ. Mapping of the p53 and MDM2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao ZX, Chen J, Levine AJ, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 5.Martin K, Trouche D, Hagemeier C, et al. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 6.Mu Z, Hachem P, Agrawal S, et al. Antisense MDM2 oligonucleotides restore the apoptotic response of prostate cancer cells to androgen deprivation. Prostate. 2004;60:187–196. doi: 10.1002/pros.20044. [DOI] [PubMed] [Google Scholar]
- 7.Mu Z, Hachem P, Agrawal S, et al. Antisense MDM2 sensitizes prostate cancer cells to androgen deprivation, radiation, and the combination. Int J Radiat Oncol Biol Phys. 2004;58:336–343. doi: 10.1016/j.ijrobp.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 8.Stoyanova R, Hachem P, Hensley H, et al. Antisense-MDM2 sensitizes LNCaP prostate cancer cells to androgen deprivation, radiation, and the combination in vivo. Int J Radiat Oncol Biol Phys. 2007;68:1151–1160. doi: 10.1016/j.ijrobp.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu Z, Hachem P, Hensley H, et al. Antisense MDM2 enhances the response of androgen insensitive human prostate cancer cells to androgen deprivation in vitro and in vivo. Prostate. 2008;68:599–609. doi: 10.1002/pros.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khor LY, Desilvio M, Al-Saleem T, et al. MDM2 as a predictor of prostate carcinoma outcome: An analysis of Radiation Therapy Oncology Group protocol 8610. Cancer. 2005;104:962–967. doi: 10.1002/cncr.21261. [DOI] [PubMed] [Google Scholar]
- 11.Che M, DeSilvio M, Pollack A, et al. Prognostic value of abnormal p53 expression in locally advanced prostate cancer treated with androgen deprivation and radiotherapy: A study based on RTOG 9202. Int J Radiat Oncol Biol Phys. 2007;69:1117–1123. doi: 10.1016/j.ijrobp.2007.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack A, DeSilvio M, Khor LY, et al. Ki-67 staining is a strong predictor of distant metastasis and mortality for men with prostate cancer treated with radiotherapy plus androgen deprivation: Radiation Therapy Oncology Group Trial 92-02. J Clin Oncol. 2004;22:2133–2140. doi: 10.1200/JCO.2004.09.150. [DOI] [PubMed] [Google Scholar]
- 13.Hanks GE, Pajak TF, Porter A, et al. Phase III trial of long-term adjuvant androgen deprivation after neoadjuvant hormonal cytoreduction and radiotherapy in locally advanced carcinoma of the prostate: The Radiation Therapy Oncology Group protocol 92-02. J Clin Oncol. 2003;21:3972–3978. doi: 10.1200/JCO.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz EM, Bae K, Hanks GE, et al. Ten-year follow-up of Radiation Therapy Oncology Group Protocol 92-02: A phase III trial of the duration of elective androgen deprivation in locally advanced prostate cancer. J Clin Oncol. 2008;26:2497–2504. doi: 10.1200/JCO.2007.14.9021. [DOI] [PubMed] [Google Scholar]
- 15.Brown RE, Lun M, Prichard JW, et al. Morphoproteomic and pharmacoproteomic correlates in hormone-receptor-negative breast carcinoma cell lines. Ann Clin Lab Sci. 2004;34:251–262. [PubMed] [Google Scholar]
- 16.Hilbe W, Gachter A, Duba HC, et al. Comparison of automated cellular imaging system and manual microscopy for immunohistochemically stained cryostat sections of lung cancer specimens applying p53, ki-67, and p120. Oncol Rep. 2003;10:15–20. [PubMed] [Google Scholar]
- 17.Khor LY, Bae K, Pollack A, et al. COX-2 expression predicts prostate-cancer outcome: Analysis of data from the RTOG 92-02 trial. Lancet Oncol. 2007;8:912–920. doi: 10.1016/S1470-2045(07)70280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakravarti A, DeSilvio M, Zhang M, et al. Prognostic value of p16 in locally advanced prostate cancer: A study based on Radiation Therapy Oncology Group protocol 9202. J Clin Oncol. 2007;25:3082–3089. doi: 10.1200/JCO.2006.08.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Statist Assoc. 1999;94:496–509. [Google Scholar]
- 20.Cronauer MV, Schulz WA, Burchardt T, et al. Inhibition of p53 function diminishes androgen receptor-mediated signaling in prostate cancer cell lines. Oncogene. 2004;23:3541–3549. doi: 10.1038/sj.onc.1207346. [DOI] [PubMed] [Google Scholar]
- 21.Lin HK, Wang L, Hu YC, et al. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require MDM2 E3 ligase. Embo J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaughan L, Logan IR, Neal DE, et al. Regulation of androgen receptor and histone deacetylase 1 by MDM2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pochampally R, Fodera B, Chen L, et al. A 60 kd MDM2 isoform is produced by caspase cleavage in non-apoptotic tumor cells. Oncogene. 1998;17:2629–2636. doi: 10.1038/sj.onc.1202206. [DOI] [PubMed] [Google Scholar]
- 24.Visakorpi T, Kallioniemi OP, Heikkinen A, et al. Small subgroup of aggressive, highly proliferative prostatic carcinomas defined by p53 accumulation. J Natl Cancer Inst. 1992;84:883–887. doi: 10.1093/jnci/84.11.883. [DOI] [PubMed] [Google Scholar]
- 25.Grignon DJ, Caplan R, Sarkar FH, et al. p53 status and prognosis of locally advanced prostatic adenocarcinoma: A study based on RTOG 8610. J Natl Cancer Inst. 1997;89:158–165. doi: 10.1093/jnci/89.2.158. [DOI] [PubMed] [Google Scholar]
- 26.Quinn DI, Henshall SM, Head DR, et al. Prognostic significance of p53 nuclear accumulation in localized prostate cancer treated with radical prostatectomy. Cancer Res. 2000;60:1585–1594. [PubMed] [Google Scholar]
- 27.Agus DB, Cordon-Cardo C, Fox W, et al. Prostate cancer cell cycle regulators: Response to androgen withdrawal and development of androgen independence. J Natl Cancer Inst. 1999;91:1869–1876. doi: 10.1093/jnci/91.21.1869. [DOI] [PubMed] [Google Scholar]
- 28.Wu CT, Chen WC, Liao SK, et al. The radiation response of hormone-resistant prostate cancer induced by long-term hormone therapy. Endocr Relat Cancer. 2007;14:633–643. doi: 10.1677/ERC-07-0073. [DOI] [PubMed] [Google Scholar]