Abstract
Through alternative splicing, multiple different transcripts can be generated from a single gene. Alternative splicing represents an important molecular mechanism of gene regulation in physiological processes such as developmental programming as well as in disease. In cancer, splicing is significantly altered. Tumors express a different collection of alternative spliceoforms than normal tissues. Many tumor-associated splice variants arise from genes with an established role in carcinogenesis or tumor progression, and their functions can be oncogenic. This raises the possibility that products of alternative splicing play a pathogenic role in cancer. Moreover, cancer-associated spliceoforms represent potential diagnostic biomarkers and therapeutic targets. G protein-coupled peptide hormone receptors provide a good illustration of alternative splicing in cancer. The wild-type forms of these receptors have long been known to be expressed in cancer and to modulate tumor cell functions. They are also recognized as attractive clinical targets. Recently, splice variants of these receptors have been increasingly identified in various types of cancer. In particular, alternative cholecystokinin type 2, secretin, and growth hormone-releasing hormone receptor spliceoforms are expressed in tumors. Peptide hormone receptor splice variants can fundamentally differ from their wild-type receptor counterparts in pharmacological and functional characteristics, in their distribution in normal and malignant tissues, and in their potential use for clinical applications.
RNA splicing is the process by which introns are removed from precursor mRNA to form a mature transcript ready for translation. Through splice site variation, multiple different transcripts can be produced from a single pre-mRNA. This so-called alternative splicing contributes to the diversity of the human proteome, which is generated from a limited genome. Alternative splicing occurs physiologically, such as during development, as well as in pathology. It has been found to be associated with a number of nonneoplastic diseases like cystic fibrosis and retinitis pigmentosa. Moreover, alternative splicing is increasingly recognized in neoplasia. Indeed, cancers express a broad collection of mRNA splice variants that may be distinct from those occurring physiologically, with some even exhibiting oncogenic functions. This raises the possibility that products of alternative splicing play a pathogenic role in cancer and modify tumor behavior, as well as may be potentially useful as diagnostic or prognostic biomarkers or as therapeutic targets.
A large variety of cellular and extracellular proteins relevant in the neoplastic process are alternatively spliced in cancer.1 Among them, G protein-coupled peptide hormone receptors represent important examples. Situated in the cell membrane, these receptors are specialized to transmit extracellular signals into the cell: On ligand binding, intracellular second messenger systems are activated through mediation of G proteins, which eventually modulate gene expression and protein activity. Peptide hormone receptors constitute the largest family of plasma membrane proteins. They are involved in the regulation of a plethora of important cell functions in physiology and disease. Accordingly, they are the targets of a large number of pharmaceutical drugs.
Alternative splicing of peptide hormone receptors has long been known to occur under physiological conditions, where it may contribute to functional receptor diversity, despite a restricted repertoire of receptor genes and receptor ligands.2,3 More recently, peptide hormone receptor splice variants have been recognized to arise also in cancer. These cancer-associated receptor spliceoforms can fundamentally differ from the wild-type receptors by exhibiting other pharmacological or functional characteristics. For instance, a peptide receptor splice variant may not bind a ligand that shows high affinity for the wild-type receptor.4 Or a peptide receptor splice variant may exhibit constitutive activity, whereas the wild-type form signals only on ligand binding.5 Moreover, peptide receptor splice variants may display a different distribution in normal and malignant tissues than wild-type receptors, with often predominant expression in cancer.6,7,8 Finally, peptide receptor splice variants may differ from wild-type forms in their potential use for clinical applications.8 Therefore, peptide hormone receptors provide a good illustration of our current understanding of alternative splicing in cancer.
We have organized this review to first describe the splicing process and general aspects of alternative splicing in cancer with respect to underlying molecular mechanisms, presumed biological significance, and clinical potential. As an illustration of these concepts, we then focus on alternative splicing of peptide hormone receptors in cancer. We analyze the impact of alternative splicing events on receptor pharmacology and functionality, and possible uses of peptide receptor spliceoforms for cancer diagnosis and therapy.
The Process of Pre-mRNA Splicing
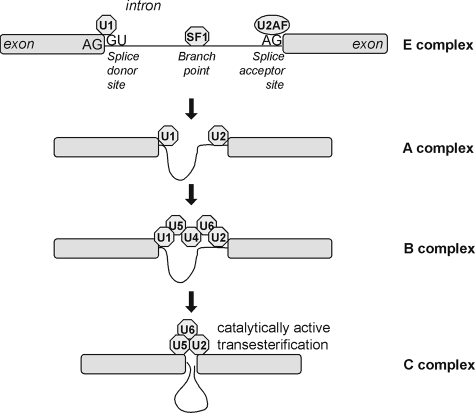
Most eukaryotic pre-mRNAs contain intervening sequences (introns) that must be removed (spliced out) to yield the correct coding frame within the concatenated exon sequences. Splicing occurs when specific nucleotide sequences at intron-exon boundaries within the pre-mRNA are recognized by the spliceosomal splicing machinery, a complex of small nuclear ribonucleoproteins (snRNPs) and proteins that assemble in a temporally and spatially specific manner to catalyze intron excision and ligation of exon ends (Figure 1).9 This process involves sequential changes in RNA–RNA, RNA–protein, and protein–protein interactions within this complex. The classical splicing process acts on an intron defined by a donor splice site at the 5′ end beginning with a GU dinucleotide, an acceptor splice site at the 3′ end ending with an AG dinucleotide, and a branch point upstream of the 3′ end. Splicing is initiated by the establishment of the E (early) complex, with U1 snRNP binding to the donor site, splicing factor 1 binding to the branch point, and U2 snRNP auxiliary factor binding to the acceptor site. Splicing initiation is followed by formation of the A complex, with U2 snRNP binding to the branch point, and formation of the B complex, with U4, U6, and U5 snRNPs joining the complex and leading to remodeling. This remodeling leads to the C complex, which is catalytically active, resulting in trans-esterification reactions leading to the cleavage and ligation of the nucleotide chain.
Figure 1.
Molecular mechanism of pre-mRNA splicing. The splicing process is initiated by recognition of the splice donor and acceptor sites by U1 snRNP and U2 snRNP auxiliary factor, respectively, and binding of splicing factor 1 to the branch point. Thereupon, the intron ends are brought in close proximity. This is followed by further assembly of the spliceosome and remodeling of the complex, which eventually becomes catalytically active to splice out the intron and join the ends of the exons.
Exonic and intronic splicing enhancer and silencer regions represent short (<10) nucleotide sequences away from the splice sites that bind a wide range of stimulatory and inhibitory regulatory splicing factors, such as serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins. These importantly contribute to splice site recognition by the splicing machinery. The activity of splicing factors is often regulated by their phosphorylation status.10
Splice site selection by the spliceosome is flexible, allowing the generation of differentially spliced mRNA isoforms of the same gene. Given the molecular mechanisms described above, it can be easily envisioned that alternative splicing may occur in response to less-than-optimal splice site sequences or variations in activity of protein kinases or phosphatases, in the stoichiometry of splicing regulatory factors, or even key core components of the spliceosome.11,12 These differences can result in the excision of an exon that is normally included in coding, the inclusion of an intron, the inclusion of an exonic cassette that is not normally recognized, or the use of alternative 5′ or 3′ splice sites. Combinations of these events are also observed. Substantial protein sequence variation can occur by a shift in the coding frame, by the inclusion of nucleotide sequences that are not normally part of the mature mRNA, or by the introduction of a premature stop codon.
Splice Variant Expression Patterns in Tumors
The splice variant expression in tumors has classically been assessed at the single gene level by PCR and sequencing. Recently, it has also increasingly been examined on a genome-wide basis with bioinformatic and microarray strategies.13 Tumoral RNA splicing can also be analyzed computationally by comparing mRNA sequences deposited in the GenBank with the human EST database. This process reveals extensive variant spliceoforms present in tumor libraries.14 Furthermore, splice variant expression signatures in tumors can be characterized with oligonucleotide arrays directed against splice sites.15 These investigations have rapidly enlarged the number of known splice variants in cancer. Moreover, they have revealed that splice variant expression patterns in tumors may be distinct from those in normal tissues.
For many genes, the expression ratio between a splice variant and the wild-type form or between various splice variants significantly differs between cancer and normal cells. In particular, many splice variants are up-regulated in tumors compared with normal tissues. Some spliceoforms even appear to arise de novo in cancer, as they are not found under normal conditions. Of note, splice variant expression patterns in tumors have usually been compared with those in normal tissues, but not with those occurring in nonneoplastic diseases such as inflammation. It is possible that some spliceoforms thought to be typical of cancer may also arise in nontumor pathology.
Molecular Mechanisms of Splicing in Tumors
The molecular mechanisms underlying the distinct splicing in cancer are incompletely understood and are by far not defined for every spliceoform. The investigation of individual genes has, however, revealed that the normal splicing process can be disrupted in cancer at various critical steps.
Point mutations and single nucleotide polymorphisms can interfere with normal splice site selection and splicing efficiency: Nucleotide changes occurring within splice site consensus sequences can reduce binding affinity of spliceosomal proteins; likewise, sequence alterations within exonic and intronic splicing enhancer and silencer regions affect binding of regulatory splicing factors. Possible results of these changes include exon skipping, such as in the phosphatase and tensin homolog (PTEN) and adenomatosis polyposis colic (APC) splice variants that underlie familial cancer syndromes,16,17 intron retention, or preferential use of a nearby cryptic splice site, such as in a Kruppel-like factor 6 splice variant associated with prostate cancer.18 Furthermore, genomic sequence alterations can create new splice sites, a mechanism that is responsible for instance for a the tyrosine kinase receptor splice variant in gastrointestinal stromal tumors,19 or new splicing enhancer or silencer regions that influence splice site choice.18 It is likely that many genomic mutations and single nucleotide polymorphisms important for missplicing in cancer are not yet detected or recognized as relevant, since splicing enhancer and silencer regions are not fully defined.
An imbalance in the levels of stimulatory and inhibitory splicing factors is likely to contribute to splicing in cancer as well. Indeed, individual splicing factors are selectively up-regulated in tumors. For example, human ovarian cancers exhibit significantly higher levels of specific SR proteins than nonneoplastic ovaries.20 Likewise, the spliceosomal protein SPF45 is overexpressed in numerous carcinomas as compared with normal tissues.21 Up-regulation of stimulatory splicing factors during the neoplastic process was found to be associated with increased splicing of genes regulated by these splicing factors, such as in a mouse model of mammary tumorigenesis and in human breast cancer.22,23
Changes in splicing factor levels, as well as sequence alterations in genomic regions critical for splicing, usually do not completely abrogate wild-type expression; instead, wild-type and splice variant forms are concomitantly expressed.
Do Splice Variants Contribute to Malignancy?
Some cancer-associated spliceoforms arise from genes with an established role in carcinogenesis or tumor progression, such as tumor suppressor genes, protooncogenes, and genes involved in cell motility. The effects of these splice variants are often favorable for cancer development. In fact, the functions of some of these splice variants may be more oncogenic than those of the corresponding wild-type forms. In particular, many splice variants of genes involved in cell death antagonize the pro-apoptotic effect of their wild-type counterparts in a dominant negative fashion and thus stimulate tumor growth. A typical example is the Kruppel-like factor 6 splice variant 1 (SV1), which is over-expressed in metastatic prostate cancer. SV1 stimulates growth and invasion, whereas wild-type Kruppel-like factor 6 suppresses proliferation of prostate cancer cells.18,24 Conversely, splice variants of proto oncogenes can show a gain of function compared with corresponding wild-type forms. For instance, a splice variant of the tyrosine kinase receptor occurring in gastrointestinal stromal tumors was found to be constitutively active, in contrast to wild-type the tyrosine kinase receptor.19 Constitutively active the tyrosine kinase receptor is known to lead to gastrointestinal stromal tumors development. Similarly, over-expression of a splice variant of the proto oncogene Mdm2 p53 binding protein homolog (MDM2) leads to malignant transformation of transfected cells in vitro and spontaneous tumor formation in vivo.25,26 Finally, splice variants of genes involved in cell migration can stimulate invasion of cancer cells, as demonstrated for a constitutively active spliceoform of Ron overexpressed in breast and colon cancer27 and various CD44 spliceoforms. For instance, the CD44 splice variant CD44v5 was found to increase tumor cell invasiveness.28 Moreover, tumors expressing the CD44 splice variant CD44v4-10 showed significantly more metastases, as well as larger primary tumors, than tumors expressing nonspliced standard CD44 in vivo.29
But what is the actual biological relevance of the oncogenic effects of tumor-associated splice variants defined in experimental models? This is indeed an unresolved issue. Functions of splice variants are generally assessed in cells engineered to express the spliceoform in question selectively and in what may be nonphysiologically high amounts.24,27 It is possible that splice variants do not modify cell functions to the same extent in naturally occurring tumors, where they are often expressed at significantly lower levels and in a different cellular context.
Moreover, it has not been clearly determined whether splice variants may play a primary role in carcinogenesis and tumor progression. Alternatively, their occurrence in cancer may reflect an epiphenomenon related to the general deregulation of cellular functions or the special tumoral microenvironment, such as hypoxia.30 While the latter scenario may explain the presence of splice variants with no apparent function in cancer, it also provides the possibility for oncogenic splice variants to contribute to the malignant phenotype through the process of clonal selection.31
Potential Clinical Use of Cancer-Associated Splice Variants
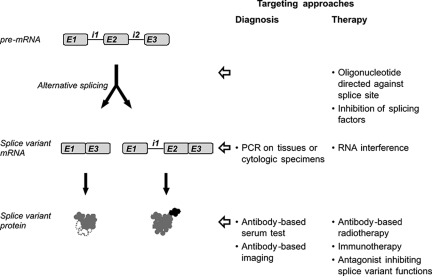
Based on their expression and functions in neoplasia, splice variants may potentially be used clinically for diagnostic and therapeutic purposes. Conceivable approaches are summarized in Figure 2. Such applications are the focus of current research, but have, however, not been established in clinical practice.
Figure 2.
Potential approaches to use cancer-associated splice variants clinically for diagnostic or therapeutic purposes. For diagnostic applications, the splice variant transcript or protein can serve as target that is detected by PCR or with an antibody, respectively. For cancer therapy, splice variants with a function promoting tumor development may be down-regulated by inhibition of splice variant synthesis at the level of the splicing process or by RNA interference at the mature mRNA level. Furthermore, tumoral splice variant proteins may serve as targets for radiotherapy or immunotherapy.
Splice Variant-Targeted Tumor Diagnosis
Diagnostic approaches aim at taking advantage of the increased production of splice variant transcripts or proteins by tumors, as compared with normal tissues, such that splice variant functions need not be known.
Tumor cells can be identified in clinical specimens with PCR on the basis of their specific splice variant expression pattern. For instance, by measuring in pleural effusion or urine samples the mRNA ratio of two CD44 splice variants (namely CD44v8-10 expressed predominantly in epithelial cancers and CD44v10 occurring in normal leukocytes) malignant conditions could be distinguished from reactive ones with high specificity. This assay was able to reveal cancer even in specimens that were cytologically negative or ambiguous.32 Furthermore, such a test may also provide prognostic information, as shown in a follow-up study: The ratio of CD44v8-10 versus standard CD44 mRNA levels in urine samples correlated with bladder cancer stage, metastasis, and survival.33 These studies provide interesting examples of how cancer cells may be detected with a highly sensitive technique, such as PCR, based on their splice variant expression pattern, despite the fact that the splice variants are present not only in tumor cells, but also in normal cells. However, these tests have not reached clinical practice to date.
Furthermore, splice variants may serve as serum markers of cancer. If the protein product of a spliceoform is soluble and secreted from tumor cells, it may be detectable in the serum with an antibody-based assay. This is the case for the aforementioned CD44 spliceoform CD44v8-10, which can be measured in the serum with an enzyme-linked immunosorbent assay (ELISA) test using an antibody directed against the unique region of this variant. CD44v8-10 serum levels were thus found to be significantly higher in patients with colon cancer than in normal controls and were associated with tumor burden, stage, and metastasis.34
Splice variant-specific antibodies may also be used for targeted tumor scintigraphy when radiolabeled. This technique was successfully performed for a fibronectin spliceoform that expresses the variable extracellular domain B and accumulates around tumoral neovascularizations.35 Fibronectin-extracellular domain B-expressing lung cancers and liver metastases as small as 4 to 6 mm could be imaged with an intravascularly injected 123I-labeled antibody specifically recognizing the extracellular domain B.36 Conversely, less encouraging results were obtained for tumors expressing the CD44 spliceoform CD44v6. With a 99mTc-labeled antibody against CD44v6, 80% of primary tumors, but only 20% of metastases of head and neck cancers, could be visualized.37 Likewise, a 186Re-labeled CD44v6 antibody did not clearly image breast cancers because of high background radioactivity.38 Indeed, success of tumor targeting with radiolabeled antibodies depends on a number of factors that include, besides antibody factors, high target expression levels in tumors, but low target levels in normal background tissues. Apparently, these prerequisites are not fulfilled for all tumor-associated splice variants considered for this application.
Splice Variant-Targeted Tumor Therapy
For therapeutic applications, either the presence or the functions of cancer-associated splice variants may be targeted. The former strategy appears timelier, as many spliceoforms are not yet functionally characterized or their biological relevance is not fully elucidated.
Specific antibodies coupled to a radionuclide or a chemotherapeutic agent may potentially be used for targeted radio- or chemotherapy, analogous to splice variant-targeted tumor imaging. This technique has been tested clinically for CD44v6-expressing tumors. Phase I trials were performed with various toxic anti-CD44v6 antibodies in patients with head and neck cancer, as well as breast cancer.39,40 However, tumor response rates were small, and fatal side effects related to antibody binding to normal tissues occurred. It is likely that for such applications the CD44v6 expression levels are not sufficient in tumors, while being too high in normal tissues.41
The presence of splice variants in cancer may furthermore be used for immunotherapy.13 The unique sequences of cancer-associated splice variants that are not shared by the wild-type forms represent tumoral neoantigens that can be recognized by the immune system. Accordingly, a number of splice variant proteins have been found to induce a cellular or humoral immune response in cancer patients.13 This represents the basis for potential immunotherapies, including stimulation of cytotoxic T-cells and vaccination, which in fact have been shown to be effective in animal tumor models. For instance, in a mouse model of mammary adenocarcinoma, active immunization with a cancer-associated CD44 splice variant resulted in significantly reduced tumor growth.29
As for the therapeutic inhibition of splice variant effects on important tumor cell functions, such as apoptosis, the main approach has been to decrease splice variant synthesis (reviewed by Garcia-Blanco42), whereas pharmaceutical antagonism of splice variant functions has been less explored in cancer. The expression of a splice variant may be specifically inhibited with an oligonucleotide interfering with splice site choice. Conventional antisense oligonucleotides sterically block splice sites. For instance, oligonucleotides directed against the anti-apoptotic splice variant of bcl-x increased the relative expression of the pro-apoptotic bcl-x form and stimulated apoptosis in cancer cells.43,44 Conversely, splicing can be induced by bifunctional oligonucleotides exhibiting an RNA binding domain directed against an exonic splicing enhancer sequence and a splicing factor recruitment domain mediating interaction with RNA binding proteins.45 Furthermore, spliceoforms can be degraded through RNA interference.42
The expression of splice variants can also be modified nonspecifically by inhibiting splicing factors either pharmacologically or by RNA interference. This approach was shown to exert a negative effect on cancer cells in vitro. For example, the topoisomerase I inhibitor NB-506 reduces phosphorylation and thus the activity of SR proteins. In cancer cells, it inhibits splicing and is cytotoxic.46 Likewise, knockdown of a SR protein kinase responsible for SR protein phosphorylation by RNA interference increased apoptosis and sensitivity to chemotherapeutic drugs of pancreatic cancer cell lines.47
Alternative Splicing of Peptide Hormone Receptors in Cancer
G protein-coupled peptide hormone receptors represent an important family of molecular targets in cancer.48 Many of them show significant overexpression in tumors. On this basis, clinical applications are being developed. Radiolabeled receptor ligands can be applied to targeted imaging and radiotherapy of receptor-expressing tumors. Moreover, peptide hormone analogs that interfere with receptor-regulated tumor cell functions, such as hormone secretion and proliferation, are used for therapy. In the last few years, it has increasingly been recognized that tumoral peptide receptors correspond not only to the wild-type forms, but also to splice variants. The functional characteristics of these splice variants often differ substantially from those of their wild-type counterparts. While some splice variants inhibit expression or ligand binding and signaling of the corresponding wild-type forms, others amplify wild-type effects. Peptide receptor spliceoforms may thus influence clinical applications based on wild-type receptor expression and functions. Furthermore, they also represent potential clinical targets on their own.
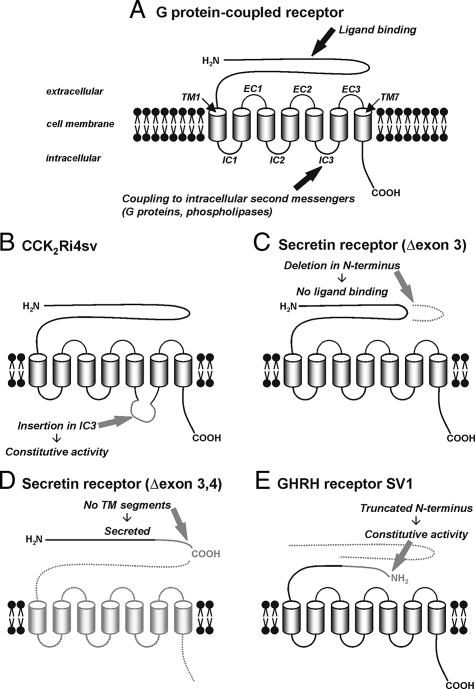
Table 1 provides a summary of the peptide hormone receptor splice variants known to occur in cancer. Figure 3, A–E shows typical examples of how the normal receptor structure can be altered in these splice variants and how this affects receptor function. In the following sections, the most important receptor spliceoforms are discussed in detail.
Table 1.
G Protein-Coupled Peptide Hormone Receptor Splice Variants in Tumors
| Receptor | Transcript | Protein | Function | Tumoral expression | In vivo targeting |
|---|---|---|---|---|---|
| CCK2 receptor | Intron 4 retention | 69 aa insertion in IC3 | Constitutive activity; stimulation of proliferation | Insulinomas, gastrointestinal stromal tumors, colorectal adenomas and adenocarcinomas, pancreatic and gastric adenocarcinomas, Barrett’s mucosa, small and non-small cell lung cancer | Antisense oligonucleotide inhibits growth of pancreatic cancer xenografts in nude mice; 90in vivo tumor binding of radiolabeled ligand in animal model |
| Secretin receptor | Exon 3 skipping | 36 aa deletion in N-terminus | No ligand binding, dominant negative activity on wild-type. Inhibition of gastrin secretion from gastrinomas? | Gastrinomas, pancreatic adenocarcinomas, cholangiocellular carcinomas | |
| Exons 3 + 4 skipping | No TMs | No ligand binding, secreted | Pancreatic adenocarcinomas, cholangiocellular carcinomas | Detectable in pancreatic cancer and chronic pancreatitis patients’ sera with ELISA | |
| Exons 2 + 3 skipping | No TMs | No ligand binding | Lung carcinoids | ||
| Exon 9 skipping | Truncation in IC2 | No signaling | Lung carcinoids | ||
| GHRH receptor | Exons 1-3 skipping, intron 3 retention (SV1) | N-terminal truncation | Constitutive activity; stimulation of proliferation | Prostate, breast, endometrial, ovarian, pancreatic, colorectal, gastric, esophageal, and small cell lung cancer, adrenal cortical carcinomas, insulinomas, gastrinomas, pituitary adenomas, glioblastomas, malignant melanomas, lymphomas | Antagonists inhibit growth of tumor xenografts in nude mice |
| Exons 1-3 and 7 skipping, intron 3 retention (SV2) | N-terminal truncation, truncation after EC1 | Prostate, breast, ovarian, and pancreatic cancer | |||
| Exons 1-3 and 5–7 skipping, intron 3 retention (SV4) | N-terminal truncation, no TMs | Pituitary adenomas | |||
| Exon 11 skipping | Truncation after IC3 | No signaling; dominant negative activity on wild-type | Pituitary adenomas | ||
| PAC1 receptor | Variable skipping of exons 4-6 and 13-16 | Deletions in N-terminus and IC3 | Variable ligand binding affinity and stimulation of intracellular signal transduction | Neuroblastomas, retinoblastomas, colorectal carcinomas | |
| VPAC1 receptor | Exons 10 + 11 skipping | Deletion of part of TM5, IC3, TM6, EC3, and TM7 | No signaling | B and T cell lymphomas, colorectal carcinomas | |
| VPAC2 receptor | Exons 10 + 11 skipping | Deletion of part of TM5, IC3, TM6, EC3, and part of TM7 | No signaling | T cell leukemia | |
| LH/hCG receptor | Exon 9 skipping | 62 aa deletion in N-terminus | No ligand binding, no cell surface expression, complexing with and degradation of wild-type LH/hCG and FSH receptors in endoplasmic reticulum | Endometrial and ovarian adenocarcinoma | |
| Exon 9 skipping, intron 10 retention89 | No TMs | Ovarian adenocarcinoma | |||
| Corticotropin releasing factor receptor 1 | Intron retention | 29 aa insertion in IC191 | Pituitary adenoma |
aa, amino acids; EC, extracellular loop; IC, intracellular loop; TM, transmembrane segment.
Figure 3.
Alternative splicing of G protein-coupled peptide hormone receptors. A: Schematic structure of G protein-coupled peptide hormone receptors with the functionally important domains. The extracellular N-terminal domain provides the ligand binding site. The seven transmembrane segments (TM) anchor the receptor in the cell membrane; they are connected by three intracellular (IC) and three extracellular (EC) loops. The intracellular loops, especially the third, couple to intracellular second messengers as a result of a conformational change of the receptor on ligand binding and are, thus, important for intracellular signal transduction. The intracellular C-terminal domain mediates receptor desensitization and internalization. B–E: Typical structural changes in receptor splice variants. B: An extension in the third intracellular loop of the CCK2 receptor, created by intron 4 retention, is associated with constitutive activation of intracellular signaling pathways in the absence of a ligand. C: An in-frame deletion within the extracellular N-terminal domain of the secretin receptor, originating from exon 3 skipping, inhibits ligand binding. D: A deletion in the N-terminal domain of the secretin receptor produced by skipping of exons 3 and 4 leads to a frame-shift, early stop codon, and truncation; in the absence of any transmembrane domain, the receptor is not anchored in the cell membrane and secreted from the cell. E: N-terminal truncation of the GHRH receptor in SV1, created by deletion of exons 1 through 3 and retention of intron 3, leads to receptor activity in the absence of ligand binding.
Cholecystokinin Receptor Type 2
The cholecystokinin receptor type 2 (CCK2 receptor) is physiologically expressed mainly in the brain and gastrointestinal tract, where it mediates the effects of gastrin on gastric acid secretion and cell growth.49 In addition, its wild-type form is expressed by a wide variety of gut and lung tumors, where it also stimulates proliferation.50,51 Several splice variants of the CCK2 receptor exist, and one of them has been found to be also present in cancer. This particular spliceoform retained intron 4 (CCK2Ri4sv). The molecular basis for this alternative splicing event was characterized to be a weak 3′ splice site of intron 4 in association with reduced levels of the splicing factor U2 snRNP auxiliary factor 35 in tumor cells.52
CCK2Ri4sv expression is strongly associated with cancer. It was described in a number of gastrointestinal and lung tumors and tumor precursor lesions (Table 1), but much more rarely in normal tissues.5,8,52,53,54,55 Of note, reports on CCK2Ri4sv expression in colorectal and gastric cancer are inconsistent.5,8,56 This variable identification of CCK2Ri4sv may at least in part be due to the very small amounts of CCK2 receptor transcripts in these tumors.8 For amplification of very low abundance transcripts, technical aspects that varied between the different studies, such as the amount of RNA used for RT-PCR or primer design, may be critical.
Intron 4 retention leads to the insertion of 69 amino acids in the third intracellular loop of the CCK2 receptor protein, a domain relevant for signal transduction (Figure 3B). Accordingly, CCK2Ri4sv was found to differ from the wild-type CCK2 receptor with respect to activation of intracellular signal transduction pathways, whereas ligand binding characteristics were unchanged: CCK2Ri4sv showed spontaneous signaling in the absence of a receptor ligand.5 Of note, this constitutive activity appears to depend on the cell type expressing CCK2Ri4sv. It was observed particularly in mesenchymal cells, but not consistently in epithelial cells.57,58 In transfected cells, CCK2Ri4sv stimulated cell proliferation in vitro and in vivo, possibly more potently than the wild-type receptor.5,58,59 CCK2Ri4sv expression was also associated with increased tumoral angiogenesis in vivo.59 However, it is unclear if these tumor-promoting effects of CCK2Ri4sv observed in experimental models are similar in naturally occurring tumors. In fact, studies assessing the functional effects of CCK2Ri4sv were performed mainly with engineered cells exhibiting excessive CCK2Ri4sv expression, while original tumors were found to express this variant at very low levels.8 Consequently, the impact of CCK2Ri4sv on tumor growth could be much smaller in original tumors. Therefore, the potential therapeutic benefit that may be obtained from the inhibition of CCK2Ri4sv with, for instance, antisense oligonucleotides in cancer patients is at present unknown.
It has also been proposed to target tumoral CCK2Ri4sv with a selective radiolabeled receptor ligand for imaging or radiotherapeutic purposes has also been proposed based on a pilot study performed in a CCK2Ri4sv-expressing animal tumor model.60 The attractive aspect of such a procedure would be the low expression of CCK2Ri4sv in normal tissues. However, selective ligands that bind only to CCK2Ri4sv but not to the wild-type CCK2 receptor, which exhibits essentially the same ligand binding domain as the splice variant and is abundantly expressed in normal tissues, have not yet been reported. Moreover, it is doubtful that the CCK2Ri4sv protein levels in original human tumors are sufficient for a successful targeting with a radioactive ligand.
Secretin Receptor
The secretin receptor occurs physiologically in pancreatic and bile ducts and the gastric mucosa, where it stimulates secretion of bicarbonate and gastrin, respectively. It is expressed by a limited number of liver, pancreatic, and lung tumors.6,7,61 All these tumors exhibit not only wild-type, but also splice variant secretin receptor expression (Table 1). Alternative splicing of the secretin receptor is characterized by several special features. In contrast to the CCK2 receptor, a number of different cancer-associated splice variants have been identified for the secretin receptor due to a more complex gene structure with more exons. Moreover, these secretin receptor spliceoforms show a high specificity for cancer. Indeed, most of them have been identified only in tumors, but not in normal tissues. Finally, the various spliceoforms are differentially expressed in tumors. Thus, secretin receptor splice variants are of particular interest for potential clinical applications.
The secretin receptor splice variant first described has exon 3 inappropriately spliced out. This variant is expressed in gastrinomas in very high incidence, as well as in a subset of pancreatic ductal adenocarcinomas and cholangiocellular carcinomas, but not in nonneoplastic pancreas or liver.4,6,7,62,63 Exon 3 skipping leads to an in-frame deletion of 36 amino acids within the N-terminal domain of the secretin receptor (Figure 3C). The variant receptor can thus not bind secretin. Though nonfunctional by itself, it inhibits wild-type secretin receptor binding and signaling through receptor heterodimerization.4,63 These data suggest that this splice variant may interfere with wild-type receptor-controlled tumor cell functions, such as stimulation of gastrin secretion from gastrinomas or cell proliferation, depending on the ratio of splice variant to wild-type receptor levels. Initial data indeed indicated that the splice variant with the exon 3 deletion may be responsible for the negative secretin stimulation test obtained in rare gastrinoma patients.4 This observation was, however, not corroborated in a recent study that included a larger number of gastrinoma patients, which may be explained by the generally very low splice variant amounts as compared with wild-type secretin receptor levels in these tumors.62 This example illustrates the importance of validating functional characteristics of splice variants obtained in in vitro models by quantitative expression analysis in original tumors and systematic clinical correlation.
A second secretin receptor spliceoform with tandem deletion of exons 3 and 4 was found expressed in pancreatic adenocarcinomas and cholangiocellular carcinomas, but not in gastrinomas or nonneoplastic pancreas or liver specimens.6,7,64 The deletion in this spliceoform is predicted to lead to a frame-shift, early truncation, and total absence of transmembrane segments in the receptor protein, whereas the leader sequence responsible for trafficking of the receptor to the cell membrane is preserved (Figure 3D). Correspondingly, this splice variant was found not to be functional, but to be secreted from cells.64 On this basis, an ELISA serum test specific for this spliceoform was developed in our laboratory and tested in a clinical pilot study.64 The results were encouraging, as the test was positive in 69% of pancreatic cancer patients, but not in healthy controls. Of note, splice variant serum levels were also elevated in chronic pancreatitis patients, although splice variant transcripts were not detected with PCR in chronic pancreatitis specimens.6 At present, it is unclear if this discrepancy between the ELISA and PCR results may be due to nonspecific reactivity of the ELISA test or if the spliceoform also arises focally in chronic pancreatitis, for instance in hyperplastic or preneoplastic ductal changes that were missing in the samples subjected to PCR analysis. These data emphasize the need to fully optimize a sensitive and specific clinical assay for a splice variant that is intended to serve as cancer biomarker and to validate this test in a spectrum of patients with relevant nonneoplastic and neoplastic diseases, as well as to thoroughly correlate test results with splicing patterns in tissues.
An entirely different secretin receptor splice variant expression pattern was present in bronchopulmonary carcinoid tumors. These tumors were found to express two spliceoforms not detected in other tumors before, one with an exon 9 deletion and another one with exon 2 and 3 deletions (Table 1). However, splice variants typical of pancreas and liver cancer were not identified.61 Both secretin receptor spliceoforms occurring in lung carcinoids are likely not to signal in response to secretin. Their significance remains to be defined.
Of note, a unique secretin receptor splicing pattern was also found in peritumoral lung exhibiting chronic inflammation. In these tissues, a splice variant missing part of exon 1 and all of exons 2 to 4 was identified, while the secretin receptor spliceoforms arising in cancer were not present.61 This example underlines that splice variants are not restricted to cancer, but can also occur selectively in nonneoplastic pathological conditions.
Growth Hormone-Releasing Hormone Receptor
The growth hormone(GH)-releasing hormone (GHRH) receptor is physiologically expressed on the somatotrophs of the anterior pituitary gland, where it mediates the effects of GHRH on GH secretion. Four different spliceoforms of this receptor have been recognized in neoplasia (Table 1). Of particular interest, these splice variants occur not only in pituitary adenomas, ie, at the site of physiological GHRH receptor expression, but also in many clinically important extra-pituitary tumors.65,66,67,68 Among these spliceoforms, splice variant 1 (SV1) has most extensively been explored, as it is the only one predicted to be functional.
SV1 and full-length, pituitary-type GHRH (pGHRH) receptor were found to be expressed, concomitantly as well as independently, in cell lines and tissues of a large variety of human epithelial, neuroendocrine, and lymphatic cancers at the mRNA and protein level (Table 1).65,69,70,71,72,73,74,75 SV1 also shows a wide-spread expression in nonneoplastic tissues, such as prostate, kidney, liver, pancreas, colon, lung, and pituitary gland.
SV1 has lost exons 1 to 3 but has intron 3 retained, which contains a new in-frame start codon. As a result, the first 89 amino acids at the N-terminus of the pGHRH receptor are replaced by a novel 25 amino acid sequence in SV1, while the rest of the spliceoform is identical to the full-length receptor (Figure 3E).65 SV1 still binds GHRH at the truncated N-terminus and signals in response to GHRH. In addition, it exhibits constitutive, ligand-independent signaling, in contrast to the pGHRH receptor.70 Both SV1 and pGHRH receptors stimulate tumor cell proliferation on GHRH activation, with SV1 being more potent than the pGHRH receptor in certain tumor cell types.70,76 Moreover, basal tumor cell proliferation in the absence of a ligand is increased in cells transfected with SV1, as compared with cells expressing the pGHRH receptor.70 GHRH receptors expressed on tumor cells stimulate tumor cell proliferation probably indirectly via multiple mechanisms, namely via secretion of the mitogens GHRH, GH, and/or insulin-like growth factors I and II.77,78,79
The proliferative effects of both SV1 and the pGHRH receptor on tumors represent the basis for potential therapeutic applications. GHRH receptor antagonists, which equally bind to SV1 and the pGHRH receptor, have been developed that potently inhibit growth and metastasis of a large number of experimental tumors expressing SV1 and pGHRH receptors in vitro and in vivo.77,79,80 Of note, it is unknown if these antiproliferative effects are mediated by inhibition mainly of SV1, pGHRH receptors, or both. As it has been shown that GHRH receptor antagonists are well tolerated by humans81 and that SV1 and pGHRH receptors are expressed in substantial amounts in original human tumors, the requirements necessary to start on clinical trials on the use of GHRH receptor antagonists in cancer patients appear largely fulfilled. However, such studies have not yet been performed.
Other G Protein-Coupled Receptors
A number of other G protein-coupled receptors have been found to be alternatively spliced in cancer (Table 1). However, these spliceoforms have been less extensively characterized with respect to expression and biological activity.
The vasoactive intestinal peptide/adenylate cyclase activating polypeptide 1 (pituitary) (VIP/PACAP) receptors are widely expressed in cancer.48 However, alternative splicing of these receptors has been assessed in only a limited number of tumors. The adenylate cyclase activity polypeptide 1 (pituitary) receptor type 1 (PAC1) receptor was investigated in selected embryonal tumors.82,83 As many as 14 different spliceoforms of this receptor were identified in neuroblastomas (summarized by Lutz et al82). These show variable combinations of alternative splicing at mainly two receptor regions, the N-terminal domain and the third intracellular loop. Accordingly, they differ in ligand binding affinity and potency to stimulate intracellular signal transduction pathways. Of particular interest, the PAC1 receptor splice variant expression pattern was significantly different in neuroblastomas as compared with the adult brain.
Splicing of the vasoactive intestinal peptide receptors 1 and 2 (VPAC 1 and 2) is different from that of the related PAC1 receptor.84 VPAC1 and VPAC2 express a similar spliceoform, lacking the third intracellular loop and the last two transmembrane segments. Despite the absence of the domain important for coupling to intracellular messengers, these spliceoforms still signal in response to ligand binding, which is, however, different from that of their wild-type counterparts. The VPAC1 and VPAC2 receptor splice variants were found to be differentially expressed in tumor cell lines derived from hematopoietic malignancies and colorectal carcinoma.84,85
VIP/PACAP receptor splice variants have presently been assessed only in selected tumor cell lines. It may be worth evaluating whether a tumor-specific splicing pattern can be observed in a larger number of normal tissues and naturally occurring tumors, such as PAC1 receptor-expressing neuroblastomas, gliomas, endometrial carcinomas, and the many VPAC1 receptor-expressing adenocarcinomas.48 In fact, the high VIP/PACAP receptor levels in nonneoplastic tissues have limited the development of in vivo tumor targeting of these receptors.48 If splice variants can be found that show a more selective expression in cancer than in wild-type cells, more specific targeting approaches may be developed for these receptors.
Furthermore, the luteinizing hormone/human chorionic gonadotropin receptor (LH/hCG) was found to exhibit alternative splicing in tumors. A splice variant with an exon 9 deletion was identified that is not expressed on the cell surface but is retained in the endoplasmic reticulum, where it complexes with wild-type LH/hCG and follicle-stimulating hormone receptors, resulting in lysosomal degradation.86,87 This splice variant appeared to be up-regulated in endometrial adenocarcinoma but lost in ovarian cancer as compared with the respective nonneoplastic tissues of tumor origin.88,89 Moreover, potential additional LH/hCG receptor spliceoforms were observed in endometrial cancer.88 The biological significance and clinical potential of LH/hCG splice variants in gynecological malignancies is undetermined.
Conclusion and Future Directions
G protein-coupled peptide hormone receptors provide a good illustration of the current knowledge of the presence, functions, and potential clinical relevance of splice variants in cancer in general. Although peptide receptors have been known to be expressed in cancer for decades, their splice variants have received attention only recently. A number of peptide receptor spliceoforms have already been detected in tumors, and, based on the large number of peptide receptors present in neoplasia, more can be expected to be discovered. Initial functional data indicate that the role of some peptide receptor splice variants in tumor cell biology may be just as relevant as that of their wild-type counterparts. Expression and functions of peptide receptor spliceoforms represent the molecular basis for clinical applications in cancer patients. Indeed, preliminary studies suggest a potential of some of these spliceoforms for tumor diagnosis and therapy.
The characterization of alternative spliceoform expression in cancer is quite incomplete. Splice variant expression should be investigated not only in cancer cell lines, but especially also in original tumor tissues. Indeed, splicing patterns arising in vitro often differ significantly from those occurring in vivo, and cancer cell lines can even totally lose the expression of a splice variant under culture conditions (personal observations).31,55,57,64 Moreover, the expression of splice variants should be quantified with respect to incidence and mRNA or protein levels in a representative number of original tumors. A splice variant is suitable for in vivo tumor targeting only if abundantly present in a large proportion of cases. In addition, if exhibiting dominant negative activity on its wild-type counterpart, it is likely to be of biological significance only when expressed in significant amounts relative to the wild-type form, as exemplified by the secretin receptor exon 3 deletion variant in gastrinomas. Furthermore, it can be relevant to ascertain which cell type expresses the splice variant in a tumor. Functional data on a splice variant obtained in transfected cell lines will be interpreted differently if the variant is present in the actual tumor cell or, as is possible with CCK2Ri4sv, in an intratumoral stromal cell.57 Finally, splice variants have to be assessed not only in tumors, but also in normal tissues of tumor origin as well as in nonneoplastic diseases. A splice variant represents a suitable clinical target only if it is substantially up-regulated in cancer and is expressed in low amounts in normal tissues and nonneoplastic pathologies, as illustrated with PCR detection of CD44v4-10 in cytological specimens,32,33 in vivo targeting of CD44v6 with toxic antibodies,39,40 and the ELISA serum test for the secretin receptor(Δexon3,4).64
A thorough characterization of the expression of splice variants in human normal and pathological tissues will importantly contribute to the interpretation of functional data obtained in cell lines, as well as allow estimations of the usefulness of spliceoforms for clinical applications and the choice of adequate tumor models for in vivo targeting studies. Thus, we may eventually be able to take clinical advantage of alternative splicing for early diagnostics, prognostics, and therapy of cancer.
Footnotes
Address reprint requests to Meike Körner, M.D., Institute of Pathology of the University of Berne, Murtenstrasse 31, CH-3010 Berne, Switzerland. E-mail: meike.koerner@pathology.unibe.ch or Laurence J. Miller, M.D., Mayo Clinic, 13400 East Shea Blvd, Scottsdale, AZ 85259. E-mail: miller@mayo.edu.
Supported by Fellowship #1267 from the Swiss Foundation for Medical-Biological Fellowships and Novartis (to M.K.), grants DK32878 and DK46577 from the National Institutes of Health (to L.J.M.).
References
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Kilpatrick GJ, Dautzenberg FM, Martin GR, Eglen RM. 7TM receptors: the splicing on the cake. Trends Pharmacol Sci. 1999;20:294–301. doi: 10.1016/s0165-6147(99)01355-3. [DOI] [PubMed] [Google Scholar]
- Minneman KP. Splice variants of G protein-coupled receptors. Mol Interv. 2001;1:108–116. [PubMed] [Google Scholar]
- Ding WQ, Kuntz S, Bohmig M, Wiedenmann B, Miller LJ. Dominant negative action of an abnormal secretin receptor arising from mRNA missplicing in a gastrinoma. Gastroenterology. 2002;122:500–511. doi: 10.1053/gast.2002.31039. [DOI] [PubMed] [Google Scholar]
- Hellmich MR, Rui XL, Hellmich HL, Fleming RY, Evers BM, Townsend CM., Jr Human colorectal cancers express a constitutively active cholecystokinin-B/gastrin receptor that stimulates cell growth. J Biol Chem. 2000;275:32122–32128. doi: 10.1074/jbc.M005754200. [DOI] [PubMed] [Google Scholar]
- Körner M, Hayes GM, Rehmann R, Zimmermann A, Friess H, Miller LJ, Reubi JC. Secretin receptors in normal and diseased human pancreas: marked reduction of receptor binding in ductal neoplasia. Am J Pathol. 2005;167:959–968. doi: 10.1016/S0002-9440(10)61186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner M, Hayes GM, Rehmann R, Zimmermann A, Scholz A, Wiedenmann B, Miller LJ, Reubi JC. Secretin receptors in the human liver: expression in biliary tract and cholangiocarcinoma, but not in hepatocytes or hepatocellular carcinoma. J Hepatol. 2006;45:825–835. doi: 10.1016/j.jhep.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Körner M, Waser B, Reubi JC, Miller LJ: CCK2 receptor splice variant with intron 4 retention in human gastrointestinal and lung tumors. J Cell Mol Med 2009, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Linē A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005:424342–57. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lo HS, Yang H, Gere S, Hu Y, Buetow KH, Lee MP. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- Li HR, Wang-Rodriguez J, Nair TM, Yeakley JM, Kwon YS, Bibikova M, Zheng C, Zhou L, Zhang K, Downs T, Fu XD, Fan JB. Two-dimensional transcriptome profiling: identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Pilarski R, Eng C. Different splicing defects lead to differential effects downstream of the lipid and protein phosphatase activities of PTEN. Hum Mol Genet. 2005;14:2459–2468. doi: 10.1093/hmg/ddi246. [DOI] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Sun Y, Pagenstecher C, Mangold E, Caspari R, Möslein G, Schulmann K, Propping P, Friedl W. Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Hum Mutat. 2004;24:370–380. doi: 10.1002/humu.20087. [DOI] [PubMed] [Google Scholar]
- Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A, McDonnell SK, Wiley KE, Jacobsen SJ, Isaacs SD, Walsh PC, Zheng SL, Chang BL, Friedrichsen DM, Stanford JL, Ostrander EA, Chinnaiyan AM, Rubin MA, Xu J, Thibodeau SN, Friedman SL, Martignetti JA. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- Chen LL, Sabripour M, Wu EF, Prieto VG, Fuller GN, Frazier ML. A mutation-created novel intra-exonic pre-mRNA splice site causes constitutive activation of KIT in human gastrointestinal stromal tumors. Oncogene. 2005;24:4271–4280. doi: 10.1038/sj.onc.1208587. [DOI] [PubMed] [Google Scholar]
- Fischer DC, Noack K, Runnebaum IB, Watermann DO, Kieback DG, Stamm S, Stickeler E. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–1090. [PubMed] [Google Scholar]
- Sampath J, Long PR, Shepard RL, Xia X, Devanarayan V, Sandusky GE, Perry WL, 3rd, Dantzig AH, Williamson M, Rolfe M, Moore RE. Human SPF45, a splicing factor, has limited expression in normal tissues, is overexpressed in many tumors, and can confer a multidrug-resistant phenotype to cells. Am J Pathol. 2003;163:1781–1790. doi: 10.1016/S0002-9440(10)63538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Watermann DO, Tang Y, Zur Hausen A, Jäger M, Stamm S, Stickeler E. Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 2006;66:4744–4780. doi: 10.1158/0008-5472.CAN-04-3294. [DOI] [PubMed] [Google Scholar]
- Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, Hod E, Leake D, Friedman SL, Hall SJ, Chinnaiyan AM, Gerald WL, Rubin MA, Martignetti JA. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, Jones SN. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem. 2004;279:4877–4886. doi: 10.1074/jbc.M305966200. [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach-Dayan SB, Rubinstein AM, Hand C, Breuer R, Naor D. DNA vaccination with CD44 variant isoform reduces mammary tumor local growth and lung metastasis. Mol Cancer Ther. 2008;7:1615–1623. doi: 10.1158/1535-7163.MCT-07-2383. [DOI] [PubMed] [Google Scholar]
- Turpin E, Dalle B, de Roquancourt A, Plassa LF, Marty M, Janin A, Beuzard Y, de Thé H. Stress-induced aberrant splicing of TSG101: association to high tumor grade and p53 status in breast cancers. Oncogene. 1999;18:7834–7837. doi: 10.1038/sj.onc.1203196. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Morisaki T, Sasaki J, Miyake H, Matsumoto MSM, Ando M, Saya H. Molecular detection of cancer cells by competitive reverse transcription-polymerase chain reaction analysis of specific CD44 variant RNAs. J Natl Cancer Inst. 1998;90:307–315. doi: 10.1093/jnci/90.4.307. [DOI] [PubMed] [Google Scholar]
- Miyake H, Eto H, Arakawa S, Kamidono S, Hara I. Over expression of CD44v8-10 in urinary exfoliated cells as an independent prognostic predictor in patients with urothelial cancer. J Urol. 2002;167:1282–1287. [PubMed] [Google Scholar]
- Yamaguchi A, Goi T, Taguchi S, Ohtaki N, Seki K, Hirose K, Nakagawara G, Urano T, Furukawa K. Clinical significance of serum levels of CD44 variant exons 8–10 protein in colorectal cancer. J Gastroenterol. 1998;33:349–353. doi: 10.1007/s005350050095. [DOI] [PubMed] [Google Scholar]
- Ebbinghaus C, Scheuermann J, Neri D, Elia G. Diagnostic and therapeutic applications of recombinant antibodies: targeting the extra-domain B of fibronectin, a marker of tumor angiogenesis. Curr Pharm Des. 2004;10:1537–1549. doi: 10.2174/1381612043384808. [DOI] [PubMed] [Google Scholar]
- Santimaria M, Moscatelli G, Viale GL, Giovannoni L, Neri G, Viti F, Leprini A, Borsi L, Castellani P, Zardi L, Neri D, Riva P. Immunoscintigraphic detection of the ED-B domain of fibronectin, a marker of angiogenesis, in patients with cancer. Clin Cancer Res. 2003;9:571–579. [PubMed] [Google Scholar]
- Colnot DR, Roos JC, de Bree R, Wilhelm AJ, Kummer JA, Hanft G, Heider KH, Stehle G, Snow GB, van Dongen GA. Safety, biodistribution, pharmacokinetics, and immunogenicity of 99mTc-labeled humanized monoclonal antibody BIWA 4 (bivatuzumab) in patients with squamous cell carcinoma of the head and neck. Cancer Immunol Immunother. 2003;52:576–592. doi: 10.1007/s00262-003-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppe M, Schaijk F, Roos J, Leeuwen P, Heider KH, Kuthan H, Bleichrodt R. Safety, pharmacokinetics, immunogenicity, and biodistribution of (186)Re-labeled humanized monoclonal antibody BIWA 4 (Bivatuzumab) in patients with early-stage breast cancer. Cancer Biother Radiopharm. 2004;19:720–729. doi: 10.1089/cbr.2004.19.720. [DOI] [PubMed] [Google Scholar]
- Börjesson PK, Postema EJ, Roos JC, Colnot DR, Marres HA, van Schie MH, Stehle G, de Bree R, Snow GB, Oyen WJ, van Dongen GA. Phase I therapy study with (186)Re-labeled humanized monoclonal antibody BIWA 4 (bivatuzumab) in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2003;9:3961S–3972S. [PubMed] [Google Scholar]
- Rupp U, Schoendorf-Holland E, Eichbaum M, Schuetz F, Lauschner I, Schmidt P, Staab A, Hanft G, Huober J, Sinn HP, Sohn C, Schneeweiss A. Safety and pharmacokinetics of bivatuzumab mertansine in patients with CD44v6-positive metastatic breast cancer: final results of a phase I study. Anticancer Drugs. 2007;18:477–485. doi: 10.1097/CAD.0b013e32801403f4. [DOI] [PubMed] [Google Scholar]
- Heider KH, Kuthan H, Stehle G, Munzert G. CD44v6: a target for antibody-based cancer therapy. Cancer Immunol Immunother. 2004;53:567–579. doi: 10.1007/s00262-003-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nature Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nature Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- Zangemeister-Wittke U, Leech SH, Olie RA, Simões-Wüst AP, Gautschi O, Luedke GH, Natt F, Häner R, Martin P, Hall J, Nalin CM, Stahel RA. A novel bispecific antisense oligonucleotide inhibiting both bcl-2 and bcl-xL expression efficiently induces apoptosis in tumor cells. Clin Cancer Res. 2000;6:2547–2555. [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Pilch B, Allemand E, Facompré M, Bailly C, Riou JF, Soret J, Tazi J. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61:6876–6884. [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Beck AM, Miller LJ. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- Reubi JC. Peptide receptors as molecular targets for cancer diagnosis and therapy. Endocr Rev. 2003;24:389–427. doi: 10.1210/er.2002-0007. [DOI] [PubMed] [Google Scholar]
- Rozengurt E, Walsh JH. Gastrin. CCK, signaling, and cancer. Annu Rev Physiol. 2001;63:49–76. doi: 10.1146/annurev.physiol.63.1.49. [DOI] [PubMed] [Google Scholar]
- Reubi JC. Targeting CCK receptors in human cancers. Curr Top Med Chem. 2007;7:1239–1242. doi: 10.2174/156802607780960546. [DOI] [PubMed] [Google Scholar]
- Baldwin GS, Shulkes A. CCK receptors and cancer. Curr Top Med Chem. 2007;7:1232–1238. doi: 10.2174/156802607780960492. [DOI] [PubMed] [Google Scholar]
- Ding WQ, Kuntz SM, Miller LJ. A misspliced form of the cholecystokinin-B/gastrin receptor in pancreatic carcinoma: role of reduced sellular U2AF35 and a suboptimal 3′-splicing site leading to retention of the fourth intron. Cancer Res. 2002;62:947–952. [PubMed] [Google Scholar]
- Smith JP, Verderame MF, McLaughlin P, Martenis M, Ballard E, Zagon IS. Characterization of the CCK-C (cancer) receptor in human pancreatic cancer. Int J Mol Med. 2002;10:689–694. [PubMed] [Google Scholar]
- Zhou J, Chen M, Zhang Q, Hu J, Wang W. Human gastric tissues simultaneously express the classical and alternative splicing cholecystokinin-B/gastrin receptors. Receptors Channels. 2004;10:185–188. doi: 10.3109/10606820490926179. [DOI] [PubMed] [Google Scholar]
- Harris JC, Clarke PA, Awan A, Jankowski J, Watson SA. An antiapoptotic role for gastrin and the gastrin/CCK-2 receptor in Barrett’s esophagus. Cancer Res. 2004;64:1915–1919. doi: 10.1158/0008-5472.can-03-2713. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Otte JM, Stechele HU, Reimann B, Banasiewicz T, Fölsch UR, Schmidt WE, Herzig KH. CCK-B/gastrin receptors in human colorectal cancer. Eur J Clin Invest. 2001;31:812–820. doi: 10.1046/j.1365-2362.2001.00870.x. [DOI] [PubMed] [Google Scholar]
- Chao C, Tallman ML, Ives KL, Townsend CM, Jr, Hellmich MR. Gastrointestinal hormone receptors in primary human colorectal carcinomas. J Surg Res. 2005;129:313–321. doi: 10.1016/j.jss.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Harikumar KG, Ding WQ, Holicky EL, Miller LJ. Analysis of the cellular and molecular mechanisms of trophic action of a misspliced form of the type B cholecystokinin receptor present in colon and pancreatic cancer. Cancer Lett. 2005;222:95–105. doi: 10.1016/j.canlet.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Chao C, Goluszko E, Lee YT, Kolokoltsov AA, Davey RA, Uchida T, Townsend CM, Jr, Hellmich MR. Constitutively active CCK2 receptor splice variant increases Src-dependent HIF-1 alpha expression and tumor growth. Oncogene. 2007;26:1013–1019. doi: 10.1038/sj.onc.1209862. [DOI] [PubMed] [Google Scholar]
- Laverman P, Roosenburg S, Gotthardt M, Park J, Oyen WJ, de Jong M, Hellmich MR, Rutjes FP, van Delft FL, Boerman OC. Targeting of a CCK(2) receptor splice variant with (111)In-labelled cholecystokinin-8 (CCK8) and (111)In-labelled minigastrin. Eur J Nucl Med Mol Imaging. 2008;35:386–392. doi: 10.1007/s00259-007-0604-1. [DOI] [PubMed] [Google Scholar]
- Körner MU, Hayes GM, Carrigan PE, Rehmann R, Miller LJ, Reubi JC. Wild-type and splice-variant secretin receptors in lung cancer: overexpression in carcinoid tumors and peritumoral lung tissue. Mod Pathol. 2008;21:387–395. doi: 10.1038/modpathol.3801005. [DOI] [PubMed] [Google Scholar]
- Long SH, Berna MJ, Thill M, Pace A, Pradhan TK, Hoffmann KM, Serrano J, Jensen RT. Secretin-receptor and secretin-receptor-variant expression in gastrinomas: correlation with clinical and tumoral features and secretin and calcium provocative test results. J Clin Endocrinol Metab. 2007;92:4394–4402. doi: 10.1210/jc.2007-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WQ, Cheng ZJ, McElhiney J, Kuntz SM, Miller LJ. Silencing of secretin receptor function by dimerization with a misspliced variant secretin receptor in ductal pancreatic adenocarcinoma. Cancer Res. 2002;62:5223–5229. [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Dong M, Reubi JC, Miller LJ. A novel secretin receptor splice variant potentially useful for early diagnosis of pancreatic carcinoma. Gastroenterology. 2007;133:853–861. doi: 10.1053/j.gastro.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Rekasi Z, Czompoly T, Schally AV, Halmos G. Isolation and sequencing of cDNAs for splice variants of growth hormone-releasing hormone receptors from human cancers. Proc Natl Acad Sci USA. 2000;97:10561–10566. doi: 10.1073/pnas.180313297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Lagacé G, Castagné J, Collu R. Identification of human growth hormone-releasing hormone receptor splicing variants. J Clin Endocrinol Metab. 1995;80:2381–2387. doi: 10.1210/jcem.80.8.7629234. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Koga M, Motomura T, Kasayama S, Kouhara H, Ohnishi T, Arita N, Hayakawa T, Sato B, Kishimoto T. Identification of alternatively spliced messenger ribonucleic acid encoding truncated growth hormone-releasing hormone receptor in human pituitary adenomas. J Clin Endocrinol Metab. 1995;80:2933–2939. doi: 10.1210/jcem.80.10.7559877. [DOI] [PubMed] [Google Scholar]
- Motomura T, Hashimoto K, Koga M, Arita N, Hayakawa T, Kishimoto T, Kasayama S. Inhibition of signal transduction by a splice variant of the growth hormone-releasing hormone receptor expressed in human pituitary adenomas. Metabolism. 1998;47:804–808. doi: 10.1016/s0026-0495(98)90116-0. [DOI] [PubMed] [Google Scholar]
- Halmos G, Schally AV, Czompoly T, Krupa M, Varga JL, Rekasi Z. Expression of growth hormone-releasing hormone and its receptor splice variants in human prostate cancer. J Clin Endocrinol Metab. 2002;87:4707–4714. doi: 10.1210/jc.2002-020347. [DOI] [PubMed] [Google Scholar]
- Kiaris H, Chatzistamou I, Schally AV, Halmos G, Varga JL, Koutselini H, Kalofoutis A. Ligand-dependent and -independent effects of splice variant 1 of growth hormone-releasing hormone receptor. Proc Natl Acad Sci USA. 2003;100:9512–9517. doi: 10.1073/pnas.1533185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havt A, Schally AV, Halmos G, Varga JL, Toller GL, Horvath JE, Szepeshazi K, Köster F, Kovitz K, Groot K, Zarandi M, Kanashiro CA. The expression of the pituitary growth hormone-releasing hormone receptor and its splice variants in normal and neoplastic human tissues. Proc Natl Acad Sci USA. 2005;102:17424–17429,. doi: 10.1073/pnas.0506844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freddi S, Arnaldi G, Fazioli F, Scarpelli M, Appolloni G, Mancini T, Kola B, Bertagna X, Mantero F, Collu R, Boscaro M. Expression of growth hormone-releasing hormone receptor splicing variants in human primary adrenocortical tumours. Clin Endocrinol (Oxf) 2005;62:533–538. doi: 10.1111/j.1365-2265.2005.02253.x. [DOI] [PubMed] [Google Scholar]
- Hohla F, Moder A, Mayrhauser U, Hauser-Kronberger C, Schally AV, Varga JL, Zarandi M, Buchholz S, Huber R, Aigner E, Ritter M, Datz C. Differential expression of GHRH receptor and its splice variant 1 in human normal and malignant mucosa of the oesophagus and colon. Int J Oncol. 2008;33:137–143. [PubMed] [Google Scholar]
- Chatzistamou I, Volakaki AA, Schally AV, Kiaris H, Kittas C. Expression of growth hormone-releasing hormone receptor splice variant 1 in primary human melanomas. Regul Pept. 2008;147:33–36. doi: 10.1016/j.regpep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Volakaki AA, Lafkas D, Kassi E, Schally AV, Papvassiliou AG, Kiaris H. Essential role of p21/waf1 in the mediation of the anti-proliferative effects of GHRH antagonis JMR-132. J Mol Endocrinol. 2008;41:389–392. doi: 10.1677/JME-08-0106. [DOI] [PubMed] [Google Scholar]
- Barabutis N, Tsellou E, Schally AV, Kouloheri S, Kalofoutis A, Kiaris H. Stimulation of proliferation of MCF-7 breast cancer cells by a transfected splice variant of growth hormone-releasing hormone receptor. Proc Natl Acad Sci USA. 2007;104:5575–5579. doi: 10.1073/pnas.0700407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiaris H, Schally AV, Varga JL, Groot K, Armatis P. Growth hormone-releasing hormone: an autocrine growth factor for small cell lung carcinoma. Proc Natl Acad Sci USA. 1999;96:14894–14898. doi: 10.1073/pnas.96.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabutis N, Schally AV. Knocking down gene expression for growth hormone-releasing hormone inhibits proliferation of human cancer cell lines. Br J Cancer. 2003;98:1790–1796. doi: 10.1038/sj.bjc.6604386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schally AV, Varga JL, Engel JB. Antagonists of growth-hormone-releasing hormone: an emerging new therapy for cancer. Nat Clin Pract Endocrinol Metab. 2008;4:33–43. doi: 10.1038/ncpendmet0677. [DOI] [PubMed] [Google Scholar]
- Hohla F, Schally AV, Szepeshazi K, Varga JL, Buchholz S, Köster F, Heinrich E, Halmos G, Rick FG, Kannadka C, Datz C, Kanashiro CA. Synergistic inhibition of growth of lung carcinomas by antagonists of growth hormone-releasing hormone in combination with docetaxel. Proc Natl Acad Sci USA. 2006;103:14513–14518. doi: 10.1073/pnas.0605309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe CA, Friberg RD, Barkan AL. Suppression of growth hormone (GH) secretion by a selective GH-releasing hormone (GHRH) antagonist. Direct evidence for involvement of endogenous GHRH in the generation of GH pulses J Clin Invest. 1993;92:695–701. doi: 10.1172/JCI116639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz EM, Ronaldson E, Shaw P, Johnson MS, Holland PJ, Mitchell R. Characterization of novel splice variants of the PAC1 receptor in human neuroblastoma cells: consequences for signaling by VIP and PACAP. Mol Cell Neurosci. 2006;31:193–209. doi: 10.1016/j.mcn.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Dautzenberg FM, Mevenkamp G, Wille S, Hauger RL. N-terminal splice variants of the type I PACAP receptor: isolation, characterization and ligand binding/selectivity determinants. J Neuroendocrinol. 1999;11:941–949. doi: 10.1046/j.1365-2826.1999.00411.x. [DOI] [PubMed] [Google Scholar]
- Bokaei PB, Ma XZ, Byczynski B, Keller J, Sakac D, Fahim S, Branch DR. Identification and characterization of five-transmembrane isoforms of human vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide receptors. Genomics. 2006;88:791–800. doi: 10.1016/j.ygeno.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Germano PM, Le SV, Oh DS, Fan R, Lieu S, Siu A, Pisegna JR. Differential coupling of the PAC1 SV1 splice variant on human colonic tumors to the activation of intracellular cAMP but not intracellular Ca2+ does not activate tumor proliferation. J Mol Neurosci. 2004;22:83–92. doi: 10.1385/JMN:22:1-2:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Yamashita S, Omori Y, Minegishi T. A splice variant of the human luteinizing hormone (LH) receptor modulates the expression of wild-type human LH receptor. Mol Endocrinol. 2004;18:1461–1470. doi: 10.1210/me.2003-0489. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Nakamura K, Omori Y, Tsunekawa K, Murakami M, Minegishi T. Association of human follitropin (FSH) receptor with splicing variant of human lutropin/choriogonadotropin receptor negatively controls the expression of human FSH receptor. Mol Endocrinol. 2005;19:2099–2111. doi: 10.1210/me.2005-0049. [DOI] [PubMed] [Google Scholar]
- Lin J, Lei ZM, Lojun S, Rao CV, Satyaswaroop PG, Day TG. Increased expression of luteinizing hormone/human chorionic gonadotropin receptor gene in human endometrial carcinomas. J Clin Endocrinol Metab. 1994;79:1483–1491. doi: 10.1210/jcem.79.5.7962347. [DOI] [PubMed] [Google Scholar]
- Mandai M, Konishi I, Kuroda H, Fukumoto M, Komatsu T, Yamamoto S, Nanbu K, Rao CV, Mori T. Messenger ribonucleic acid expression of LH/hCG receptor gene in human ovarian carcinomas. Eur J Cancer. 1997;33:1501–1507. doi: 10.1016/s0959-8049(97)00166-4. [DOI] [PubMed] [Google Scholar]
- Smith JP, Stanley WB, Verderame MF, Zagon IS. The functional significance of the cholecystokinin-C (CCK-C) receptor in human pancreatic cancer. Pancreas. 2004;29:271–277. doi: 10.1097/00006676-200411000-00005. [DOI] [PubMed] [Google Scholar]
- Chen R, Lewis KA, Perrin MH, Vale WW. Expression cloning of a human corticotropin-releasing-factor receptor. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]