Abstract
Purpose
To determine the prognostic importance of the meningioma 1 (MN1) gene expression levels in the context of other predictive molecular markers, and to derive MN1 associated gene– and microRNA–expression profiles in cytogenetically normal acute myeloid leukemia (CN-AML).
Patients and Methods
MN1 expression was measured in 119 untreated primary CN-AML adults younger than 60 years by real-time reverse-transcriptase polymerase chain reaction. Patients were also tested for FLT3, NPM1, CEBPA, and WT1 mutations, MLL partial tandem duplications, and BAALC and ERG expression. Gene- and microRNA-expression profiles were attained by performing genome-wide microarray assays. Patients were intensively treated on two first-line Cancer and Leukemia Group B clinical trials.
Results
Higher MN1 expression associated with NPM1 wild-type (P < .001), increased BAALC expression (P = .004), and less extramedullary involvement (P = .01). In multivariable analyses, higher MN1 expression associated with a lower complete remission rate (P = .005) after adjustment for WBC; shorter disease-free survival (P = .01) after adjustment for WT1 mutations, FLT3 internal tandem duplications (FLT3-ITD), and high ERG expression; and shorter survival (P = .04) after adjustment for WT1 and NPM1 mutations, FLT3-ITD, and WBC. Gene- and microRNA-expression profiles suggested that high MN1 expressers share features with high BAALC expressers and patients with wild-type NPM1. Higher MN1 expression also appears to be associated with genes and microRNAs that are active in aberrant macrophage/monocytoid function and differentiation.
Conclusion
MN1 expression independently predicts outcome in CN-AML patients. The MN1 gene- and microRNA-expression signatures suggest biologic features that could be exploited as therapeutic targets.
INTRODUCTION
Nonrandom cytogenetic abnormalities are among the most important prognostic factors in acute myeloid leukemia (AML).1–4 However, approximately 45% of adults younger than 60 years of age with primary AML have cytogenetically normal (CN) disease at diagnosis and thus lack informative chromosome markers for risk stratification.1–4 Recently, this large cytogenetic group was shown to be composed of subsets differing for the presence of distinct submicroscopic genetic alterations.5
The meningioma 1 (MN1) gene is located at chromosome band 22q12 and encodes a protein that participates in a gene transcription regulator complex with the nuclear receptor RAR-RXR or the vitamin D receptor.6,7 The involvement of this gene in human neoplasia was initially discovered in a case of meningioma carrying t(4;22)8 and also found in myeloid malignancies with t(12;22).9 High levels of MN1 expression were recently associated with inv16 AML,10 and shown, in a mouse model, to cooperate with CBFB-MYH11 gene fusion in the development of AML.11 However, the mechanisms through which aberrant expression of MN1 contributes to malignant transformation remain to be elucidated.12,13
Recently, Heuser et al14 reported that overexpression of MN1 predicted worse outcome in CN-AML patients. To date, however, these results have not been independently corroborated or tested in the context of several other established prognostic markers in CN-AML. Thus, to validate MN1 expression's prognostic importance in CN-AML, we measured the MN1 expression in diagnostic bone marrow (BM) samples from younger adult CN-AML patients that were also comprehensively characterized for other molecular markers associated with outcome. Furthermore, to gain insight into MN1-mediated leukemogenesis, we derived gene- and microRNA-expression signatures associated with changes in MN1 expression levels.
PATIENTS AND METHODS
Patients, Treatment, Cytogenetic, and Molecular Analyses
One hundred nineteen adults younger than 60 years of age with untreated, primary CN-AML with material available for analyses were included. Patients were treated similarly on Cancer and Leukemia Group B (CALGB) protocols 9621 (n = 38) and 19808 (n = 81) with intensive induction chemotherapy and consolidation with autologous peripheral blood stem cell transplantation (SCT; Appendix, online only).15,16 No differences in outcome (complete remission rate [CR], P = .86; disease-free survival [DFS], P = .37; overall survival [OS], P = .33) were observed between the patients studied for MN1 expression and the remaining CN-AML patients not included (n = 121).
Pretreatment BM cytogenetic analyses were performed by CALGB-approved institutional cytogenetic laboratories on CALGB 8461, a prospective cytogenetic companion, and centrally reviewed.17 MN1 copy numbers normalized to ABL copy numbers were measured in BM samples by real-time reverse transcriptase polymerase chain reaction quantification (Appendix). The presence or absence of additional molecular markers such as FLT3 internal tandem duplication (FLT3-ITD),18,19 FLT3 tyrosine kinase domain mutations (FLT3-TKD),20,21 mutations in the NPM1,22 CEBPA,23 and WT124 genes, MLL partial tandem duplication (MLL-PTD),25,26 and ERG27,28 and BAALC29,30 expression levels were assessed centrally. All patients gave informed consent for the research use of their specimens, in accordance with the Declaration of Helsinki.
Gene-Expression and MicroRNA-Expression Profiling
RNA samples from 75 of 81 patients studied for MN1 expression enrolled on CALGB 19808 were analyzed for genome-wide gene expression using Affymetrix U133 plus 2.0 GeneChips (Affymetrix, Santa Clara, CA), as previously reported (Appendix).10,31
Of the 75 samples analyzed for genome-wide gene expression, 73 were also analyzed for genome-wide microRNA expression. Biotinylated first strand cDNA from total RNA extracted from pretreatment BM or blood mononuclear cell samples was synthesized using biotin-labeled random octamer primers and was hybridized onto microRNA microarray chips, as previously reported.32 Images of the microRNA microarrays were acquired as previously reported.33
Statistical Methods
The main objective was to evaluate the impact of MN1 expression on clinical outcome. We defined CR as BM cellularity ≥ 20% and fewer than 5% blasts, and recovery of leukocyte (≥ 1,500/μL) and platelet (> 100,000/μL) counts; relapse as ≥ 5% of BM, leukemic blasts, circulating blasts, or extramedullary leukemia; DFS as the interval from CR achievement until relapse or death, regardless of cause; OS as the date on study until death. Patients alive at last follow-up were censored for both DFS and OS. MN1 expression values were calculated as the natural log transformation of the normalized MN1 copy numbers; this continuous variable was used for all statistical analyses. Pretreatment CNS, spleen, liver, skin, nodes, gum, or mediastinal mass involvement constituted extramedullary disease.
The associations of MN1 expression with baseline clinical, demographic, and molecular features, and achievement of CR were analyzed using one-way analysis of variance. Kaplan-Meier plots were generated for each time-to-event outcome measure (DFS and OS) using MN1 expression quartiles. The corresponding tests for trend were calculated for each survival end point.34 Comparisons between cases analyzed for MN1 v those not analyzed were tested using the Fisher's exact test for CR rates and the log-rank test for the OS and DFS end points.
Multivariable logistic regression models were constructed to analyze factors related to the probability of achieving CR and multivariable Cox proportional hazards models were constructed to analyze factors important for the survival end points, OS and DFS. Factors examined for model inclusion were MN1 expression, FLT3-ITD, FLT3-TKD, NPM1 and WT1 mutational status, age, hemoglobin, platelet count, WBC, percentages of BM and blood blasts, sex, race, and extramedullary involvement, and for survival end points only, MLL-PTD, CEBPA mutational status, and ERG and BAALC expression levels. For the multivariable Cox models, the proportional hazards assumption was checked for each variable individually. If the proportional hazards assumption was not met for a particular variable for a given end point, an artificial time-dependent covariate was included in the model for that end point. Variables considered for inclusion in the logistic and Cox multivariable models were those significant at α = .20 from the univariable models. All models were constructed using a limited backwards selection procedure. Variables remaining in the final models were significant at α = .05. For achievement of CR, estimated odds ratios (OR), and for survival end points, hazard ratios (HR) with their corresponding 95% CIs were obtained for each significant prognostic factor.
For microarray analyses, summary measures of gene and microRNA expression were computed, normalized, and filtered (Appendix).35 Pearson correlation coefficients were computed between the resulting expression values of 24,183 Affymetrix probe sets and the natural log transformation of MN1 expression, and between the resulting expression values of 305 microRNA probes and the natural log transformation of MN1 expression values; significant Affymetrix probe sets (P < .001) and microRNA probes (P < .005) comprised the MN1 gene- and microRNA-expression signatures, respectively. GenMAPP version 2.1 and MAPPFinder version 2.136 (Gladstone Institutes, the University of California, San Francisco, CA; http://www.genmapp.org/) were used to assess over-represented gene ontology (GO) terms within the identified gene-expression signature (Appendix).
All statistical analyses were performed by the CALGB Statistical Center.
RESULTS
Association of MN1 Expression With Molecular and Clinical Characteristics and Outcome
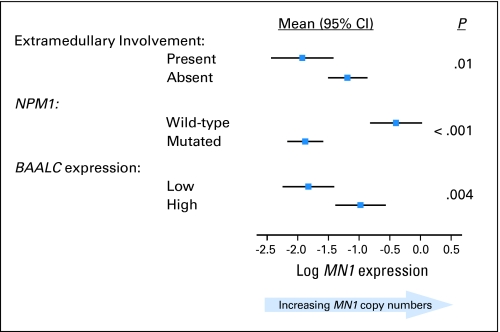
At diagnosis, higher MN1 expression (MN1/ABL copy number range, 0.007 to 7.317) was associated with lower frequency of NPM1 mutations (P < .001) and higher BAALC expression (P = .004) and less extramedullary disease (P = .01; Table 1; Fig 1). No other molecular or clinical characteristics were significantly associated with MN1 expression.
Table 1.
Relationship of Clinical and Molecular Characteristics With MN1 Expression Levels in Patients With Cytogenetically Normal Acute Myeloid Leukemia at Diagnosis (N = 119)
| Characteristic | Summary Statistics |
P * | |
|---|---|---|---|
| No. | % | ||
| Median age, years | 43 | .64 | |
| Range | 18-59 | ||
| Sex | .52 | ||
| Female | 62 | 52 | |
| Male | 57 | 48 | |
| Race | .15 | ||
| White | 104 | 88 | |
| Nonwhite | 14 | 12 | |
| Median hemoglobin, g/L | 92 | .21 | |
| Range | 48-136 | ||
| Median platelet count, ×109/L | 55 | .78 | |
| Range | 8-395 | ||
| Median WBC, ×109/L | 27.3 | .94 | |
| Range | 1.4-273.0 | ||
| Median blood blasts, % | 59 | .78 | |
| Range | 0-95 | ||
| Median bone marrow blasts, % | 67 | .20 | |
| Range | 21-99 | ||
| Extramedullary involvement | .01 | ||
| No | 85 | 72 | |
| Yes | 33 | 28 | |
| FLT3-ITD | .30 | ||
| Negative | 66 | 55 | |
| Positive | 53 | 45 | |
| FLT3-TKD | .06 | ||
| Negative | 109 | 92 | |
| Positive | 10 | 8 | |
| NPM1 | < .001 | ||
| Wild-type | 39 | 33 | |
| Mutated | 80 | 67 | |
| CEBPA | .15 | ||
| Wild-type | 97 | 83 | |
| Mutated | 20 | 17 | |
| ERG expression | .86 | ||
| Low | 50 | 56 | |
| High | 39 | 44 | |
| BAALC expression | .004 | ||
| Low | 46 | 50 | |
| High | 46 | 50 | |
| WT1 | .23 | ||
| Wild-type | 101 | 90 | |
| Mutated | 11 | 10 | |
| MLL-PTD | .81 | ||
| Negative | 111 | 93 | |
| Positive | 8 | 7 | |
NOTE. Not all 119 patients were evaluated for all the molecular markers. For each molecular marker, the number of patients negative or positive, wild-type or mutated or low or high is reported in the Summary Statistics column.
Abbreviations: FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation of the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene.
P values are from the one-way analysis of variance overall F-test, evaluating the presence of any linear relationship between the MN1 expression and the variable tested. For tests with a P value < .20, the characteristic associated with higher MN1 expression appears in bold.
Fig 1.
Clinical and molecular variables significantly associated with the meningioma 1 (MN1) gene expression. The direction of the correlation is shown by displaying the mean values and corresponding 95% CIs of MN1 expression for each category of the clinical and molecular variables.
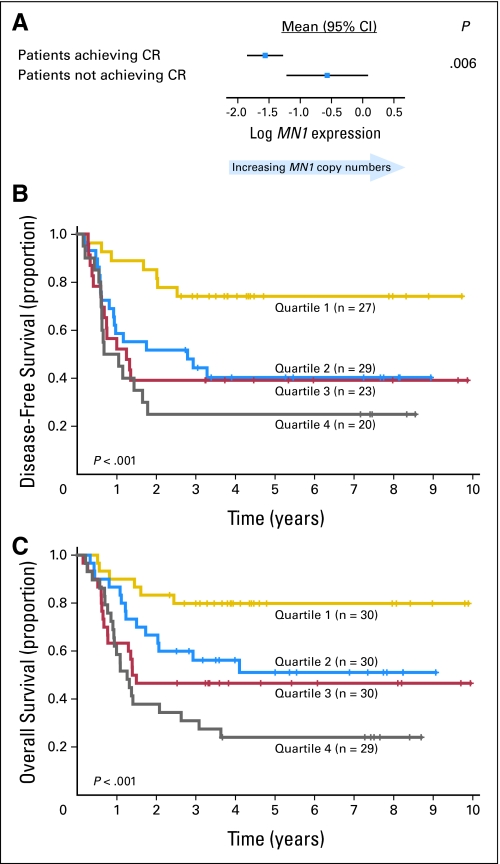
The overall CR rate of the patients analyzed for MN1 expression was 83%. Patients who failed to achieve CR had higher MN1 levels (P = .006; Fig 2A). No interaction between MN1 levels and induction treatment (ie, with or without PSC-833) was found for CR achievement. On multivariable analysis, patients with higher MN1 expression were less likely to achieve CR (P = .005) after adjustment for WBC (P = .005; Table 2).
Fig 2.
Outcome of cytogenetically normal acute myeloid leukemia (CN-AML) patients according to the meningioma 1 (MN1) gene expression levels. (A) Comparison of MN1 expression in patients who achieved a complete remission (CR) compared with patients who did not achieve a CR. The direction of the correlation is shown by displaying the mean MN1 expression and corresponding 95% CIs. (B) Disease-free survival of CN-AML patients according to quartile value of MN1 expression levels. CR rates for each quartile are as follows: 90%, 97%, 77%, 69% for Q1, Q2, Q3, Q4, respectively. (C) Overall survival of CN-AML patients according to quartile value of MN1 expression levels. For display purposes, MN1 expression was treated as a categoric variable (patients were grouped according to the MN1 copy quartiles from the lowest [quartile 1] to the highest [quartile 4] and Kaplan-Meier plots were generated). P values evaluate the trend in survival across MN1 expression quartiles.
Table 2.
Multivariable Analyses for Clinical Outcome
| Variables in Final Model by End Point | HR/OR | 95% CI | P |
|---|---|---|---|
| CR* | |||
| MN1 expression | 0.54 | 0.35 to 0.83 | .005 |
| WBC | 0.52 | 0.33 to 0.82 | .005 |
| DFS† | |||
| MN1 expression | 1.35 | 1.06 to 1.72 | .01 |
| WT1, mutated v wild-type | 3.16 | 1.28 to 7.81 | .01 |
| FLT3-ITD, positive v negative | 2.18 | 1.03 to 4.61 | .02‡ |
| ERG expression, high v low | 1.99 | 1.03 to 3.84 | .04 |
| OS§ | |||
| MN1 expression | 1.27 | 1.01 to 1.58 | .04 |
| WT1, mutated v wild-type | 6.00 | 2.80 to 12.86 | < .001‡ |
| NPM1, wild-type v mutated | 2.23 | 1.04 to 4.76 | .04 |
| FLT3-ITD, positive v negative | 2.70 | 1.41 to 5.17 | .01‡ |
| WBC | 1.75 | 1.34 to 2.28 | < .001 |
NOTE. ORs < 1.0 mean lower CR rate for the higher values of the continuous variables. HRs > 1.0 indicate higher risk for an event for the higher values of the continuous variables and the first category listed for the categorical variables.
Abbreviations: CR, complete remission; DFS, disease-free survival; OS, overall survival; HR, hazard ratio; OR, odds ratio; MN1, natural log transformation of normalized MN1 copy numbers; WBC, white blood count in 50 unit increments; FLT3-ITD, internal tandem duplication of the FLT3 gene.
Variables considered in the model based on univariable analyses were MN1, FLT3-ITD (positive v negative), age, hemoglobin, WBC (50 unit increments), and race.
Variables considered in the model based on univariable analyses were MN1, ERG expression (high v low), WT1 (mutated v wild-type), FLT3-ITD (positive v negative), WBC (50 unit increments), race, and NPM1 (wild-type v mutated).
Does not meet the proportional hazards assumption. For DFS, the hazard ratio for FLT3-ITD is reported at 9 months (was not significant after this time point). for OS, the hazard ratio for WT1 is reported at 9 months (not significant before this time point), FLT3-ITD is reported at 1 year (not significant after this time point).
Variables considered in the model based on univariable analyses were MN1, ERG expression (high v low), FLT3-ITD (positive v negative), WT1 (mutated v wild-type), BAALC expression (high v low), WBC (50 unit increments), age, hemoglobin, percentage of blood blasts, extramedullary involvement, and NPM1 (wild-type v mutated).
The median follow-up for patients with no event (ie, failure to achieve CR, relapse, or death) was 5.1 years (range, 2.7 to 9.9 years). Higher MN1 expression was associated with shorter DFS (P < .001) and OS (P < .001). An interaction between MN1 levels and variations in the consolidation or maintenance treatments could not be evaluated because of sample size limitations. In multivariable models, higher MN1 expression was associated with shorter DFS (P = .01) after adjusting for WT1 mutations (P = .01), FLT3-ITD (P = .02), and high ERG expression (P = .04). Likewise, shorter OS (P = .04) was associated with higher MN1 expression when controlling for WT1 (P < .001) and NPM1 mutations (P = .04), FLT3-ITD (P = .01), and WBC (P < .001; Table 2). Similar results were observed when the FLT3-ITD/FLT3 wild-type allelic ratio (no FLT3-ITD v FLT3-ITD/FLT3 wild-type < .7 v FLT3-ITD/FLT3 wild-type ≥ .7) rather than presence compared with absence of FLT3-ITD, was utilized as a factor in the multivariable models.
To graphically display the relationship between MN1 expression and clinical outcome, patients were divided into four groups corresponding to the quartile (Q) values of MN1 expression (Figs 2B, 2C). The 5-year DFS and OS estimates were progressively lower from Q1 (ie, patients with the lowest 25% of MN1 expression values) to Q4 (ie, patients with the highest 25% of MN1 expression values; P < .001, test for trend for both DFS and OS). Patients in Q1 had remarkably favorable outcomes, with expected 5-year DFS and OS rates of 74% and 80%, respectively, compared with only 36% and 40%, respectively, for the remaining patients.
Biologic Insights
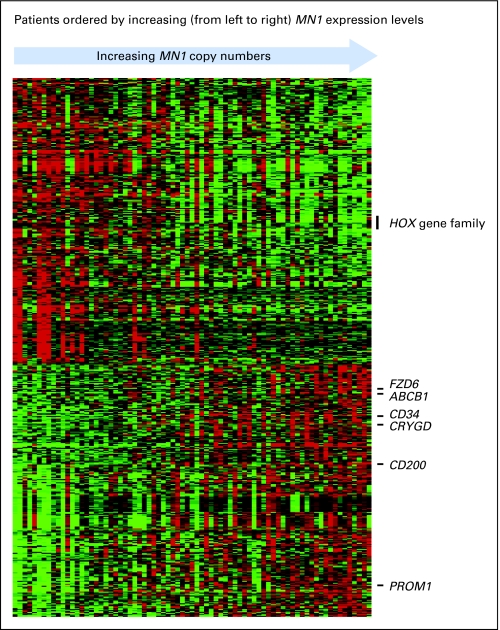
To gain insight into leukemogenic mechanisms associated with changes in MN1 expression, we derived both gene- and microRNA-expression signatures using microarray assays. The MN1-associated gene-expression signature consisted of 555 probes (Appendix Table A1, online only; Fig 3). Expression of 261 probe sets positively correlated with MN1 expression levels, and expression of 294 probe sets negatively correlated with MN1 expression levels. The probe set for MN1 had the highest positive coefficient of correlation (r = .87), corroborating the quantification of MN1 expression obtained by real-time RT-PCR. Furthermore, we found MN1 expression levels to be directly correlated with BAALC expression levels and with the expression of genes recently reported as associated with a BAALC expression signature,30 specifically, PROM1, CD34, FZD6, CRYGD, CD200, and ABCB1 (MDR1). MN1 expression levels were negatively associated with expression of HOX genes (ie, HOXA2, HOXA3, HOXA4, HOXA5, and MEIS1) that have also been reported to be expressed at lower levels in NPM1 wild-type patients.37 Thus, the microarray data were consistent with the association between higher MN1 levels and high BAALC expresser and NPM1 wild-type status observed at diagnosis in our patients (Table 1; Fig 1).
Fig 3.
Heat map of gene probe sets that correlated significantly with the meningioma 1 (MN1) gene expression. Expression values of the probe sets are represented by color, with green indicating expression below and red expression above the median value for the given probe set. For display purposes, the expression values of the probe sets were centered so that each probe set has the same median expression value. Rows represent probe sets and columns represent patients. Patients are ordered according to MN1 expression levels measured by real-time reverse transcriptase polymerase chain reaction.
Using GO (www.geneontology.org), a project that groups together genes (referred to as members) participating in specific biologic processes (referred to as terms), we tested separately which terms were over-represented among the genes positively and negatively correlated with MN1 expression levels. An over-represented term is one for which more members assigned to that term are found in the microarray signature than expected by chance. Thus, over-represented terms may provide insight into the biologic functions of the gene-expression signature associated with MN1 expression changes. Sixteen GO terms were over-represented among the 261 gene probes positively correlated with MN1 expression (Appendix Table A2, online only). Most of the 16 GO terms were related to the macrophage immune function of antigen processing and presentation.38 Twenty-nine GO terms were over-represented among the 294 probe sets that negatively correlated with MN1 expression (Appendix Table A2). Among those 29 GO terms, most were related to DNA, chromatin or chromosome organization, and tissue and organ development.
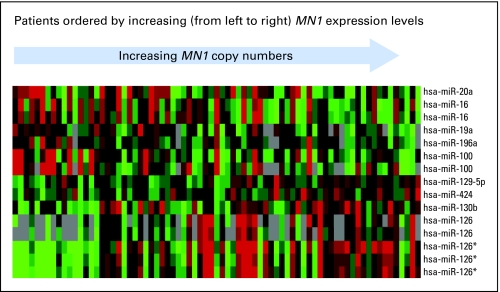
We derived an MN1-associated microRNA-expression signature comprising 15 microRNAs (Appendix Table A3, online only; Fig 4). Of the 15 microRNA probes, expression of 8 was positively and expression of 7 negatively correlated with MN1 expression. Five of 8 microRNA probes positively associated with MN1 expression corresponded to the hsa-miR-126 family (including both hsa-miR-126 and hsa-miR-126*). This microRNA family was recently reported to enhance the proangiogenic activity of VEGF and regulate new blood vessel formation.39,40 We also noted upregulation of hsa-miR-424, a regulator of monocyte and macrophage differentiation.41 Among the microRNA probes negatively correlated with MN1, we found microRNAs involved in apoptosis (ie, hsa-miR-16)42 or malignant transformation (ie, hsa-miR-19a and hsa-miR-20a members of the miR-17-92 polycistron)43,44 in addition to other microRNAs with unknown gene targets (ie, hsa-miR-100 and hsa-miR-196a).
Fig 4.
Heat map of microRNA probes that correlated significantly with the meningioma 1 (MN1) gene expression. Expression values of the probes are represented by color, with green indicating expression below and red expression above the median value for the given probe, and gray indicating a missing value. For display purposes, the expression values of the probes were centered so that each probe has the same median expression value. Rows represent probes and columns represent patients. Patients are ordered according to MN1 expression levels measured by real-time reverse transcriptase polymerase chain reaction.
DISCUSSION
High levels of MN1 expression were recently reported to negatively impact on outcome of CN-AML patients.14 To our knowledge, these results have not yet been independently validated. Thus, we tested the prognostic value of MN1 expression levels in younger CN-AML patients enrolled on similar CALGB first-line treatment protocols. We showed that the levels of MN1 expression directly correlated with the risk of failing remission induction chemotherapy, relapse, and/or death, and predicted outcome independently of other clinical and molecular variables, thereby confirming the initial observation by Heuser et al.14 All patients survived ≥ 30 days and were assessable for disease response after treatment. Therefore, the value of MN1 expression to predict treatment-related mortality was not assessed.
The two studies presented several methodologic differences. We analyzed exclusively patients diagnosed with primary AML, whereas Heuser et al14 also included patients with secondary AML. The patients in our study were similarly treated on two CALGB protocols that included consolidation treatment with autologous SCT (ASCT) or, in those few cases where ASCT was not possible, intensive consolidation chemotherapy. Patients who underwent allogeneic SCT in first CR were excluded. The German study included patients who underwent allogeneic SCT in addition to those receiving consolidation with ASCT or intensive chemotherapy.14 In the German study,14 MN1 expression levels were measured using both BM and blood samples, and comparison of outcome was performed between higher and lower MN1 expressers, dichotomized at the median value of MN1 expression. In our study, only BM samples were analyzed, and we considered MN1 expression as a continuous variable to avoid the need to adjust for different tissue types and eliminate the necessity of choosing arbitrary cutoff values to define groups of patients for comparison. Finally, Heuser et al14 analyzed patients only for FLT3-ITD, FLT3-TKD, MLL-PTD, and NPM1 mutations along with MN1 expression. In addition to these molecular markers, we also analyzed WT1 and CEBPA mutations, and ERG and BAALC expression levels. Despite these differences, the two studies were remarkably similar in their conclusions regarding the association of higher MN1 levels with wild-type NPM1 and poor outcome. However, while Heuser et al14 showed that MN1 expression was the only molecular marker that remained predictive of outcome in the final bivariable and multivariable models, we found that MN1 expression provided prognostic information additional to that provided by FLT3-ITD and WT1 mutations (for DFS and OS), high ERG expression (for DFS), and NPM1 mutations (for OS).
Previous studies reported MN1 as a fusion partner in the MN1/ETV6 chimeric gene in t(12;22), and to be overexpressed in inv16 AML.11,12 MN1 overexpression was shown to confer resistance to the differentiation activity of all-trans-retinoic acid (ATRA) in AML.13 Although murine models have in part recapitulated the ATRA-resistant phenotype of human MN1-associated AML, little is known about the mechanism through which aberrant expression of MN1 drives myeloid leukemogenesis.11,13 Thus, to gain insight into the functional significance of MN1 expression in AML, we derived gene and microRNA profiles that correlated with MN1 expression levels.
The gene-expression signature associated with MN1 expression comprised 555 probes. Notably, BAALC was among genes that correlated most strongly with MN1 expression. At diagnosis, high BAALC expressers indeed had higher levels of MN1 expression (Table 1; Fig 1). Consistent with this finding, we observed similarities between a signature associated with BAALC expression that we recently reported30 and the signature associated with MN1 expression. Associated with higher MN1 and BAALC expression were PROM1, CD34, FZD6, and CRYGD (genes expressed in noncommitted hematopoietic precursors), CD200 (associated with poor outcome in AML), and ABCB1 (involved in chemoresistance). Furthermore, in a comparative GO analysis (not shown), eight GO terms related to DNA, chromatin, and chromosome assembly, and organization were over-represented among the genes downregulated in both the BAALC and MN1 gene-expression signatures. These findings suggest a potential functional interplay between MN1 and BAALC in their contribution to myeloid leukemogenesis.
Despite the aforementioned similarities, the leukemogenic mechanisms associated with aberrant expression of MN1 and BAALC are unlikely to be identical. Using GO analysis, we showed that genes involved in antigen processing and presentation were positively associated with the MN1, but not BAALC, gene-expression signature. Among those, there were genes encoding both MHC class I and class II proteins and CD74 that are central to the mechanisms of antigen processing and presentation for T-cell activation by macrophage and dendritic cells.38 Interestingly, higher MN1 expression was also associated with higher expression of hsa-miR-424, which is transactivated by SPI1 (PU.1) and upregulated during monocyte/macrophage differentiation.41 We have recently published data suggesting that overexpression of certain microRNAs that potentially target genes encoding Toll-like receptors and IL1B, which also participate in macrophage and dendritic cell activation, are associated with worse prognosis.33 Altogether, these data suggest that aberrant activation of mechanisms involved in both native and acquired immunologic response may play a role in sustaining myeloblast proliferation and survival.
Among the eight microRNA probes whose higher expression was associated with higher MN1 expression, five corresponded to miR-126 family members. hsa-miR-126 and hsa-miR-126* are generated from the splicing and processing of intron 7 of the EGFL7 gene.40,45 Consistent with these data, we observed that MN1 expression positively correlated with expression of both EGFL7 and hsa-miR-126. A leukemogenic role for hsa-miR-126 has hitherto not been reported. However, two recent studies have shown that hsa-miR-126 regulates vascular integrity and angiogenesis by repressing negative regulators of the VEGF pathways.39,40 Whether aberrant activation of these mechanisms can contribute to leukemogenesis and impact the treatment response and outcome of CN-AML patients remains to be determined. Finally, since hsa-miR-16 targets the antiapoptotic BCL2 gene and is downregulated in cancer patients with poor outcome,42 it is not surprising that lower hsa-miR-16 expression was associated with higher MN1 expression predicting treatment resistance and worse outcome. It was somewhat surprising, however, that lower expression levels of hsa-miR-19a and hsa-miR-20, both part of the hsa-miR17-92 cluster, were associated with higher MN1 levels as this cluster was previously reported to be overexpressed in aggressive neoplasms (ie, B-cell lymphoma and lung cancer) and function as an oncogene.43,44
In summary, we show that higher MN1 expression is associated with wild-type NPM1, higher BAALC expression and worse outcome in CN-AML independent of other prognostic molecular markers. Patients with higher MN1 expression appear to share biologic features with patients with higher BAALC expression, namely upregulation of genes involved in chemoresistance in noncommitted hematopoietic precursors, and/or those with wild-type NPM1 (ie, lower expression of HOX genes). Aberrant MN1 expression seemingly contributes to leukemogenesis by affecting mechanisms of monocytic/macrophage function and differentiation. Validation of these findings in preclinical models and larger clinical studies may lead to the designing of novel therapies targeting activation of these potentially leukemogenic mechanisms by MN1 overexpression.
Supplementary Material
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study: The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, and Nyla A. Heerema (grant no. CA77658); North Shore–Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R.K. Koduru (grant no. CA35279); Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Wendy L. Flejter, and Mark J. Pettenati (grant no. CA03927); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett, Michael S. Watson, and Jaime Garcia-Heras (grant no. CA77440); University of Massachusetts Medical Center, Worcester, MA: William W. Walsh, Vikram Jaswaney, Michael J. Mitchell, and Patricia Miron (grant no. CA37135); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (grant no. CA02599); Eastern Maine Medical Center, Bangor, ME: Harvey M. Segal and Laurent J. Beauregard (grant no. CA35406); University of Puerto Rico School of Medicine, San Juan, Puerto Rico: Eileen I. Pacheco, Leonard L. Atkins, Cynthia C. Morton, and Paola Dal Cin; Dana-Farber Cancer Institute, Boston, MA: Eric P. Winer, Paola Dal Cin, and Cynthia C. Morton (grant no. CA32291); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff and Thuluvancheri K. Mohandas (grant no. CA04326); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Mazin B. Qumsiyeh (grant no. CA47577); University of Chicago Medical Center, Chicago, IL: Gini Fleming, Diane Roulston, Katrin M. Carlson, Yanming Zhang, and Michelle M. Le Beau (grant no. CA41287); University of Iowa Hospitals, Iowa City, IA: Gerald H. Clamon and Shivanand R. Patil (grant no. CA47642); University of North Carolina, Chapel Hill, NC: Thomas C. Shea and Kathleen W. Rao (grant no. CA47559); University of California at San Diego: Barbara A. Parker, Renée Bernstein, and Marie L. Dell'Aquila (grant no. CA11789); Christiana Care Health Services Inc, Newark, DE: Stephen S. Grubbs and Jeanne M. Meck (grant no. CA45418); Ft Wayne Medical Oncology/Hematology, Ft Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Georgetown University Medical Center, Washington, DC: Minnetta C. Liu and Jeanne M. Meck (grant no. CA77597); Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Paola Dal Cin, and Cynthia C. Morton (grant no. CA 12,449); Rhode Island Hospital, Providence, RI: William Sikov, Shelly L. Kerman, and Aurelia Meloni-Ehrig (grant no. CA08025); State University of New York Upstate Medical University, Syracuse, NY: Stephen L. Graziano and Constance K. Stein (grant no. CA21060); Virginia Commonwealth University Minority Based Community Clinical Oncology Program, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (grant no. CA52784); Weill Medical College of Cornell University, New York, NY: John Leonard, Prasad R.K. Koduru, and Andrew J. Carroll (grant no. CA07968); Western Pennsylvania Hospital, Pittsburgh, PA: John Lister and Gerard R. Diggans; Vermont Cancer Center, Burlington, VT: Hyman B. Muss and Mary Tang (grant no. CA77406); Long Island Jewish Medical Center CCOP, Lake Success, NY: Kanti R. Rai and Prasad R.K. Koduru (grant no. CA11028); Medical University of South Carolina, Charleston, SC: Mark R. Green and G. Shashidhar Pai (grant no. CA03927); Minneapolis VA Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (grant no. CA47555); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (grant no. CA04457); Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John A. Ellerton and Marie L. Dell'Aquila (grant no. CA35421); University of California at San Francisco: Charles J. Ryan and Kathleen E. Richkind (grant no. CA60138); University of Illinois at Chicago: David J. Peace and Maureen M. McCorquodale (grant no. CA74811); University of Minnesota, Minneapolis, MN: Bruce A. Peterson and Betsy A. Hirsch (grant no. CA16450); University of Nebraska Medical Center, Omaha, NE: Anne Kessinger and Warren G. Sanger (grant no. CA77298); Walter Reed Army Medical Center, Washington, DC: Brendan M. Weiss and Digamber S. Borgaonkar (grant no. CA26806).
Treatment
Patients enrolled in Cancer and Leukemia Group B (CALGB) study 19808 were randomly assigned to receive induction chemotherapy with cytarabine, daunorubicin, and etoposide with or without PSC-833 (valspodar), a multidrug resistance protein inhibitor (Kolitz JE, George SL, Marcucci G, et al: Blood 106:122a-123a, 2005 [abstr 407]). On achievement of complete remission (CR), patients were assigned to intensification with high-dose cytarabine and etoposide for stem cell mobilization followed by myeloablative treatment with busulfan and etoposide supported by autologous peripheral blood stem cell transplantation. Patients enrolled in CALGB 9621 were treated similarly to those in CALGB 19808, as previously reported (Kolitz JE, George SL, Dodge RK, et al: J Clin Oncol 22:4290-4301, 2004). The only difference was that CALGB 9621 tested dose escalation of daunorubicin and etoposide during induction treatment, whereas the doses of these drugs were the same for all patients enrolled onto CALGB 19808. In addition, all patients on CALGB 9621 who achieved CR were assigned to receive interleukin-2, whereas on CALGB 19808, patients were randomly assigned to either receive interleukin-2 or observation.
Criteria for Response, Relapse, and Definition of Clinical End Points
CR was defined by bone marrow (BM) cellularity of at least 20%, lower than 5% leukemic blasts, no Auer rods, and maturation in all cell lineages and blood recovery of leukocyte (≥ 1,500/μL) and platelet (> 100,000/μL) counts. Relapse was defined as reoccurrence of ≥ 5% of leukemic blasts in BM, reappearance of circulating blasts, or the development of extramedullary leukemia. Disease-free survival was defined as the interval from the date of CR until removal from study due to relapse or death from any cause, censoring for patients alive at last follow-up. Overall survival was defined as the date on study until death, censoring for patients alive at last follow-up.
Clinical Outcome
The CR rate for the 119 cases analyzed for the meningioma 1 (MN1) gene expression was not different from those 121 that were not analyzed (83% v 84%; P = .86). Likewise, the time to event end points were similar between the two groups (3 year DFS: 47% v 46%; P = .37; 3 years OS: 54% v 49%; P = .33).
MN1 Analysis
Mononuclear cells from pretreatment BM were enriched by Ficoll-Hypaque gradient and cryopreserved in liquid nitrogen until they were thawed at 37°C for this analysis. Total RNA extraction was performed using Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using MMLV reverse transcriptase (Invitrogen) and random hexamers. Quantitative real-time RT-PCR assays were carried out in a final reaction volume of 10 μL using 1 μL of cDNA, 1x universal master mix (Applied Biosystems, Foster City, CA) and 250 nmol MN1 probe (5′-FAM AACAGCAAAGAAGCCCACGACCTCC-TAMRA) with 900 nmol MN1 forward (5′-GAAGGCCAAACCCCAGAAC) and reverse (5′-GATGCTGAGGCCTTGTTTGC) primers. Primers and probe were designed using Primer Express software v2.0 (Applied Biosystems). For ABL, used here as an internal control, the previously described primers and probes were used (Beillard E, Pallisgaard N, van der Velden VHJ, et al: Leukemia 17:2474-2486, 2003). Samples were tested in duplicates on the 7900HT Fast Real-Time PCR System (Applied Biosystems). Positive controls (cDNA from the MN1 expressing cell line KG1a), negative controls (water control of the cDNA synthesis), and standard curves (serial dilutions of plasmids containing MN1 or ABL cloned fragments) were included in each run. MN1 copy numbers were measured and normalized to the copy numbers of ABL using standard curves constructed as reported previously (Marcucci G, Caligiuri MA, Döhner H, et al: Leukemia 15:1072-1080, 2001).
The independent prognostic value of MN1 expression was evaluated in the context of other prognostic clinical and molecular markers, as detailed in the statistical section of the article. For the statistical analyses, we did not impute missing data. Patients with available data on all variables were used in each step of the multivariable analyses.
Microarray Data Analysis
RNA samples from patients enrolled on CALGB 19808 and studied for MN1 expression were analyzed for genome-wide gene expression using Affymetrix U133 plus 2.0 GeneChips (Affymetrix). Double-stranded cDNA was prepared (Invitrogen, Carlsbad, CA) from total RNA using T7-Oligo(dT) primer (Affymetrix). In vitro transcription was performed with the BioArray HighYield RNA Transcript Labeling Kit (T7) (Enzo Life Science, Farmingdale, NY). Fragmented, biotinylated RNA samples were hybridized to the U133 plus 2.0 GeneChip for 16 hours at 45°C. Scanned images were converted to CEL files using GCOS software (Affymetrix).
For the gene expression microarrays, summary measures of the expression levels were computed for each probe set using the robust multichip average method, which incorporates quantile normalization of arrays (Irizarry RA, Bolstad BM, Collin F, et al: Nucleic Acids Res 31:e15, 2003). Expression values were logged (base 2) before analysis. A filtering step was performed to remove probe sets that did not display significant variation in expression across arrays. In this procedure, a χ2 test was used to test whether the observed variance in expression of a probe set was significantly larger than the median observed variance in expression for all probe sets using α = .01 as the significance level. A total of 24,183 probe sets passed the filtering criterion and were included in subsequent analyses.
RNA samples from patients enrolled on CALGB 19808 and studied for MN1 expression were also analyzed for genome-wide microRNA expression with microRNA microarray chips as previously reported (Marcucci G, Radmacher MD, Maharry K, et al: N Engl J Med 358:1919-1928, 2008). For microRNA microarrays, the signal intensity was calculated for each spot without adjusting for local background. Spots with a low signal-to-noise ratio were considered as missing values. Intensities were log-transformed and log-intensities from replicate spots were averaged. A median-centering normalization was performed based on all human microRNA probes represented on the array. MicroRNA probes with a low signal-to-noise ratio on 50% or more of arrays were excluded from subsequent analyses, reducing the number of examined human microRNA probes in the training set to 305. For each microRNA probe, an adjustment was made for batch effects (ie, differences in expression related to the batch in which arrays were hybridized). The batch adjustment was made by fitting a linear model for the expression values of each microRNA probe with array batch as the factor. A correction to the expression values was then made for the measured batch effects.
Microarray expression analyses were performed using BRB-ArrayTools version 3.7.0 (R. Simon, A.P. Lam, National Cancer Institute, Bethesda, MD) and using the R version 2.5.1 (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org/).
Gene Ontology Analysis
We tested separately which gene ontology terms for biologic processes were over-represented among the genes that positively and negatively correlated with MN1 expression levels. An over-represented term is one for which more members assigned to that term are found in the microarray signature than would be expected by chance. In our analysis, we only considered gene ontology terms for biologic processes for which at least 5 members (ie, genes) of the term were included in our microarray analysis. GenMAPP version 2.1 and MAPPFinder version 2.1 (Dahlquist KD, Salomonis N, Vranizan K, et al: Nat Genet 31:19-20, 2002) were used to assess over-represented gene ontologies among the genes comprising the identified signature. MAPPFinder uses a permutation procedure to determine the over-represented gene ontologies; a permutation P value of < .005 was considered significant.
Table A1.
Signature of 555 Affymetrix Probe Sets Significantly Correlated With MN1 Expression Level, Grouped by Direction of Correlation and Ordered Alphabetically by Gene Symbol
| Probe Set | Gene Symbol | Name | Pearson Correlation | P |
|---|---|---|---|---|
| Positively correlated with MN1 expression | ||||
| 209993_at | ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 0.468 | 2.29E-05 |
| 209994_s_at | ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 0.458 | 3.66E-05 |
| 243951_at | ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 0.446 | 6.14E-05 |
| 202850_at | ABCD3 | ATP-binding cassette, sub-family D (ALD), member 3 | 0.373 | .000987 |
| 232081_at | ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 0.491 | 7.70E-06 |
| 204567_s_at | ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 0.381 | .000755 |
| 1570432_at | ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 0.373 | .000968 |
| 204638_at | ACP5 | Acid phosphatase 5, tartrate resistant | 0.453 | 4.49E-05 |
| 1554974_at | ACY3 | Aspartoacylase (aminocyclase) 3 | 0.412 | .00024 |
| 230481_at | ACY3 | Aspartoacylase (aminocyclase) 3 | 0.375 | .000912 |
| 209321_s_at | ADCY3 | Adenylate cyclase 3 | 0.463 | 2.84E-05 |
| 209320_at | ADCY3 | Adenylate cyclase 3 | 0.378 | .000833 |
| 221718_s_at | AKAP13 | A kinase (PRKA) anchor protein 13 | 0.432 | .000108 |
| 224884_at | AKAP13 | A kinase (PRKA) anchor protein 13 | 0.388 | .000585 |
| 208325_s_at | AKAP13 | A kinase (PRKA) anchor protein 13 | 0.380 | .000786 |
| 200602_at | APP | Amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease) | 0.410 | .000258 |
| 237571_at | APP | Amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease) | 0.385 | .000639 |
| 239567_at | ARHGAP10 | Rho GTPase activating protein 10 | 0.450 | 5.13E-05 |
| 225166_at | ARHGAP18 | Rho GTPase activating protein 18 | 0.470 | 2.09E-05 |
| 222780_s_at | BAALC | Brain and acute leukemia, cytoplasmic | 0.569 | 1E-07 |
| 218899_s_at | BAALC | Brain and acute leukemia, cytoplasmic | 0.561 | 2E-07 |
| 210201_x_at | BIN1 | Bridging integrator 1 | 0.382 | .00073 |
| 214439_x_at | BIN1 | Bridging integrator 1 | 0.379 | .000801 |
| 229801_at | C10orf47 | Chromosome 10 open reading frame 47 | 0.439 | 8.08E-05 |
| 230051_at | C10orf47 | Chromosome 10 open reading frame 47 | 0.416 | .000207 |
| 233138_at | C18orf1 | Chromosome 18 open reading frame 1 | 0.439 | 8.10E-05 |
| 242551_at | C18orf1 | Chromosome 18 open reading frame 1 | 0.438 | 8.64E-05 |
| 210785_s_at | C1orf38 | Chromosome 1 open reading frame 38 | 0.455 | 4.10E-05 |
| 207571_x_at | C1orf38 | Chromosome 1 open reading frame 38 | 0.451 | 4.83E-05 |
| 223039_at | C22orf13 | Chromosome 22 open reading frame 13 | 0.418 | .000187 |
| 221823_at | C5orf30 | Chromosome 5 open reading frame 30 | 0.378 | .000814 |
| 1554486_a_at | C6orf114 | Chromosome 6 open reading frame 114 | 0.378 | .000839 |
| 223075_s_at | C9orf58 | Chromosome 9 open reading frame 58 | 0.403 | .000337 |
| 209583_s_at | CD200 | CD200 molecule | 0.514 | 2.40E-06 |
| 209582_s_at | CD200 | CD200 molecule | 0.470 | 2.08E-05 |
| 203593_at | CD2AP | CD2-associated protein | 0.466 | 2.55E-05 |
| 209933_s_at | CD300A | CD300a molecule | 0.442 | 7.18E-05 |
| 217078_s_at | CD300A | CD300a molecule | 0.392 | .000511 |
| 209543_s_at | CD34 | CD34 molecule | 0.431 | .000114 |
| 209619_at | CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | 0.389 | .00057 |
| 218451_at | CDCP1 | CUB domain containing protein 1 | 0.387 | .000611 |
| 239317_at | CEACAM21 | Carcinoembryonic antigen-related cell adhesion molecule 21 | 0.563 | 1E-07 |
| 213618_at | CENTD1 | Centaurin, delta 1 | 0.444 | 6.55E-05 |
| 206210_s_at | CETP | Cholesteryl ester transfer protein, plasma | 0.436 | 9.35E-05 |
| 219161_s_at | CKLF | Chemokine-like factor | 0.483 | 1.15E-05 |
| 221058_s_at | CKLF | Chemokine-like factor | 0.433 | .000104 |
| 231219_at | CKLF | Chemokine-like factor | 0.415 | .000216 |
| 1556209_at | CLEC2B | C-type lectin domain family 2, member B | 0.544 | 4.00E-07 |
| 209732_at | CLEC2B | C-type lectin domain family 2, member B | 0.444 | 6.50E-05 |
| 226425_at | CLIP4 | CAP-GLY domain containing linker protein family, member 4 | 0.399 | .000399 |
| 229967_at | CMTM2 | CKLF-like MARVEL transmembrane domain containing 2 | 0.387 | .000608 |
| 225009_at | CMTM4 | CKLF-like MARVEL transmembrane domain containing 4 | 0.415 | .000212 |
| 227953_at | CMTM6 | CKLF-like MARVEL transmembrane domain containing 6 | 0.417 | .000199 |
| 203642_s_at | COBLL1 | COBL-like 1 | 0.536 | 7.00E-07 |
| 203641_s_at | COBLL1 | COBL-like 1 | 0.437 | 8.88E-05 |
| 202119_s_at | CPNE3 | Copine III | 0.422 | .000163 |
| 202118_s_at | CPNE3 | Copine III | 0.400 | .000381 |
| 205984_at | CRHBP | Corticotropin releasing hormone binding protein | 0.527 | 1.20E-06 |
| 201380_at | CRTAP | Cartilage associated protein | 0.391 | .00053 |
| 207532_at | CRYGD | Crystallin, gamma D | 0.387 | .000599 |
| 207030_s_at | CSRP2 | Cysteine and glycine-rich protein 2 | 0.459 | 3.42E-05 |
| 211126_s_at | CSRP2 | Cysteine and glycine-rich protein 2 | 0.442 | 7.08E-05 |
| 215785_s_at | CYFIP2 | Cytoplasmic FMR1 interacting protein 2 | 0.385 | .000642 |
| 222134_at | DDO | D-aspartate oxidase | 0.411 | .000246 |
| 1558742_at | DEXI | Dexamethasone-induced transcript | 0.382 | .000731 |
| 202481_at | DHRS3 | Dehydrogenase/reductase (SDR family) member 3 | 0.429 | .00012 |
| 212888_at | DICER1 | Dicer1, Dcr-1 homolog (Drosophila) | 0.373 | .000995 |
| 232252_at | DUSP27 | Dual specificity phosphatase 27 (putative) | 0.379 | .000802 |
| 239574_at | ECHDC3 | Enoyl Coenzyme A hydratase domain containing 3 | 0.424 | .000153 |
| 218825_at | EGFL7 | EGF-like-domain, multiple 7 | 0.453 | 4.55E-05 |
| 225159_s_at | ELK4 | ELK4, ETS-domain protein (SRF accessory protein 1) | 0.376 | .000881 |
| 201325_s_at | EMP1 | Epithelial membrane protein 1 | 0.470 | 2.13E-05 |
| 201324_at | EMP1 | Epithelial membrane protein 1 | 0.443 | 6.85E-05 |
| 228256_s_at | EPB41L4A | Erythrocyte membrane protein band 4.1 like 4A | 0.393 | .000482 |
| 202609_at | EPS8 | Epidermal growth factor receptor pathway substrate 8 | 0.381 | .00076 |
| 32259_at | EZH1 | Enhancer of zeste homolog 1 (Drosophila) | 0.472 | 1.87E-05 |
| 213506_at | F2RL1 | Coagulation factor II (thrombin) receptor-like 1 | 0.602 | < 1e-07 |
| 206429_at | F2RL1 | Coagulation factor II (thrombin) receptor-like 1 | 0.513 | 2.60E-06 |
| 228678_at | FAM116B | Family with sequence similarity 116, member B | 0.395 | .000454 |
| 217967_s_at | FAM129A | Family with sequence similarity 129, member A | 0.497 | 5.80E-06 |
| 217966_s_at | FAM129A | Family with sequence similarity 129, member A | 0.484 | 1.08E-05 |
| 208229_at | FGFR2 | Fibroblast growth factor receptor 2 (bacteria-expressed kinase, keratinocyte growth factor receptor, craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer syndrome, Jackson-Weiss syndrome) | 0.411 | .000246 |
| 1562433_at | FLJ10489 | Hypothetical protein FLJ10489 | 0.582 | < 1e-07 |
| 1555486_a_at | FLJ14213 | Hypothetical protein FLJ14213 | 0.524 | 1.40E-06 |
| 219383_at | FLJ14213 | Hypothetical protein FLJ14213 | 0.518 | 1.90E-06 |
| 233379_at | FLJ14213 | Hypothetical protein FLJ14213 | 0.501 | 4.80E-06 |
| 236322_at | FLJ31951 | Hypothetical protein FLJ31951 | 0.434 | 9.83E-05 |
| 238949_at | FLJ31951 | Hypothetical protein FLJ31951 | 0.398 | .000406 |
| 226077_at | FLJ31951 | Hypothetical protein FLJ31951 | 0.379 | .000788 |
| 215330_at | FLJ43663 | Hypothetical protein FLJ43663 | 0.457 | 3.72E-05 |
| 228702_at | FLJ43663 | Hypothetical protein FLJ43663 | 0.447 | 5.84E-05 |
| 242768_at | FLJ43663 | Hypothetical protein FLJ43663 | 0.435 | 9.60E-05 |
| 239901_at | FLJ43663 | Hypothetical protein FLJ43663 | 0.398 | .000411 |
| 238619_at | FLJ43663 | Hypothetical protein FLJ43663 | 0.385 | .000647 |
| 218084_x_at | FXYD5 | FXYD domain containing ion transport regulator 5 | 0.413 | .000233 |
| 224252_s_at | FXYD5 | FXYD domain containing ion transport regulator 5 | 0.412 | .000238 |
| 203987_at | FZD6 | Frizzled homolog 6 (Drosophila) | 0.512 | 2.60E-06 |
| 1557030_at | GAB1 | GRB2-associated binding protein 1 | 0.415 | .000216 |
| 227428_at | GABPA | GA binding protein transcription factor, alpha subunit 60kDa | 0.397 | .000422 |
| 203765_at | GCA | Grancalcin, EF-hand calcium binding protein | 0.422 | .000164 |
| 228376_at | GGTA1 | Glycoprotein, alpha-galactosyltransferase 1 | 0.405 | .000309 |
| 209276_s_at | GLRX | Glutaredoxin (thioltransferase) | 0.407 | .00029 |
| 207987_s_at | GNRH1 | Gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) | 0.617 | < 1e-07 |
| 235540_at | GNRH1 | Gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) | 0.473 | 1.79E-05 |
| 219313_at | GRAMD1C | GRAM domain containing 1C | 0.381 | .000759 |
| 200696_s_at | GSN | Gelsolin (amyloidosis, Finnish type) | 0.396 | .000439 |
| 1557915_s_at | GSTO1 | Glutathione S-transferase omega 1 | 0.453 | 4.43E-05 |
| 201470_at | GSTO1 | Glutathione S-transferase omega 1 | 0.449 | 5.40E-05 |
| 217436_x_at | HLA-A | Major histocompatibility complex, class I, A | 0.437 | 8.74E-05 |
| 211911_x_at | HLA-B | Major histocompatibility complex, class I, B | 0.431 | .000111 |
| 209140_x_at | HLA-B | Major histocompatibility complex, class I, B | 0.414 | .000223 |
| 208729_x_at | HLA-B | Major histocompatibility complex, class I, B | 0.405 | .000317 |
| 208812_x_at | HLA-C | Major histocompatibility complex, class I, C | 0.454 | 4.23E-05 |
| 214459_x_at | HLA-C | Major histocompatibility complex, class I, C | 0.426 | .000137 |
| 211799_x_at | HLA-C | Major histocompatibility complex, class I, C | 0.403 | .000342 |
| 216526_x_at | HLA-C | Major histocompatibility complex, class I, C | 0.400 | .000384 |
| 217478_s_at | HLA-DMA | Major histocompatibility complex, class II, DM alpha | 0.452 | 4.77E-05 |
| 226878_at | HLA-DOA | Major histocompatibility complex, class II, DO alpha | 0.400 | .000375 |
| 211991_s_at | HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 0.438 | 8.49E-05 |
| 211990_at | HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 0.426 | .000139 |
| 201137_s_at | HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | 0.480 | 1.29E-05 |
| 244485_at | HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | 0.469 | 2.21E-05 |
| 208894_at | HLA-DRA | Major histocompatibility complex, class II, DR alpha | 0.425 | .000145 |
| 210982_s_at | HLA-DRA | Major histocompatibility complex, class II, DR alpha | 0.411 | .000245 |
| 209312_x_at | HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 | 0.420 | .000179 |
| 215193_x_at | HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 | 0.415 | .000212 |
| 208306_x_at | HLA-DRB4 | Major histocompatibility complex, class II, DR beta 4 | 0.421 | .000167 |
| 204670_x_at | HLA-DRB5 | Major histocompatibility complex, class II, DR beta 5 | 0.431 | .000114 |
| 217362_x_at | HLA-DRB6 | Major histocompatibility complex, class II, DR beta 6 (pseudogene) | 0.423 | .000157 |
| 200904_at | HLA-E | Major histocompatibility complex, class I, E | 0.391 | .000533 |
| 217456_x_at | HLA-E | Major histocompatibility complex, class I, E | 0.377 | .000845 |
| 204806_x_at | HLA-F | Major histocompatibility complex, class I, F | 0.400 | .000375 |
| 221875_x_at | HLA-F | Major histocompatibility complex, class I, F | 0.379 | .000798 |
| 211529_x_at | HLA-G | HLA-G histocompatibility antigen, class I, G | 0.465 | 2.69E-05 |
| 210514_x_at | HLA-G | HLA-G histocompatibility antigen, class I, G | 0.462 | 3.03E-05 |
| 211530_x_at | HLA-G | HLA-G histocompatibility antigen, class I, G | 0.454 | 4.39E-05 |
| 211528_x_at | HLA-G | HLA-G histocompatibility antigen, class I, G | 0.434 | 9.96E-05 |
| 211597_s_at | HOP | Homeodomain-only protein | 0.375 | .00093 |
| 210253_at | HTATIP2 | HIV-1 Tat interactive protein 2, 30kDa | 0.497 | 5.70E-06 |
| 239704_at | IBRDC2 | IBR domain containing 2 | 0.497 | 5.60E-06 |
| 228153_at | IBRDC2 | IBR domain containing 2 | 0.376 | .000879 |
| 208966_x_at | IFI16 | Interferon, gamma-inducible protein 16 | 0.438 | 8.43E-05 |
| 208965_s_at | IFI16 | Interferon, gamma-inducible protein 16 | 0.432 | .000107 |
| 206332_s_at | IFI16 | Interferon, gamma-inducible protein 16 | 0.423 | .000154 |
| 204439_at | IFI44L | Interferon-induced protein 44-like | 0.380 | .000786 |
| 226267_at | JDP2 | Jun dimerization protein 2 | 0.487 | 9.40E-06 |
| 239835_at | KBTBD8 | Kelch repeat and BTB (POZ) domain containing 8 | 0.401 | .000369 |
| 224316_at | KCTD9 | Potassium channel tetramerisation domain containing 9 | 0.471 | 1.96E-05 |
| 218823_s_at | KCTD9 | Potassium channel tetramerisation domain containing 9 | 0.436 | 9.22E-05 |
| 229878_at | KIAA1731 | KIAA1731 | 0.427 | .000133 |
| 221221_s_at | KLHL3 | Kelch-like 3 (Drosophila) | 0.437 | 8.77E-05 |
| 200650_s_at | LDHA | Lactate dehydrogenase A | 0.379 | .000809 |
| 212658_at | LHFPL2 | Lipoma HMGIC fusion partner-like 2 | 0.406 | .000301 |
| 219541_at | LIME1 | Lck interacting transmembrane adaptor 1 | 0.374 | .000937 |
| 223925_s_at | LOC767558 | Myeloproliferative disease-associated SEREX antigen | 0.424 | .000151 |
| 210102_at | LOH11CR2A | Loss of heterozygosity, 11, chromosomal region 2, gene A | 0.466 | 2.54E-05 |
| 205011_at | LOH11CR2A | Loss of heterozygosity, 11, chromosomal region 2, gene A | 0.461 | 3.15E-05 |
| 240338_at | LRAP | Leukocyte-derived arginine aminopeptidase | 0.383 | .000694 |
| 203523_at | LSP1 | Lymphocyte-specific protein 1 | 0.442 | 7.25E-05 |
| 202145_at | LY6E | Lymphocyte antigen 6 complex, locus E | 0.398 | .000403 |
| 224480_s_at | MAG1 | Lung cancer metastasis-associated protein | 0.420 | .000178 |
| 1569136_at | MGAT4A | Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | 0.539 | 6.00E-07 |
| 1558166_at | MGC16275 | Hypothetical protein MGC16275 | 0.406 | .000297 |
| 206247_at | MICB | MHC class I polypeptide-related sequence B | 0.394 | .000477 |
| 239272_at | MMP28 | Matrix metallopeptidase 28 | 0.501 | 4.80E-06 |
| 205330_at | MN1 | Meningioma (disrupted in balanced translocation) 1 | 0.866 | < 1e-07 |
| 219648_at | MREG | Melanoregulin | 0.499 | 5.30E-06 |
| 218027_at | MRPL15 | Mitochondrial ribosomal protein L15 | 0.374 | .000931 |
| 219363_s_at | MTERFD1 | MTERF domain containing 1 | 0.403 | .000336 |
| 225111_s_at | NAPB | N-ethylmaleimide-sensitive factor attachment protein, beta | 0.399 | .000395 |
| 243246_at | NAT12 | N-acetyltransferase 12 | 0.422 | .000161 |
| 236197_at | NCBP1 | Nuclear cap binding protein subunit 1, 80kDa | 0.491 | 7.70E-06 |
| 240824_at | OBFC1 | Oligonucleotide/oligosaccharide-binding fold containing 1 | 0.493 | 7.10E-06 |
| 223259_at | ORMDL3 | ORM1-like 3 (S. cerevisiae) | 0.402 | .000345 |
| 228966_at | PANK2 | Pantothenate kinase 2 (Hallervorden-Spatz syndrome) | 0.423 | .000158 |
| 232140_at | PGM5P1 | Phosphoglucomutase 5 pseudogene 1 | 0.393 | .000494 |
| 235389_at | PHF20 | PHD finger protein 20 | 0.393 | .000486 |
| 209780_at | PHTF2 | Putative homeodomain transcription factor 2 | 0.400 | .000382 |
| 206370_at | PIK3CG | Phosphoinositide-3-kinase, catalytic, gamma polypeptide | 0.395 | .000447 |
| 235230_at | PLCXD2 | Phosphatidylinositol-specific phospholipase C, X domain containing 2 | 0.396 | .000443 |
| 201136_at | PLP2 | Proteolipid protein 2 (colonic epithelium-enriched) | 0.384 | .000684 |
| 213241_at | PLXNC1 | Plexin C1 | 0.460 | 3.34E-05 |
| 206470_at | PLXNC1 | Plexin C1 | 0.418 | .000188 |
| 206471_s_at | PLXNC1 | Plexin C1 | 0.414 | .000223 |
| 209799_at | PRKAA1 | Protein kinase, AMP-activated, alpha 1 catalytic subunit | 0.426 | .00014 |
| 222582_at | PRKAG2 | Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | 0.389 | .000555 |
| 214203_s_at | PRODH | Proline dehydrogenase (oxidase) 1 | 0.422 | .000163 |
| 204304_s_at | PROM1 | Prominin 1 | 0.406 | .000301 |
| 241133_at | PRSS1 | Protease, serine, 1 (trypsin 1) | 0.431 | .000113 |
| 240766_at | PRSS1 | Protease, serine, 1 (trypsin 1) | 0.399 | .000388 |
| 209040_s_at | PSMB8 | Proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) | 0.470 | 2.05E-05 |
| 204279_at | PSMB9 | Proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) | 0.406 | .000298 |
| 201087_at | PXN | Paxillin | 0.446 | 6.01E-05 |
| 230405_at | RAD50 | RAD50 homolog (S. cerevisiae) | 0.467 | 2.42E-05 |
| 232253_at | RAD50 | RAD50 homolog (S. cerevisiae) | 0.415 | .000211 |
| 235846_at | RAD54B | RAD54 homolog B (S. cerevisiae) | 0.413 | .000234 |
| 204070_at | RARRES3 | Retinoic acid receptor responder (tazarotene induced) 3 | 0.465 | 2.62E-05 |
| 221827_at | RBCK1 | RanBP-type and C3HC4-type zinc finger containing 1 | 0.401 | .000362 |
| 218117_at | RBX1 | Ring-box 1 | 0.384 | .000682 |
| 227425_at | REPS2 | RALBP1 associated Eps domain containing 2 | 0.449 | 5.39E-05 |
| 242571_at | REPS2 | RALBP1 associated Eps domain containing 2 | 0.430 | .000117 |
| 220570_at | RETN | Resistin | 0.404 | .000323 |
| 214000_s_at | RGS10 | Regulator of G-protein signalling 10 | 0.425 | .000144 |
| 204319_s_at | RGS10 | Regulator of G-protein signalling 10 | 0.411 | .000247 |
| 219045_at | RHOF | ras homolog gene family, member F (in filopodia) | 0.414 | .000219 |
| 243178_at | RNF149 | Ring finger protein 149 | 0.373 | .000989 |
| 229543_at | RP1-93H18.5 | Hypothetical protein LOC441168 | 0.561 | 2E-07 |
| 228362_s_at | RP1-93H18.5 | Hypothetical protein LOC441168 | 0.543 | 5.00E-07 |
| 229391_s_at | RP1-93H18.5 | Hypothetical protein LOC441168 | 0.525 | 1.30E-06 |
| 229390_at | RP1-93H18.5 | Hypothetical protein LOC441168 | 0.524 | 1.40E-06 |
| 226335_at | RPS6KA3 | Ribosomal protein S6 kinase, 90kDa, polypeptide 3 | 0.428 | .000127 |
| 1554876_a_at | S100Z | S100 calcium binding protein Z | 0.472 | 1.89E-05 |
| 226169_at | SBF2 | SET binding factor 2 | 0.470 | 2.08E-05 |
| 242935_at | SBF2 | SET binding factor 2 | 0.440 | 7.88E-05 |
| 233914_s_at | SBF2 | SET binding factor 2 | 0.421 | .000171 |
| 206995_x_at | SCARF1 | Scavenger receptor class F, member 1 | 0.374 | .000935 |
| 41220_at | SEPT9 | Septin 9 | 0.382 | .000723 |
| 211474_s_at | SERPINB6 | Serpin peptidase inhibitor, clade B (ovalbumin), member 6 | 0.458 | 3.54E-05 |
| 1556950_s_at | SERPINB6 | Serpin peptidase inhibitor, clade B (ovalbumin), member 6 | 0.416 | .000202 |
| 209723_at | SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 0.453 | 4.53E-05 |
| 242814_at | SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 0.402 | .000354 |
| 218346_s_at | SESN1 | Sestrin 1 | 0.384 | .000662 |
| 241245_at | SFRS4 | Splicing factor, arginine/serine-rich 4 | 0.406 | .000297 |
| 201811_x_at | SH3BP5 | SH3-domain binding protein 5 (BTK-associated) | 0.377 | .000868 |
| 219256_s_at | SH3TC1 | SH3 domain and tetratricopeptide repeats 1 | 0.386 | .000622 |
| 203124_s_at | SLC11A2 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | 0.374 | .000951 |
| 226601_at | SLC30A7 | Solute carrier family 30 (zinc transporter), member 7 | 0.377 | .000859 |
| 212944_at | SLC5A3 | Solute carrier family 5 (inositol transporters), member 3 | 0.405 | .00031 |
| 206543_at | SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 0.388 | .000586 |
| 219109_at | SPAG16 | Sperm associated antigen 16 | 0.440 | 7.87E-05 |
| 212667_at | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 0.476 | 1.57E-05 |
| 200665_s_at | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 0.460 | 3.30E-05 |
| 213820_s_at | STARD5 | START domain containing 5 | 0.404 | .000323 |
| 201061_s_at | STOM | Stomatin | 0.461 | 3.23E-05 |
| 201060_x_at | STOM | Stomatin | 0.393 | .000489 |
| 226117_at | TIFA | TRAF-interacting protein with a forkhead-associated domain | 0.431 | .000112 |
| 241844_x_at | TMEM156 | Transmembrane protein 156 | 0.376 | .000893 |
| 231775_at | TNFRSF10A | Tumor necrosis factor receptor superfamily, member 10a | 0.426 | .00014 |
| 209354_at | TNFRSF14 | Tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | 0.373 | .000972 |
| 214581_x_at | TNFRSF21 | Tumor necrosis factor receptor superfamily, member 21 | 0.413 | .00023 |
| 218856_at | TNFRSF21 | Tumor necrosis factor receptor superfamily, member 21 | 0.403 | .000335 |
| 235973_at | TRIP11 | Thyroid hormone receptor interactor 11 | 0.431 | .000114 |
| 225775_at | TSPAN33 | Tetraspanin 33 | 0.515 | 2.30E-06 |
| 213172_at | TTC9 | Tetratricopeptide repeat domain 9 | 0.388 | .00059 |
| 1553183_at | UMODL1 | Uromodulin-like 1 | 0.444 | 6.73E-05 |
| 208844_at | VDAC3 | Voltage-dependent anion channel 3 | 0.394 | .000477 |
| 205672_at | XPA | Xeroderma pigmentosum, complementation group A | 0.456 | 3.93E-05 |
| 230913_at | 0.496 | 5.90E-06 | ||
| 226550_at | 0.482 | 1.19E-05 | ||
| 239184_at | 0.478 | 1.44E-05 | ||
| 240153_at | 0.477 | 1.49E-05 | ||
| 1558105_a_at | 0.452 | 4.74E-05 | ||
| 231431_s_at | 0.446 | 5.99E-05 | ||
| 237315_at | 0.435 | 9.54E-05 | ||
| 210824_at | 0.422 | .000159 | ||
| 233867_at | 0.415 | .000213 | ||
| 229380_at | 0.393 | .000481 | ||
| 232227_at | 0.387 | .000606 | ||
| 229011_at | 0.385 | .00064 | ||
| 225567_at | 0.382 | .000726 | ||
| 244561_at | 0.380 | .000766 | ||
| Negatively correlated with MN1 expression | ||||
| 218434_s_at | AACS | Acetoacetyl-CoA synthetase | −0.427 | 1.32E-04 |
| 1553605_a_at | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | −0.393 | .000485 |
| 210377_at | ACSM3 | Acyl-CoA synthetase medium-chain family member 3 | −0.413 | 2.34E-04 |
| 205942_s_at | ACSM3 | Acyl-CoA synthetase medium-chain family member 3 | −0.417 | 2.00E-04 |
| 201792_at | AEBP1 | AE binding protein 1 | −0.437 | 8.81E-05 |
| 212173_at | AK2 | Adenylate kinase 2 | −0.486 | 9.9E-06 |
| 212747_at | ANKS1A | Ankyrin repeat and sterile alpha motif domain containing 1A | −0.533 | 9E-07 |
| 225286_at | ARSD | Arylsulfatase D | −0.374 | .000955 |
| 223695_s_at | ARSD | Arylsulfatase D | −0.375 | .000918 |
| 230131_x_at | ARSD | Arylsulfatase D | −0.384 | .000673 |
| 204608_at | ASL | Argininosuccinate lyase | −0.381 | .00075 |
| 218908_at | ASPSCR1 | Alveolar soft part sarcoma chromosome region, candidate 1 | −0.395 | .000461 |
| 240747_at | ATP8B4 | ATPase, Class I, type 8B, member 4 | −0.377 | .00086 |
| 220416_at | ATP8B4 | ATPase, Class I, type 8B, member 4 | −0.423 | 1.55E-04 |
| 227877_at | AXIIR | Similar to annexin II receptor | −0.416 | 2.06E-04 |
| 203304_at | BAMBI | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | −0.509 | .000003 |
| 218332_at | BEX1 | Brain expressed, X-linked 1 | −0.428 | 1.27E-04 |
| 202265_at | BMI1 | BMI1 polycomb ring finger oncogene | −0.388 | .000588 |
| 213578_at | BMPR1A | Bone morphogenetic protein receptor, type IA | −0.412 | 2.38E-04 |
| 240772_at | C10orf11 | Chromosome 10 open reading frame 11 | −0.487 | 9.4E-06 |
| 223703_at | C10orf11 | Chromosome 10 open reading frame 11 | −0.511 | 2.8E-06 |
| 219988_s_at | C1orf164 | Chromosome 1 open reading frame 164 | −0.404 | .000322 |
| 230381_at | C1orf186 | Chromosome 1 open reading frame 186 | −0.394 | .000473 |
| 223063_at | C1orf198 | Chromosome 1 open reading frame 198 | −0.373 | .000974 |
| 219951_s_at | C20orf12 | Chromosome 20 open reading frame 12 | −0.388 | .000579 |
| 238767_at | C4orf36 | Chromosome 4 open reading frame 36 | −0.394 | .000476 |
| 201309_x_at | C5orf13 | Chromosome 5 open reading frame 13 | −0.396 | .000442 |
| 201310_s_at | C5orf13 | Chromosome 5 open reading frame 13 | −0.410 | 2.62E-04 |
| 238465_at | C5orf35 | Chromosome 5 open reading frame 35 | −0.430 | 1.19E-04 |
| 219261_at | C7orf26 | Chromosome 7 open reading frame 26 | −0.379 | .000811 |
| 232668_at | C8orf72 | Chromosome 8 open reading frame 72 | −0.462 | 3.07E-05 |
| 228790_at | C8orf72 | Chromosome 8 open reading frame 72 | −0.482 | .000012 |
| 221959_at | C8orf72 | Chromosome 8 open reading frame 72 | −0.498 | 5.5E-06 |
| 207129_at | CA5B | Carbonic anhydrase VB, mitochondrial | −0.397 | .000418 |
| 214082_at | CA5B | Carbonic anhydrase VB, mitochondrial | −0.413 | 2.33E-04 |
| 243416_at | CACHD1 | Cache domain containing 1 | −0.445 | 6.40E-05 |
| 225627_s_at | CACHD1 | Cache domain containing 1 | −0.464 | 2.77E-05 |
| 210817_s_at | CALCOCO2 | Calcium binding and coiled-coil domain 2 | −0.374 | .000958 |
| 220162_s_at | CARD9 | Caspase recruitment domain family, member 9 | −0.411 | 2.52E-04 |
| 201432_at | CAT | Catalase | −0.495 | 6.3E-06 |
| 211922_s_at | CAT | Catalase | −0.510 | .000003 |
| 228061_at | CCDC126 | Coiled-coil domain containing 126 | −0.379 | .000792 |
| 237305_at | CDH2 | Cadherin 2, type 1, N-cadherin (neuronal) | −0.403 | .000338 |
| 222755_s_at | CHD7 | Chromodomain helicase DNA binding protein 7 | −0.504 | .000004 |
| 226123_at | CHD7 | Chromodomain helicase DNA binding protein 7 | −0.509 | 3.1E-06 |
| 218829_s_at | CHD7 | Chromodomain helicase DNA binding protein 7 | −0.511 | 2.9E-06 |
| 205131_x_at | CLEC11A | C-type lectin domain family 11, member A | −0.388 | .000587 |
| 210783_x_at | CLEC11A | C-type lectin domain family 11, member A | −0.403 | .000336 |
| 227209_at | CNTN1 | Contactin 1 | −0.398 | .000402 |
| 212489_at | COL5A1 | Collagen, type V, alpha 1 | −0.377 | .000857 |
| 212488_at | COL5A1 | Collagen, type V, alpha 1 | −0.419 | 1.83E-04 |
| 213622_at | COL9A2 | Collagen, type IX, alpha 2 | −0.446 | 5.96E-05 |
| 223457_at | COPG2 | Coatomer protein complex, subunit gamma 2 | −0.400 | .000384 |
| 205624_at | CPA3 | Carboxypeptidase A3 (mast cell) | −0.453 | 4.49E-05 |
| 205653_at | CTSG | Cathepsin G | −0.385 | .000642 |
| 229415_at | CYCS | Cytochrome c, somatic | −0.388 | .000584 |
| 1567101_at | DACH1 | Dachshund homolog 1 (Drosophila) | −0.374 | .000934 |
| 217025_s_at | DBN1 | Drebrin 1 | −0.376 | .000893 |
| 210397_at | DEFB1 | Defensin, beta 1 | −0.398 | .000414 |
| 207147_at | DLX2 | Distal-less homeobox 2 | −0.384 | .000674 |
| 228598_at | DPP10 | Dipeptidyl-peptidase 10 | −0.373 | .000985 |
| 238784_at | DPY19L2 | dpy-19-like 2 (C. elegans) | −0.393 | .000491 |
| 204750_s_at | DSC2 | Desmocollin 2 | −0.424 | 1.52E-04 |
| 204751_x_at | DSC2 | Desmocollin 2 | −0.445 | 6.28E-05 |
| 226817_at | DSC2 | Desmocollin 2 | −0.462 | 3.03E-05 |
| 205741_s_at | DTNA | Dystrobrevin, alpha | −0.467 | 2.38E-05 |
| 210091_s_at | DTNA | Dystrobrevin, alpha | −0.477 | 1.49E-05 |
| 227084_at | DTNA | Dystrobrevin, alpha | −0.481 | 1.24E-05 |
| 1557803_at | DTNA | Dystrobrevin, alpha | −0.536 | 7E-07 |
| 219469_at | DYNC2H1 | Dynein, cytoplasmic 2, heavy chain 1 | −0.383 | .000688 |
| 1565149_at | DYNC2H1 | Dynein, cytoplasmic 2, heavy chain 1 | −0.410 | 2.55E-04 |
| 205107_s_at | EFNA4 | Ephrin-A4 | −0.399 | .000397 |
| 1558871_at | EPGN | Epithelial mitogen homolog (mouse) | −0.522 | 1.5E-06 |
| 203349_s_at | ETV5 | ets variant gene 5 (ets-related molecule) | −0.389 | .000553 |
| 201828_x_at | FAM127A | Family with sequence similarity 127, member A | −0.396 | .000444 |
| 224973_at | FAM46A | Family with sequence similarity 46, member A | −0.383 | .000685 |
| 221766_s_at | FAM46A | Family with sequence similarity 46, member A | −0.423 | 1.53E-04 |
| 229546_at | FAM84A | Family with sequence similarity 84, member A | −0.398 | .000412 |
| 228459_at | FAM84A | Family with sequence similarity 84, member A | −0.399 | .000395 |
| 234331_s_at | FAM84A | Family with sequence similarity 84, member A | −0.469 | 2.24E-05 |
| 225667_s_at | FAM84A | Family with sequence similarity 84, member A | −0.479 | 1.38E-05 |
| 233087_at | FBXL17 | F-box and leucine-rich repeat protein 17 | −0.402 | .000348 |
| 227203_at | FBXL17 | F-box and leucine-rich repeat protein 17 | −0.424 | 1.50E-04 |
| 238174_at | FBXL17 | F-box and leucine-rich repeat protein 17 | −0.466 | 2.49E-05 |
| 242034_at | FBXL17 | F-box and leucine-rich repeat protein 17 | −0.477 | .000015 |
| 215000_s_at | FEZ2 | Fasciculation and elongation protein zeta 2 (zygin II) | −0.406 | .000301 |
| 229280_s_at | FLJ22536 | Hypothetical locus LOC401237 | −0.409 | .000271 |
| 212288_at | FNBP1 | Formin binding protein 1 | −0.374 | .000961 |
| 209702_at | FTO | Fatso | −0.511 | 2.8E-06 |
| 204452_s_at | FZD1 | Frizzled homolog 1 (Drosophila) | −0.383 | .000689 |
| 214106_s_at | GMDS | GDP-mannose 4,6-dehydratase | −0.407 | .000294 |
| 204983_s_at | GPC4 | Glypican 4 | −0.422 | 1.64E-04 |
| 232453_at | GPC6 | Glypican 6 | −0.387 | .000613 |
| 220773_s_at | GPHN | Gephyrin | −0.384 | .00068 |
| 234941_s_at | GPHN | Gephyrin | −0.417 | 1.96E-04 |
| 221892_at | H6PD | Hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) | −0.451 | 4.96E-05 |
| 206643_at | HAL | Histidine ammonia-lyase | −0.532 | 9E-07 |
| 238021_s_at | hCG_1815491 | hCG1815491 | −0.473 | 1.84E-05 |
| 238022_at | hCG_1815491 | hCG1815491 | −0.492 | 7.5E-06 |
| 219687_at | HHAT | Hedgehog acyltransferase | −0.377 | .000855 |
| 237466_s_at | HHIP | Hedgehog interacting protein | −0.375 | .000911 |
| 1556037_s_at | HHIP | Hedgehog interacting protein | −0.419 | 1.86E-04 |
| 207982_at | HIST1H1T | Histone cluster 1, H1t | −0.423 | 1.55E-04 |
| 214522_x_at | HIST1H2AD | Histone cluster 1, H2ad | −0.393 | .000491 |
| 239669_at | HIST1H2AD | Histone cluster 1, H2ad | −0.529 | 1.1E-06 |
| 214644_at | HIST1H2AK | Histone cluster 1, H2ak | −0.394 | .000464 |
| 239041_at | HIST1H2AK | Histone cluster 1, H2ak | −0.435 | 9.54E-05 |
| 214455_at | HIST1H2BC | Histone cluster 1, H2bc | −0.439 | 8.06E-05 |
| 209911_x_at | HIST1H2BD | Histone cluster 1, H2bd | −0.378 | .000836 |
| 222067_x_at | HIST1H2BD | Histone cluster 1, H2bd | −0.380 | .000781 |
| 208527_x_at | HIST1H2BE | Histone cluster 1, H2be | −0.403 | .000341 |
| 208490_x_at | HIST1H2BF | Histone cluster 1, H2bf | −0.385 | .000659 |
| 236193_at | HIST1H2BG | Histone cluster 1, H2bg | −0.464 | 2.73E-05 |
| 208523_x_at | HIST1H2BI | Histone cluster 1, H2bi | −0.404 | .000331 |
| 214502_at | HIST1H2BJ | Histone cluster 1, H2bj | −0.383 | .000704 |
| 207226_at | HIST1H2BN | Histone cluster 1, H2bn | −0.458 | 3.65E-05 |
| 214472_at | HIST1H3D | Histone cluster 1, H3d | −0.433 | 1.04E-04 |
| 208076_at | HIST1H4D | Histone cluster 1, H4d | −0.449 | 5.29E-05 |
| 202708_s_at | HIST2H2BE | Histone cluster 2, H2be | −0.446 | 6.08E-05 |
| 1554453_at | HNRPLL | Heterogeneous nuclear ribonucleoprotein L-like | −0.380 | .000772 |
| 225385_s_at | HNRPLL | Heterogeneous nuclear ribonucleoprotein L-like | −0.456 | 3.88E-05 |
| 204647_at | HOMER3 | Homer homolog 3 (Drosophila) | −0.443 | 6.76E-05 |
| 215489_x_at | HOMER3 | Homer homolog 3 (Drosophila) | −0.476 | 1.62E-05 |
| 222222_s_at | HOMER3 | Homer homolog 3 (Drosophila) | −0.484 | .000011 |
| 214457_at | HOXA2 | Homeobox A2 | −0.495 | 6.4E-06 |
| 208604_s_at | HOXA3 | Homeobox A3 | −0.394 | .000476 |
| 206289_at | HOXA4 | Homeobox A4 | −0.382 | .000717 |
| 213844_at | HOXA5 | Homeobox A5 | −0.374 | .000956 |
| 235521_at | HOXA5 | Homeobox A5 | −0.400 | .000373 |
| 223963_s_at | IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 | −0.379 | .000814 |
| 218847_at | IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 | −0.383 | .000695 |
| 203006_at | INPP5A | Inositol polyphosphate-5-phosphatase, 40kDa | −0.406 | .000298 |
| 203331_s_at | INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | −0.374 | .000946 |
| 213392_at | IQCK | IQ motif containing K | −0.386 | .000622 |
| 224572_s_at | IRF2BP2 | Interferon regulatory factor 2 binding protein 2 | −0.464 | 2.77E-05 |
| 224570_s_at | IRF2BP2 | Interferon regulatory factor 2 binding protein 2 | −0.465 | 2.67E-05 |
| 229638_at | IRX3 | Iroquois homeobox protein 3 | −0.463 | 2.83E-05 |
| 210239_at | IRX5 | Iroquois homeobox protein 5 | −0.531 | .000001 |
| 226246_at | KCTD1 | Potassium channel tetramerisation domain containing 1 | −0.384 | .000665 |
| 226245_at | KCTD1 | Potassium channel tetramerisation domain containing 1 | −0.395 | .000453 |
| 228683_s_at | KCTD15 | Potassium channel tetramerisation domain containing 15 | −0.397 | .000426 |
| 230249_at | KHDRBS3 | KH domain containing, RNA binding, signal transduction associated 3 | −0.489 | 8.4E-06 |
| 209781_s_at | KHDRBS3 | KH domain containing, RNA binding, signal transduction associated 3 | −0.528 | 1.1E-06 |
| 1556425_a_at | KIAA0802 | KIAA0802 | −0.416 | 2.04E-04 |
| 239033_at | KIAA1958 | KIAA1958 | −0.405 | .000315 |
| 235112_at | KIAA1958 | KIAA1958 | −0.432 | 1.09E-04 |
| 213623_at | KIF3A | Kinesin family member 3A | −0.390 | .000546 |
| 236565_s_at | LARP6 | La ribonucleoprotein domain family, member 6 | −0.384 | .000683 |
| 207348_s_at | LIG3 | Ligase III, DNA, ATP-dependent | −0.426 | 1.36E-04 |
| 204123_at | LIG3 | Ligase III, DNA, ATP-dependent | −0.483 | 1.12E-05 |
| 240027_at | LIN7A | Lin-7 homolog A (C. elegans) | −0.434 | 1.01E-04 |
| 206440_at | LIN7A | Lin-7 homolog A (C. elegans) | −0.488 | 8.9E-06 |
| 241652_x_at | LIN7A | Lin-7 homolog A (C. elegans) | −0.511 | 2.8E-06 |
| 233336_at | LOC142893 | Hypothetical protein LOC142893 | −0.411 | 2.52E-04 |
| 230648_at | LOC283663 | Hypothetical protein LOC283663 | −0.395 | .000461 |
| 241370_at | LOC286052 | Hypothetical protein LOC286052 | −0.375 | .000916 |
| 227547_at | LOC388795 | Similar to CG40449-PA.3 | −0.451 | 4.98E-05 |
| 232113_at | LOC399959 | Hypothetical gene supported by BX647608 | −0.404 | .000331 |
| 240423_at | LOC441204 | Hypothetical locus LOC441204 | −0.444 | 6.75E-05 |
| 229429_x_at | LOC728855 | Hypothetical protein LOC728855 | −0.373 | .000985 |
| 202728_s_at | LTBP1 | Latent transforming growth factor beta binding protein 1 | −0.497 | 5.8E-06 |
| 202729_s_at | LTBP1 | Latent transforming growth factor beta binding protein 1 | −0.518 | .000002 |
| 228150_at | LZTR2 | Leucine zipper transcription regulator 2 | −0.399 | .000388 |
| 241607_at | LZTR2 | Leucine zipper transcription regulator 2 | −0.430 | 1.16E-04 |
| 1564423_a_at | LZTR2 | Leucine zipper transcription regulator 2 | −0.450 | 5.09E-05 |
| 242172_at | MEIS1 | Meis homeobox 1 | −0.431 | 1.12E-04 |
| 1559477_s_at | MEIS1 | Meis homeobox 1 | −0.442 | 7.12E-05 |
| 204069_at | MEIS1 | Meis homeobox 1 | −0.462 | 3.01E-05 |
| 238347_at | MGC15523 | Hypothetical protein MGC15523 | −0.376 | .000902 |
| 242942_at | MGC15523 | Hypothetical protein MGC15523 | −0.390 | .000551 |
| 210254_at | MS4A3 | Membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | −0.475 | 1.66E-05 |
| 1554892_a_at | MS4A3 | Membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | −0.476 | 1.58E-05 |
| 202247_s_at | MTA1 | Metastasis associated 1 | −0.379 | .000803 |
| 204798_at | MYB | v-myb myeloblastosis viral oncogene homolog (avian) | −0.434 | 9.98E-05 |
| 209550_at | NDN | Necdin homolog (mouse) | −0.391 | .000532 |
| 1552736_a_at | NETO1 | Neuropilin (NRP) and tolloid (TLL)-like 1 | −0.416 | 2.03E-04 |
| 209706_at | NKX3-1 | NK3 transcription factor related, locus 1 (Drosophila) | −0.477 | .000015 |
| 232478_at | NR6A1 | Nuclear receptor subfamily 6, group A, member 1 | −0.502 | 4.4E-06 |
| 220110_s_at | NXF3 | Nuclear RNA export factor 3 | −0.504 | 4.1E-06 |
| 227492_at | OCLN | Occluding | −0.407 | .000287 |
| 209925_at | OCLN | Occluding | −0.455 | 4.05E-05 |
| 223464_at | OSBPL5 | Oxysterol binding protein-like 5 | −0.449 | 5.38E-05 |
| 204082_at | PBX3 | Pre-B-cell leukemia homeobox 3 | −0.391 | .000526 |
| 209361_s_at | PCBP4 | Poly(rC) binding protein 4 | −0.392 | .000515 |
| 219737_s_at | PCDH9 | Protocadherin 9 | −0.375 | .000907 |
| 222860_s_at | PDGFD | Platelet derived growth factor D | −0.427 | 1.35E-04 |
| 219304_s_at | PDGFD | Platelet derived growth factor D | −0.490 | .000008 |
| 225829_at | PDZD8 | PDZ domain containing 8 | −0.382 | .000709 |
| 225830_at | PDZD8 | PDZ domain containing 8 | −0.402 | .000347 |
| 209438_at | PHKA2 | Phosphorylase kinase, alpha 2 (liver) | −0.463 | 2.94E-05 |
| 1559705_s_at | PHKA2 | Phosphorylase kinase, alpha 2 (liver) | −0.489 | 8.5E-06 |
| 209439_s_at | PHKA2 | Phosphorylase kinase, alpha 2 (liver) | −0.521 | 1.6E-06 |
| 225903_at | PIGU | Phosphatidylinositol glycan anchor biosynthesis, class U | −0.389 | .000568 |
| 215807_s_at | PLXNB1 | Plexin B1 | −0.428 | 1.28E-04 |
| 228383_at | PNPLA7 | Patatin-like phospholipase domain containing 7 | −0.477 | 1.49E-05 |
| 222238_s_at | POLM | Polymerase (DNA directed), mu | −0.395 | .000448 |
| 204117_at | PREP | Prolyl endopeptidase | −0.426 | 1.37E-04 |
| 37950_at | PREP | Prolyl endopeptidase | −0.464 | 2.74E-05 |
| 220798_x_at | PRG2 | Plasticity-related gene 2 | −0.633 | < 1e-07 |
| 209158_s_at | PSCD2 | Pleckstrin homology, Sec7 and coiled-coil domains 2 (cytohesin-2) | −0.414 | 2.24E-04 |
| 211373_s_at | PSEN2 | Presenilin 2 (Alzheimer disease 4) | −0.478 | 1.43E-05 |
| 213933_at | PTGER3 | Prostaglandin E receptor 3 (subtype EP3) | −0.430 | 1.19E-04 |
| 1569830_at | PTPRC | Protein tyrosine phosphatase, receptor type, C | −0.391 | .000523 |
| 1558290_a_at | PVT1 | Pvt1 oncogene homolog, MYC activator (mouse) | −0.433 | 1.03E-04 |
| 212013_at | PXDN | Peroxidasin homolog (Drosophila) | −0.394 | .000471 |
| 201481_s_at | PYGB | Phosphorylase, glycogen; brain | −0.449 | 5.30E-05 |
| 222810_s_at | RASAL2 | RAS protein activator like 2 | −0.581 | < 1e-07 |
| 244526_at | RASGRP3 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | −0.423 | 1.59E-04 |
| 205801_s_at | RASGRP3 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) | −0.454 | 4.22E-05 |
| 208031_s_at | RFX2 | Regulatory factor X, 2 (influences HLA class II expression) | −0.449 | 5.35E-05 |
| 226872_at | RFX2 | Regulatory factor X, 2 (influences HLA class II expression) | −0.458 | 3.53E-05 |
| 91816_f_at | RKHD1 | Ring finger and KH domain containing 1 | −0.384 | .000676 |
| 206851_at | RNASE3 | Ribonuclease, RNase A family, 3 (eosinophil cationic protein) | −0.375 | .00093 |
| 1556590_s_at | SAPS3 | SAPS domain family, member 3 | −0.418 | 1.93E-04 |
| 1556589_at | SAPS3 | SAPS domain family, member 3 | −0.426 | 1.38E-04 |
| 228497_at | SLC22A15 | Solute carrier family 22 (organic cation transporter), member 15 | −0.451 | 4.85E-05 |
| 1563453_at | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | −0.481 | 1.22E-05 |
| 57588_at | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | −0.552 | 3E-07 |
| 234199_at | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | −0.562 | 2E-07 |
| 219090_at | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | −0.568 | 1E-07 |
| 233773_at | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | −0.572 | 1E-07 |
| 219932_at | SLC27A6 | Solute carrier family 27 (fatty acid transporter), member 6 | −0.376 | .0009 |
| 244018_at | SLC2A9 | Solute carrier family 2 (facilitated glucose transporter), member 9 | −0.472 | 1.89E-05 |
| 1556551_s_at | SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | −0.389 | .00057 |
| 1555460_a_at | SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | −0.405 | .000318 |
| 202089_s_at | SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | −0.412 | 2.40E-04 |
| 237833_s_at | SNCAIP | Synuclein, alpha interacting protein (synphilin) | −0.416 | 2.04E-04 |
| 219511_s_at | SNCAIP | Synuclein, alpha interacting protein (synphilin) | −0.429 | 1.25E-04 |
| 243486_at | SND1 | Staphylococcal nuclease and tudor domain containing 1 | −0.380 | .000786 |
| 242736_at | SORBS1 | Sorbin and SH3 domain containing 1 | −0.394 | .000475 |
| 213668_s_at | SOX4 | SRY (sex determining region Y)-box 4 | −0.385 | .000653 |
| 201418_s_at | SOX4 | SRY (sex determining region Y)-box 4 | −0.409 | 2.67E-04 |
| 242086_at | SPATA6 | Spermatogenesis associated 6 | −0.396 | .00044 |
| 1563595_at | SRGAP3 | SLIT-ROBO Rho GTPase activating protein 3 | −0.412 | 2.43E-04 |
| 215550_at | SRGAP3 | SLIT-ROBO Rho GTPase activating protein 3 | −0.429 | 1.21E-04 |
| 209794_at | SRGAP3 | SLIT-ROBO Rho GTPase activating protein 3 | −0.467 | 2.42E-05 |
| 204548_at | STAR | Steroidogenic acute regulator | −0.494 | 6.6E-06 |
| 208425_s_at | TANC2 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | −0.459 | 3.52E-05 |
| 224952_at | TANC2 | Tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | −0.459 | 3.48E-05 |
| 205513_at | TCN1 | Transcobalamin I (vitamin B12 binding protein, R binder family) | −0.467 | 2.39E-05 |
| 209215_at | TETRAN | Tetracycline transporter-like protein | −0.487 | 9.6E-06 |
| 226050_at | TMCO3 | Transmembrane and coiled-coil domains 3 | −0.395 | .00046 |
| 224916_at | TMEM173 | Transmembrane protein 173 | −0.376 | .000895 |
| 241342_at | TMEM65 | Transmembrane protein 65 | −0.403 | .000334 |
| 238045_at | TMEM65 | Transmembrane protein 65 | −0.465 | 2.62E-05 |
| 225802_at | TOP1MT | Topoisomerase (DNA) I, mitochondrial | −0.424 | 1.49E-04 |
| 224836_at | TP53INP2 | Tumor protein p53 inducible nuclear protein 2 | −0.480 | 1.31E-05 |
| 210882_s_at | TRO | Trophinin | −0.389 | .000558 |
| 235737_at | TSLP | Thymic stromal lymphopoietin | −0.401 | .00037 |
| 230625_s_at | TSPAN12 | Tetraspanin 12 | −0.414 | 2.19E-04 |
| 230626_at | TSPAN12 | Tetraspanin 12 | −0.422 | 1.60E-04 |
| 244248_at | TTC27 | Tetratricopeptide repeat domain 27 | −0.457 | 3.79E-05 |
| 218710_at | TTC27 | Tetratricopeptide repeat domain 27 | −0.497 | 5.8E-06 |
| 235749_at | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 | −0.510 | .000003 |
| 1558466_at | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 | −0.525 | 1.3E-06 |
| 1555561_a_at | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 | −0.565 | 1E-07 |
| 1558467_a_at | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 | −0.572 | 1E-07 |
| 218801_at | UGCGL2 | UDP-glucose ceramide glucosyltransferase-like 2 | −0.586 | < 1e-07 |
| 224048_at | USP44 | Ubiquitin specific peptidase 44 | −0.499 | 5.2E-06 |
| 201557_at | VAMP2 | Vesicle-associated membrane protein 2 (synaptobrevin 2) | −0.373 | .000995 |
| 219330_at | VANGL1 | Vang-like 1 (van gogh, Drosophila) | −0.378 | .000838 |
| 229997_at | VANGL1 | Vang-like 1 (van gogh, Drosophila) | −0.382 | .000727 |
| 208626_s_at | VAT1 | Vesicle amine transport protein 1 homolog (T. californica) | −0.541 | 6E-07 |
| 239429_at | ZNF323 | Zinc finger protein 323 | −0.426 | 1.41E-04 |
| 240449_at | ZNF341 | Zinc finger protein 341 | −0.387 | .000599 |
| 1561002_at | ZNF521 | Zinc finger protein 521 | −0.439 | 8.01E-05 |
| 230106_at | ZXDC | ZXD family zinc finger C | −0.373 | .000982 |
| 218639_s_at | ZXDC | ZXD family zinc finger C | −0.380 | .000768 |
| 228919_at | −0.372 | .000997 | ||
| 240865_at | −0.374 | .000944 | ||
| 230135_at | −0.378 | .000821 | ||
| 232833_at | −0.381 | .000758 | ||
| 235681_at | −0.386 | .000638 | ||
| 235456_at | −0.389 | .000563 | ||
| 243806_at | −0.390 | .000538 | ||
| 228740_at | −0.391 | .000532 | ||
| 243742_at | −0.393 | .00048 | ||
| 1557443_s_at | −0.394 | .000473 | ||
| 239488_at | −0.399 | .000385 | ||
| 1556580_a_at | −0.404 | .000331 | ||
| 240566_at | −0.409 | .000267 | ||
| 243082_at | −0.416 | 2.09E-04 | ||
| 1555967_at | −0.422 | 1.61E-04 | ||
| 228349_at | −0.425 | 1.44E-04 | ||
| 1554599_x_at | −0.426 | 1.38E-04 | ||
| 236104_at | −0.437 | 8.91E-05 | ||
| 1554597_at | −0.438 | 8.37E-05 | ||
| 231233_at | −0.449 | 5.36E-05 | ||
| 220898_at | −0.460 | 3.33E-05 | ||
| 1559117_at | −0.461 | 3.09E-05 | ||
| 227929_at | −0.479 | .000014 | ||
| 1555968_a_at | −0.506 | 3.7E-06 | ||
| 217521_at | −0.515 | 2.3E-06 | ||
| 227036_at | −0.591 | < 1e-07 |
Table A2.
GO Terms of Biological Processes Significantly Over-Represented in the Gene-Expression Signature Associated With MN1 Expression
| GO ID | GO Term | Members of the GO Term Present in the Gene Expression Signature (%) | P |
|---|---|---|---|
| GO terms over-represented among the genes positively correlated with MN1 expression | |||
| 2504 | Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 47.06 | < .001 |
| 2478 | Antigen processing and presentation of exogenous peptide antigen | 42.86 | .002 |
| 2495 | Antigen processing and presentation of peptide antigen via MHC class II | 42.86 | .002 |
| 19,886 | Antigen processing and presentation of exogenous peptide antigen via MHC class II | 42.86 | .002 |
| 7618 | Mating | 40.00 | .005 |
| 19,884 | Antigen processing and presentation of exogenous antigen | 37.50 | .002 |
| 19,882 | Antigen processing and presentation | 32.20 | < .001 |
| 48,002 | Antigen processing and presentation of peptide antigen | 25.00 | < .001 |
| 2474 | Antigen processing and presentation of peptide antigen via MHC class I | 20.00 | < .001 |
| 6935 | Chemotaxis | 6.12 | .005 |
| 42,330 | Taxis | 6.12 | .005 |
| 6955 | Immune response | 5.22 | < .001 |
| 7610 | Behavior | 5.14 | .002 |
| 2376 | Immune system process | 4.08 | < .001 |
| 42,221 | Response to chemical stimulus | 3.83 | .005 |
| 50,896 | Response to stimulus | 3.48 | < .001 |
| GO terms over-represented among the genes negatively correlated with MN1 expression | |||
| 35,272 | Exocrine system development | 33.33 | .005 |
| 9953 | Dorsal/ventral pattern formation | 30.77 | < .001 |
| 42,384 | Cilium biogenesis | 22.22 | .004 |
| 42,742 | Defense response to bacterium | 17.95 | < .001 |
| 48,736 | Appendage development | 16.00 | < .001 |
| 35,108 | Limb morphogenesis | 16.00 | < .001 |
| 35,107 | Appendage morphogenesis | 16.00 | < .001 |
| 6334 | Nucleosome assembly | 15.38 | < .001 |
| 9617 | Response to bacterium | 15.22 | .001 |
| 31,497 | Chromatin assembly | 14.12 | < .001 |
| 35,113 | Embryonic appendage morphogenesis | 13.64 | .003 |
| 30,326 | Embryonic limb morphogenesis | 13.64 | .003 |
| 6333 | Chromatin assembly or disassembly | 10.92 | < .001 |
| 65,004 | Protein-DNA complex assembly | 10.57 | < .001 |
| 3002 | Regionalization | 10.34 | .002 |
| 7389 | Pattern specification process | 9.76 | .001 |
| 51,707 | Response to other organism | 8.41 | < .001 |
| 9607 | Response to biotic stimulus | 6.12 | .002 |
| 6325 | Establishment and/or maintenance of chromatin architecture | 5.98 | < .001 |
| 6323 | DNA packaging | 5.93 | < .001 |
| 7001 | Chromosome organization and biogenesis (sensu Eukaryota) | 5.19 | < .001 |
| 51,276 | Chromosome organization and biogenesis | 5.07 | < .001 |
| 65,003 | Macromolecular complex assembly | 3.82 | .001 |
| 6259 | DNA metabolic process | 3.64 | .004 |
| 7275 | Multicellular organismal development | 2.93 | .003 |
| 48,856 | Anatomical structure development | 2.83 | .005 |
| 32,502 | Developmental process | 2.76 | < .001 |
| 48,869 | Cellular developmental process | 2.63 | .005 |
| 30,154 | Cell differentiation | 2.63 | .005 |
Abbreviation: GO, gene ontology.
Table A3.
Signature of 15 microRNA Probes Significantly Correlated With MN1 Expression Level, Grouped by Direction of Correlation
| Target microRNA | Probe Sequence | Correlation Coefficient | P | |
|---|---|---|---|---|
| Probes positively correlated with MN1 expression | ||||
| hsa-miR-126 | ACACTTCAAACTCGTACCGTGAGTAATAATGCGCCGTCCA | 0.405 | .002893 | |
| hsa-miR-126 | ACACTTCAAACTCGTACCGTGAGTAATAATGCGCCGTCCA | 0.388 | .004862 | |
| hsa-miR-126* | CGCTGGCGACGGGACATTATTACTTTTGGTACGCGCTGTG | 0.541 | .000001 | |
| hsa-miR-126* | GCTGGCGACGGGACATTATTACTTTTGGTACGCGCTGTGA | 0.502 | .000005 | |
| hsa-miR-126* | GACGGGACATTATTACTTTTGGTACGCGCTGTGACACTTC | 0.482 | .000012 | |
| hsa-miR-129-5p | CCCTTCGCGAATCTTTTTGCGGTCTGGGCTTGCTGTACAT | 0.391 | .000517 | |
| hsa-miR-130b | CTGGGAAGCAGTGCAATGATGAAAGGGCATCGGTCAGGTC | 0.340 | .002874 | |
| hsa-miR-424 | GAGGGGATACAGCAGCAATTCATGTTTTGAAGTGTTCTAA | 0.346 | .002367 | |
| Probes negatively correlated with MN1 expression | ||||
| hsa-miR-16 | GTTCCACTCTAGCAGCACGTAAATATTGGCGTAGTGAAAT | −0.332 | .003575 | |
| hsa-miR-16 | CAATGTCAGCAGTGCCTTAGCAGCACGTAAATATTGGCGT | −0.339 | .002957 | |
| hsa-miR-19a | TGTAGTTGTGCAAATCTATGCAAAACTGATGGTGGCCTGC | −0.342 | .004595 | |
| hsa-miR-20a | TAAAGTGCTTATAGTGCAGGTAGTGTTTAGTTATCTACTG | −0.324 | .004543 | |
| hsa-miR-100 | TGAGGCCTGTTGCCACAAACCCGTAGATCCGAACTTGTGG | −0.411 | .000250 | |
| hsa-miR-100 | CCTGTTGCCACAAACCCGTAGATCCGAACTTGTGGTATTA | −0.486 | .000040 | |
| hsa-miR-196a | AGCTGATCTGTGGCTTAGGTAGTTTCATGTTGTTGGGATT | −0.335 | .004960 |
Footnotes
Supported in part by Grants No. CA101140, CA114725, CA31946, CA33601, CA16058, CA77658, CA35279, CA03927, and CA41287 from the National Cancer Institute, Bethesda, MD, and The Coleman Leukemia Research Foundation.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Guido Marcucci, Michael D. Radmacher, Clara D. Bloomfield
Financial support: Michael A. Caligiuri, Clara D. Bloomfield
Administrative support: Guido Marcucci, Michael A. Caligiuri, Clara D. Bloomfield
Provision of study materials or patients: Guido Marcucci, Ravi Vij, Bayard L. Powell, Jonathan E. Kolitz, Michael A. Caligiuri, Richard A. Larson, Clara D. Bloomfield
Collection and assembly of data: Guido Marcucci, Peter Paschka, Susan P. Whitman, Claudia D. Baldus, Andrew J. Carroll
Data analysis and interpretation: Christian Langer, Guido Marcucci, Kelsi B. Holland, Michael D. Radmacher, Kati Maharry, Peter Paschka, Susan P. Whitman, Krzysztof Mrózek, Claudia D. Baldus, Andrew J. Carroll, Clara D. Bloomfield
Manuscript writing: Christian Langer, Guido Marcucci, Kelsi B. Holland, Michael D. Radmacher, Kati Maharry, Peter Paschka, Susan P. Whitman, Krzysztof Mrózek, Clara D. Bloomfield
Final approval of manuscript: Christian Langer, Guido Marcucci, Kelsi B. Holland, Michael D. Radmacher, Kati Maharry, Peter Paschka, Susan P. Whitman, Krzysztof Mrózek, Claudia D. Baldus, Ravi Vij, Bayard L. Powell, Andrew J. Carroll, Jonathan E. Kolitz, Michael A. Caligiuri, Richard A. Larson, Clara D. Bloomfield
REFERENCES
- 1.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 3.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 4.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 5.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Wely KHM, Molijn AC, Buijs A, et al. The MN1 oncoprotein synergizes with coactivators RAC3 and p300 in RAR-RXR-mediated transcription. Oncogene. 2003;22:699–709. doi: 10.1038/sj.onc.1206124. [DOI] [PubMed] [Google Scholar]
- 7.Sutton ALM, Zhang X, Ellison TI, et al. The 1,25(OH)2D3-regulated transcription factor MN1 stimulates vitamin D receptor-mediated transcription and inhibits osteoblastic cell proliferation. Mol Endocrinol. 2005;19:2234–2244. doi: 10.1210/me.2005-0081. [DOI] [PubMed] [Google Scholar]
- 8.Lekanne Deprez RH, Riegman PHJ, Groen NA, et al. Cloning and characterization of MN1, a gene from chromosome 22q11, which is disrupted by a balanced translocation in a meningioma. Oncogene. 1995;10:1521–1528. [PubMed] [Google Scholar]
- 9.Buijs A, Sherr S, van Baal S, et al. Translocation (12;22)(p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q1. Oncogene. 1995;10:1511–1519. [PubMed] [Google Scholar]
- 10.Valk PJM, Verhaak RGW, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 11.Carella C, Bonten J, Sirma S, et al. MN1 overexpression is an important step in the development of inv16 AML. Leukemia. 2007;21:1679–1690. doi: 10.1038/sj.leu.2404778. [DOI] [PubMed] [Google Scholar]
- 12.Carella C, Bonten J, Rehg J, et al. MN1-TEL, the product of the t(12;22) in human myeloid leukemia, immortalizes murine myeloid cells and causes myeloid malignancy in mice. Leukemia. 2006;20:1582–1592. doi: 10.1038/sj.leu.2404298. [DOI] [PubMed] [Google Scholar]
- 13.Heuser M, Argiropoulos B, Kuchenbauer F, et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood. 2007;110:1639–1647. doi: 10.1182/blood-2007-03-080523. [DOI] [PubMed] [Google Scholar]
- 14.Heuser M, Beutel G, Krauter J, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 15.Kolitz JE, George SL, Dodge RK, et al. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B study 9621. J Clin Oncol. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- 16.Kolitz JE, George SL, Marcucci G, et al. A randomized comparison of induction therapy for untreated acute myeloid leukemia (AML) in patients < 60 years using P-glycoprotein (Pgp) modulation with Valspodar (PSC833): Preliminary results of Cancer and Leukemia Group B study 19808. Blood. 2005;106(abstr 407):122a–123a. [Google Scholar]
- 17.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 18.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 19.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 21.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 23.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paschka P, Marcucci G, Ruppert AS, et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 26.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 28.Marcucci G, Maharry K, Whitman SP, et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 29.Baldus CD, Tanner SM, Ruppert AS, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: A Cancer and Leukemia Group B study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 30.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: A Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radmacher MD, Marcucci G, Ruppert AS, et al. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: A Cancer and Leukemia Group B study. Blood. 2006;108:1677–1683. doi: 10.1182/blood-2006-02-005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 33.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 34.Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. New York, NY: Springer-Verlag; 1997. [Google Scholar]
- 35.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahlquist KD, Salomonis N, Vranizan K, et al. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 37.Verhaak RGW, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 38.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Donnell KA, Wentzel EA, Zeller KI, et al. C-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 44.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 45.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.