Abstract
Macroautophagy is an essential degradative pathway that can be induced to clear aggregated proteins, such as those found in Parkinson’s disease and dementia with Lewy bodies, a form of Parkinsonism. This study found that both LC3-II and beclin were significantly increased in brains from humans with Dementia with Lewy bodies and transgenic mice overexpressing mutant α-synuclein, as compared with respective controls, suggesting that macroautophagy is induced to remove α-syn, particularly oligomeric or mutant forms. Aged mutant animals had higher autophagy biomarker levels relative to younger animals, suggesting that with aging, autophagy is less efficient and requires more stimulation to achieve the same outcome. Disruption of autophagy by RNA interference significantly increased α-syn oligomer accumulation in vitro, confirming the significance of autophagy in α-syn clearance. Finally, rotenone-induced α-syn aggregates were cleared following rapamycin stimulation of autophagy. Chronic rotenone exposure and commensurate reduction of metabolic activity limited the efficacy of rapamycin to promote autophagy, suggesting that cellular metabolism is critical for determining autophagic activity. Cumulatively, these findings support the concept that neuronal autophagy is essential for protein homeostasis and, in our system, reduction of autophagy increased the accumulation of potentially pathogenic α-synuclein oligomers. Aging and metabolic state were identified as important determinants of autophagic activity. This study provides therapeutic and pathological implications for both synucleinopathy and Parkinson’s disease, identifying conditions in which autophagy may be insufficient to degrade α-syn aggregates.
Parkinson’s disease (PD) is a major neurodegenerative disease that affects approximately 1 million people in the United States. The disease affects the motor system due to loss of dopaminergic neurons. The common pathological hallmark of the disease includes the formation of intracellular inclusions called Lewy bodies,1,2,3,4 principally composed of the protein α-synuclein (α-syn). The protein misfolds and accumulates, initially as oligomers and ultimately as aggregates.5,6,7,8 The first mutation linked to PD was found in the α-synuclein gene at position A53T,9 and mutant variants of the protein are more prone to aggregate compared with wild-type protein.10,11,12,13
Other genes linked to PD include Parkin,14 UCHL1,15 DJ-1,16 PINK1,17,18 LrrK2,19,20 and ATP13a2.21 Mutations in these genes are not all associated with synucleinopathy however. Some involve mitochondrial dysfunction (PINK1, parkin, DJ-1), aberrant proteasomal activity (UCHL1), or altered autophagic or lysosomal function (LrrK2 and ATP13a2).21,22 Recently, mutations of the glucocerebrosidase gene, encoding a lysosomal enzyme, have been shown to be an important risk factor for PD23,24,25,26 and dementia with Lewy bodies (DLB), another form of Parkinsonism.24,27 The mechanism by which glucocerebrosidase mutations cause disease is currently unknown but alterations in glucocerebrosidase may affect lysosomal degradation and increase the likelihood of aberrant α-syn accumulation into Lewy bodies, and neurodegeneration.
Previously, wild-type α-syn was shown to undergo chaperone-mediated autophagy,28 a lysosomal process that degrades 40% of all proteins, while the mutant variant blocks this lysosomal action.29,30 As a result, mutant forms of α-synuclein (especially the A53T variant) are more prone to oligomerization and aggregation and are less readily degraded than the wild-type. Thus, they are more likely to accumulate as pathological intracellular inclusions due to decreased lysosomal degradation.
Macroautophagy (autophagy) is critical to neuronal health as impairment leads to aberrant protein accumulation, neuronal dysfunction, and cell death.31,32 In addition to its constitutive activity, macroautophagy is inducible and dually regulated by the mTOR33 and PI3kinase/beclin/vsp3434,35 signaling pathways. Induction of autophagy results in the recruitment of multiple Atg proteins36,37 that initiate formation of double-membrane, autophagic vacuoles (AVs) that envelop proximal cytoplasmic material including aggregated proteins and organelles.38 The AVs subsequently acquire lysosomal hydrolases or fuse with lysosomes to complete the degradative process.39,40 Normally, this constitutive process is highly efficient in neurons.
Up-regulation of autophagy has been previously reported to reverse the pathogenic accumulation of intracellular inclusions. In Huntington’s disease, induction of autophagy promotes the clearance of polyglutamine huntingtin aggregates that are a hallmark of the disease.41,42 However, induction is not always a beneficial process. In Alzheimer’s disease, it has been suggested that the accumulation of AVs results in increased production of the toxic peptide β-amyloid and may be attributed to reduced fusion or lysosomal degradation.43,44 Thus, although it appears that cells may up-regulate autophagy to attenuate dysfunction, it cannot be assumed that this is the case and the opposite may, in fact, be true.
These studies provide comprehensive pathological and biochemical evidence that macroautophagy is essential for maintaining the homeostasis of α-synuclein levels. Our studies show that autophagy was stimulated in human DLB, and in mutant α-syn-expressing mice to clear aberrant α-syn. In addition, this work provides evidence that blockage of autophagy results in the promotion of the pathogenic α-syn oligomers,45,46 particularly the mutant variant of α-syn. Finally, clearance of α-synuclein via small molecule induction of macroautophagy is dependent on the metabolic state of the cells, as cells were able to clear oligomeric α-syn when not metabolically compromised following acute exposure to rotenone, but with chronic exposure to rotenone the autophagic capacity to degrade α-synuclein was significantly diminished.
Materials and Methods
Tissue Homogenization
Human autopsy samples from Brodmann area 9 of the cerebral cortex were obtained from the Columbia University Brain Tissue Bank. A summary of tissue used can be found in Table 1. Brain tissue was weighed and homogenized in a ×10 v/w of tissue homogenizing buffer (20 mmol/L Tris, pH 7.4; 5% sucrose; 1 mmol/L phenylmethylsulfonyl fluoride; 1 mmol/L Na3VO4; 1 mmol/L NaF; and 10 μl/ml of protease inhibitor cocktail [Sigma]; EDTA; EGTA). Twenty-microgram aliquots were loaded per lane for quantitative immunoblot analysis. Brains from mutant α-syn mice (12c/SKO; murine α-synuclein knockout [SKO] expressing human mutant α-syn (A53T; 12c) under the control of the tyrosine hydoxylase promoter)47 at 6 and 18 to 22 months of age, and from mice overexpressing equivalent levels of wild-type human, or mutant (A53T; 12c) α-synuclein were used to compare the effect of normal and mutant protein overexpression on autophagy. Following cervical dislocation, mouse brains were quickly dissected and the cortex and substantia nigra were harvested for analysis using the same method as used for human tissue.
Table 1.
Information for Human Brains Used in This Study
| Sample ID | Diagnosis | Age | Sex | PMI |
|---|---|---|---|---|
| T-174 | Ctrl | 72 | M | 4:30 |
| T-305 | Ctrl | 74 | F | 18:45 |
| T-202 | Ctrl | 74 | M | 4:15 |
| T-55 | Ctrl | 81 | M | 12:00 |
| T-198 | Ctrl | 82 | F | 3:50 |
| T-171 | Ctrl | 89 | M | 4:55 |
| T-296 | DLB | 74 | M | 9:55 |
| T-227 | DLB | 76 | F | 6:44 |
| T-156 | DLB | 76 | M | 18:55 |
| T-301 | DLB | 80 | M | 7:15 |
| T-189 | DLB | 82 | F | 3:30 |
| T-251 | DLB | 83 | M | 6:28 |
PMI = Post-mortem interval, hrs:min.
Tissue Culture
M17 cells transfected with either wild-type or mutant (A53T) α-synuclein were cultured in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% bovine growth serum (Hyclone). For biochemical analysis of autophagic markers by immunoblotting, cells were grown to 90% to 95% and harvested. Rapamycin (100 nmol/L) was added to cells 4 hours before harvesting. Cells were washed 3× in PBS (pH = 7.4) and radioimmunoprecipitation assay buffer (50 mmol/L Tris-HCl, pH 7.4; 1% NP-40; 0.25% Na-deoxycholate; 150 mmol/L NaCl; 1 mmol/L EDTA; and protease and phosphatase inhibitors) was added to lyse the cells. Cell extracts were collected from the plates and sonicated using a Sonic Dismembranator (Fisher) set to a 5-second pulse. The lysate was spun at 20,000 × g for 15 minutes and the protein concentration was determined from the supernatant. For analysis of oligomer and α-syn aggregates, the Lee et al (2004)48 protocol was used.
For RNA interference (RNAi) experiments, a cell suspension containing 50,000 cells was added to a pre-made mixture of 5 μl of RNAiMax (Invitrogen) plus 20 nmol/L of RNAi targeting Atg5 or beclin (sequences from Qiagen), plated onto a 6 well plate, and incubated for 48 hours before harvesting. Rotenone (Sigma) treatments (50 nmol/L) were performed on cells at 80% confluency (24 hours rotenone treatment) or 10% confluency (7 days treatment). The human-specific Atg5 RNAi target sequence 5′-AACCTTTGGCCTAAGAAGAAA-3′ was used, and 5′-ACCGACTTGTTCCTTACGGAA-3′ was used for beclin RNAi. 3-methyladenine (3MA, 10 mmol/L) was also used over a 24 hour period as an additional method of inhibiting autophagy.
Rotenone (5 to 100 nmol/L) was used to inhibit mitochondrial activity. Briefly, cells were initially treated for 24 hours using a range of rotenone from 5 to 100 nmol/L, with a concentration of 50 nmol/L rotenone being the standard dose. Cells were plated at 10% confluency and rotenone was added to the plates designated for 7 day treatment on day 0. Media was replaced every 2 days (day 0, 2, 4, 6) with rotenone being added to the cells treated for 24 hour (80% confluency) on day 6. Four hours before harvesting, rapamycin (100 nmol/L) was added to designated 7 day and 24 hour plates.
Immunoblotting
For LC3-II (1:1000; Novus), proteins were separated through a 16% Tris-glycine gel then transferred onto 0.2 μmol/L nitrocellulose (Whatman) at 100mA for 8 hours. For beclin (1:2000), tubulin (1:4000), phospho-P70S6 kinase, and total P70S6 kinase (both 1:1000), 4% to 12% bis-Tris gels with 1× 3-(N-morpholino)propanesulfonic acid buffer were used. For α-synuclein (1:1000), 4% to 20% Tris-glycine and 4% to 12% bis-Tris gels with 1× 2-(N-morpholino)ethanesulfonic acid buffer were used. For the α-syn oligomer studies, a positive control (gift of Dr. Paul Fraser, University of Toronto) containing 100 ng of recombinant α-syn oligomer ladder was loaded alongside tissue samples. To create the ladder, 1 mg of recombinant α-syn purified from bacterial stock was dissolved in 10 μl dimethyl sulfoxide to monomerize the α-syn, the protein was diluted in PBS (100 μg/ml) and incubated at room temperature for 7 days to generate the α-synuclein oligomer ladder. The source of antibodies was as follows: beclin, BD/Pharmingen; phospho-P70S6 kinase and total P70S6 kinase, Cell Signaling Technology; tubulin, Sigma; and α-synuclein (LB509), Covance.
Immunohistochemistry
Cells were plated on 12-mm coverslips and treated as detailed above. Following treatment, cells were fixed in 2% paraformaldehyde, PBS, pH 7.4, 5% sucrose for 15 minutes, before washing twice in PBS. Cells were blocked in 5% horse/10% bovine serum containing 0.01% Triton X-100 for 15 minutes before the addition of primary antibodies (LC3 – 1:200 and α-synuclein 1:250 or A11 1:100 and α-synuclein), which was incubated overnight, washed twice in PBS and incubated in secondary antibodies conjugated to fluorescein isothiocyanate (rabbit polyclonal, 1:500; Invitrogen) or tetramethylrhodamine B isothiocyanate (mouse monoclonal, 1:500; Invitrogen). Following a final wash, sections were mounted and the coverslip was affixed to a glass slide using gelvatol. Sections were imaged using a Perkin-Elmer Spinning Disk Confocal microscope with a ×63 oil immersion lens. Images displayed were dual-channel merged and stacked acquisitions (5 μmol/L). α-Syn was shown in red, A11 or LC3 was in green, and colocalized signal was indicated by yellow. From the RGB image, colocalization analysis was performed using Photoshop CS3.
Biochemical Assays
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay was used to identify NADH reductase activity. Ten microliters of 10 mmol/L MTT dye (Sigma) per ml media was added to the treated cells and incubated under tissue culture conditions for 2 hours. Cells were then washed and the sample was solubilized overnight using 10% SDS with 0.1 N HCl. The following day, the sample signal was assessed using a BioTek ELx800 microplate reader at 562 nm (and 630 nm for reference). ATP levels were assessed using a Bioluminescence kit (Promega) and analyzed on a Tecan Infinite M200 fluorescent plate reader. Cytochrome c oxidase activity was determined both immunocytochemically, as well as using a microplate assay (490 nm, 630 nm reference). In summary, cells grown on plastic dishes were washed and air-dried before pre-incubation in 50 mmol/L Tris, pH = 7.6, 12 mmol/L cobalt chloride, 10% sucrose. The cells were then washed in 0.1 M sodium phosphate buffer (pH = 7.6) and incubated for 4 hours in the filtered reaction buffer containing 0.1 M sodium phosphate buffer, 10% sucrose, 1% cytochrome C, 1% diaminobenzidine, 2% catalase (all from Sigma). For immunochemistry, cells were imaged using an Olympus Bx600 microscope (×40 lens) equipped with a Insight Spot 4 digital camera and then solubilized in 10% SDS, 0.1 N HCl for spectrophotometric analysis. Lactate dehydrogenase activity was monitored in serum-free media to identify cell death. Briefly, using the commercial kit provided (Takara), cells were incubated in serum-free media for 4 hours and the media was collected and reacted according to the protocol. Signal was monitored at 490 nm, (reference 630 nm) using the microplate reader, with cells treated in 1% Triton X-100 as a positive control.
Electron Microscopy
Cells were prepared for electron microscopy as previously described.49 For mouse brain preparation, the animals were perfused with 4% paraformaldehyde, 2% glutaraldehyde, 0.1 M sodium cacodylate before removal of the brain. Subsequently, the brains were postfixed in 2% glutaraldehyde and 0.1 M sodium cacodylate. After washing with cacodylate buffer, the samples were postfixed in 1% osmium tetroxide, progressively dehydrated in ethanol (50% to 100%), and embedded in Epon. Thin sections (1 μm) of the substantia nigra were cut from the polymer, followed by ultrathin sections (70 to 80 nm) using a Recheirt Ultracut S microtome and placed on copper grids for structural analysis. Grids were briefly stained with uranyl acetate and lead citrate before being examined with a Philips electron microscope (model CM 10). Images were captured on a digital camera (Hamamatsu; model C4742-95) using Advantage CCD Camera System software (Advanced Microscopy Techniques Corporation). Antibody LB509 (1:5) was used to label tyroisine hydroxlase-positive neurons that express human α-syn. From these cells, AV quantification was performed using electron microscopy images at ×10, ×500 magnification (64 μm2 area) and AVs were distinguished based on morphological features, including size (>0.5μ), presence of cytoplasmic material (autophagolysosome, AL, also referred to as AVd) and identification of double membranes (AP, also referred to as AVi).50 For AV counts in murine brains, n = 3 per group, and 12 images were acquired from 3 to 4 grids. For cells, an area of 64 μm2 was assessed and all of the AVs (AL and AP) were counted within this field. A total of 50 images were surveyed for this analysis. Surface area was also calculated by measuring the area of AVs using NIH Image (v.1.63) and dividing by the total cellular surface area in the image.
Data Analysis
Western blot images using exposures that were within the linear range of pixel intensity were analyzed using ImageGauge 4.2 software. Any statistical comparison between samples was performed using a Student’s t-test with two-tailed, equal variance. For multiple comparisons, Bonferroni multiple comparison test was performed to normalize P values within the groups.
Results
Autophagy Is Induced in Dlb Brains
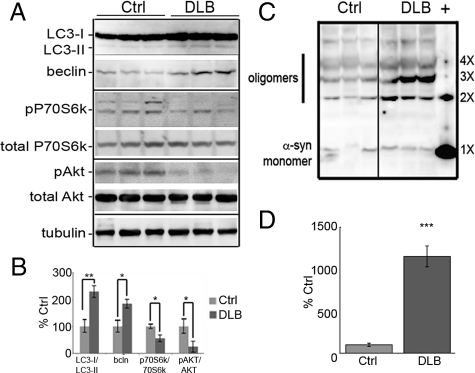
The analysis of brain homogenates from patients diagnosed with DLB indicated that there was an increase in the autophagy biomarkers beclin, phospho-P70S6 kinase (Thr389), phospho-Akt (Thr308), and LC3-II (Figure 1A). As LC3-II is a constituent of the autophagic vacuole membrane, it is a reliable marker of the level of autophagy present at the time of sample collection. Beclin is a marker of induction of autophagy. DLB cases were compared with age and post-mortem interval-matched pathologically normal patients and a 1.8-fold increase in beclin (P < 0.01) and a 2.4× increase in LC3-II levels (P < 0.001; Figure 1B) was observed. Although it was not possible to discern the levels of LC3-II in neurons compared with glial cells, it has been previously reported that the levels of LC3-I or LC3-II in glial cells is minimal,51 leading us to believe that the predominant form in our studies is derived from the neuronal population. The level of phospho-P70S6kinase (a second marker of autophagic induction), was half the level in the DLB cases when compared with age-matched controls, indicating that mammalian target of rapamycin kinase was not active50 and further confirming that autophagy was activated in DLB brains. Akt was significantly less phosphorylated,52 again favoring an increase in autophagy. Overall, our analysis of human brain material indicates a strong presence/induction of autophagy. This induction was not sufficient to clear aggregates as more α-synuclein positive oligomers were observed in the DLB brain (+; Figure 1C). On average, there was a 12-fold increase in oligomers observed in the DLB cases than in nondementia controls (Figure 1D).
Figure 1.
Macroautophagy is up-regulated in DLB. A: Homogenates of frozen human brain tissue (see Table 1) Brodmann’s area 9 (dorsolateral prefrontal cortex) from nonaffected controls (Ctrl; n = 6; 78.7 ± 4.5 years) and DLB cases (n = 6; 78.5 ± 3.7 years) were analyzed for LC3, beclin, phospho-P70S6 kinase (T389), total P70S6 kinase, phospho-Akt (T308), total Akt, and tubulin. Representative blots show three samples from each group. Overall, tubulin levels were consistent between the groups, whereas there was a significant increase in beclin (P < 0.01) and LC3 (P < 0.001) along with a significant decrease in p-P70S6k/P70S6k and p-Akt/Akt (P < 0.05) profiles between patients with nonaffected and DLB brains (B). C: Oligomeric α-syn could also be identified in the human autopsy brains, as detected by the LB509 antibody and ran alongside α-synuclein oligomers (+) to verify the MW of the nX α-synuclein species. All values are expressed as mean ± SEM; *P < 0.05; **P < 0.01. D: Densitometric analysis of oligomers in DLB and control cases indicates that there is a significant increase in oligomer formations in DLB brains (***P < 0.001).
Autophagic Induction Is Observed in Mutant Mice and Increases with Aging
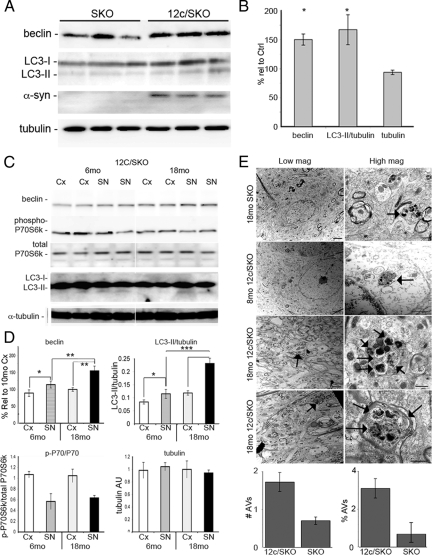
We next examined a transgenic mouse model where the murine α-synuclein gene has been ablated (SKO) and replaced with a transgene expressing mutant (A53T) human α-synuclein (12c/SKO) exclusively in the TH-positive regions. In the substantia nigra of 6-month-old (6mo) 12c/SKO mice, beclin levels were increased by 1.5 times (P < 0.05), and LC3-II/tubulin levels were 1.72 times higher (P < 0.05) than in the SKO animals (Figure 2, A and B). α-Synuclein oligomers, aggregates, and Lewy bodies were not observed in the mice, suggesting that the up-regulation of autophagy effectively clears excess α-syn from the neurons. There was an increase in the autophagic markers with aging. In the 12c/SKO mice, there was a 1.4× increase in beclin (P < 0.01) and a 2.2× increase in LC3-II/tubulin levels (P < 0.001) in the substantia nigra (SN) in mice at 18 to 24 months of age, as compared with 12c/SKO mice at 6 months of age (Figure 2, C and D). This change was only observed in the SN where α-synuclein was expressed, and not the cortex where it was not expressed. Regional comparisons demonstrated elevated expression of beclin and LC3-II in the SN relative to the cortex within each age group. The magnitude was greater in the aged mice (P < 0.01 and P < 0.001, respectively) than in the 6mo mice (P < 0.05 for beclin and P < 0.05 for LC3-II/LC3-I). No appreciable change in p-P70S6 kinase levels was observed with aging in the SN or cortex between mice at 6 months and 18+ months (18mo) of age. These results were confirmed using Bonferroni multiple comparisons, where there was a significant difference, P < 0.001, in comparisons between LC3-II/LC3-I and beclin levels for 6mo SN and 18mo SN, as well as between 18mo Cx and 18mo SN. Bonferroni multiple comparisons of the p-P70S6k/P70S6k samples were only significant (P < 0.001) within the regions in one age group (ie, 6mo Cx vs 6mo SN; 18mo Cx and 18mo SN). There was no significant difference in the levels of the loading control, tubulin. Virtually all of the AVs were autophagolysosomes—AL, or AVd and the number of AVs in the cell bodies and axons of 8-minth-old and 18mo 12c/SKO mice was compared to identify if aging is a factor in AV accumulation. At 18 months of age, a small number of AVs were observed in the SKO mice (Figure 2E), though they were not abundant in frequency. In the 8-month-old 12c/SKO mice, single AV structures (see arrows) were observed in the cell bodies but, in general, the cells appeared normal and AV distribution was of similar frequency to the 18mo SKO mice. By 18mo however, the 12c/SKO mice had numerous dystrophic neurites and axons (Figure 2E; lower half). Although a twofold increase in the abundance of AVs in the 18mo 12c/SKO mice was observed, the increase was not statistically significant when compared with the age-matched SKO group. In addition, the total surface area of AVs in the SKO sample was 0.7%, whereas it was 3.1% in the 12c/SKO line, though it was not statistically significant. Our data suggest that the number of AVs increase in response to the expression of mutant α-syn, and this induction is exacerbated with aging.
Figure 2.
Macroautophagy is up-regulated by A53T mutant α-synuclein. A: Beclin (P < 0.05) and LC3-II (P < 0.05) levels were increased in the substantia nigra (SN) of A53T mutant α-synuclein expressing mice (12c/SKO; n = 12) as compared with age-matched controls (SKO; n = 12) (6 to 8 months old). B: Densitometric quantification of results for beclin, LC3-II/tubulin, and tubulin levels. C: A comparison between SKO and 12c/SKO mice from two regions (Cortex – no α-syn expression; SN – α-syn expression only in 12c/SKO) at 6 months and 18 months of age shows that there is an increase in beclin and LC3-II levels in the SN of 12c/SKO mice (P < 0.05 vs P < 0.01 for beclin and P < 0.001 for LC3-II/tubulin). There was no relative change in tubulin or phospho-P70S6 kinase/P70S6 kinase levels. Graphical representation of the densitometric results are shown in (D). E: Electron microscopy of the SN of the 12c/SKO and SKO mice was performed to provide definitive identification of autophagic vacuoles in the substantia nigra. AVs were not identified in 8mo SKO mice, though there were some at 18 months. In the 12c/SKO mice, AVs (arrows) were identified beginning at 8 months of age, and there was an abundance of AVs in dystrophic neuritis (right panel, second from bottom) and in axons (bottom right panel) in older animals. Scale bar for low magnification = 2 μm; scale bar for high magnification panels = 500 nm. Quantification of AVs showed that there was a twofold increase in numbers of AVs and a fivefold increase in surface area in the 12c/SKO group compared with the SKO group. All values are expressed as mean ± SEM; *P < 0.01; **P < 0.01; ***P < 0.001.
Induction of Autophagy Is Greater in Mutant than in Wild-Type Mice
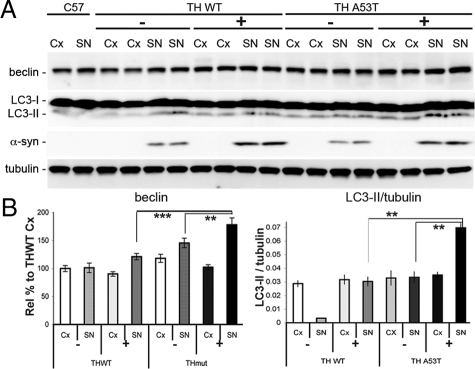
To assess whether autophagy was induced to a greater extent by mutant α-synuclein than the wild-type protein, we compared 8- to 10-month-old mice expressing either human wild-type or human A53T α-syn. A significantly higher level of beclin (1.6×; P < 0.001) and LC3-II (2.4×; P < 0.01) was observed in the mutant A53T animals relative to the wild-type α-syn mice (Figure 3, A and B). The increase in human α-synuclein was limited to the SN—there was no appreciable increase in the cortex, as expected given that transgene expression is limited to tyrosine hydroxlase-positive areas. A comparison between high-level expressing wild-type mice (+) and mice with low level expression of wild-type α-synuclein (−) showed no difference in beclin or LC3-II in the SN. However, a significant increase in beclin (P < 0.01) and LC3-II/tubulin (P < 0.01) levels was observed in the high-expressing 12c mutant line relative to the low-expressing mutant line. As has been shown by Cuervo et al (2004), the A53T variant blocks chaperone-mediated autophagy,28 and it has been suggested that this may result in an increase in alternate degradative pathways such as macroautophagy. This may partially explain why we observe an increase in LC3-II and beclin in the A53T α-syn mice. In addition, macroautophagy can be stimulated to degrade proteins that undergo oligomerization and aggregation as it is a prime target for autophagy.
Figure 3.
Macroautophagy is preferentially increased in A53T mutant variant mice compared with wild-type (WT) animals. A: Beclin and LC3-II levels were higher in the substantia nigra of mice expressing the A53T variant compared with WT mice (8 to 12 months of age; n = 8 per group). Overall, beclin was higher with increased α-synuclein expression, but not as significant as expression of the mutant variant (A53T), which produced significant increases in beclin expression. B: High level expressing (+) A53T mice had increased autophagy markers compared with the low level expressors (−), whereas there was no change in the levels between high and low level expressing WT mice. LC3-II (and LC3-II/tubulin) levels were significantly higher in SN of the A53T/+ mice but were relatively unchanged in all other groups when compared with nontransgenics, or compared with the cortex of the WT. All values are expressed as mean ± SEM; **P < 0.01; ***P < 0.001.
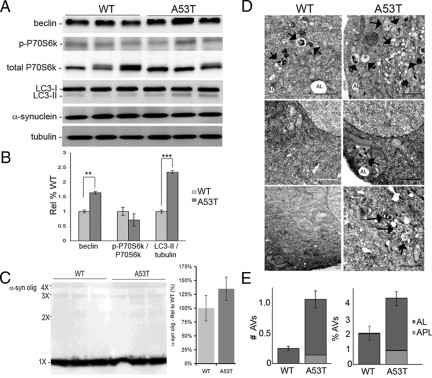
To further confirm that mutant α-syn induces autophagy and to examine the effect on oligomeric synuclein, M17 human neuroblastoma cells transfected with either wild-type or mutant (A53T) α-synuclein were compared for levels of autophagic markers. Transfected cell lines had similar levels of α-syn (Figure 4A), but there were significantly higher levels of beclin (1.62×; P < 0.01) and LC3-II/tubulin (2.3×; P < 0.001) and lower levels of phospho-P70S6 kinase in the A53T-transfected cells (Figure 4, A–B) although this difference did not reach statistical significance. Prolonged exposure of immunoblots revealed low levels of oligomers (dimers-tetramers) in the A53T cell line that comigrated with oligomers formed in vitro from purified α-synuclein (Figure 4C). Although the A53T cell line had 32% more oligomers, the difference was not significant (n = 6). This data supports our findings in animals where we observed increased autophagy in the mutant α-syn animals strengthening the idea that autophagy is enhanced to counter the accumulation of α-syn aggregates/oligomers.
Figure 4.
Macroautophagy is increased in A53T expressing cells relative to the wild-type. A: Immunoblots of representative cell harvests of wild-type (WT) and A53T-mutant cells illustrate that there are higher levels of beclin and LC3-II levels in the mutant cells than the WT cells, despite similar expression levels of the respective α-synuclein protein. There was no observable difference in the phospho-P70S6 kinase, total P70S6 kinase, or tubulin levels. B: Densitometric analysis confirms that there is a significant increase in beclin (P < 0.01) and LC3-II/tubulin (P < 0.001), but not p-P70S6k/P70S6k (sample size = nine per group). C: Although there was an abundance of monomeric α- synuclein in both WT and A53T cell lines, prolonged exposure shows a slight increase in oligomeric α-synuclein in the A53T cells, and lower levels in the WT cells. Densitometric analysis did not identify a significant difference between the two groups. D: Electron microscopic images of WT and A53T cells shows the presence of AVs (arrows) and ALs in both lines, but higher prevalence in the A53T cells (Scale bar = 2 μm). E: Quantification of AVs shows that there is a threefold increase in AVs in the A53T cells, as compared with WT cells, with approximately 10% of these AVs being nascent AVs (APL - autophagasomal with double membrane), whereas all of the AVs in WT cells were ALs. Based on surface area measurements, there was a twofold increase in total # AVs, with 26% of all AVs represented by APLs that were not evident in the WT cells. All values are expressed as mean ± SEM; **P < 0.01; ***P < 0.001.
This evidence was confirmed by electron microscopy analysis of the cells (Figure 4D) as we found that there was a significant increase in the number of AVs (arrows) in the mutant cells (P < 0.001; Figure 4E) relative to the wild-type transfected cells. Our analyses included ALs and APLs, which were classified based on published criteria.50 The most abundant type of vacuoles noted (<0.5 μm) were autolysosomes, which contained mostly degraded material. Interestingly, in the A53T-transfected cells, 8% of the AV population were newer-formed APLs (Figure 4E). In terms of cellular surface area, APLs in mutant cells accounted for 1.3% of total area and total AVs accounted for 3.5% of surface area in mutant cells (P < 0.01, as compared with wild-type cells), while APLs were not present in the wild-type cells and ALs were less than 2% of the total surface area.
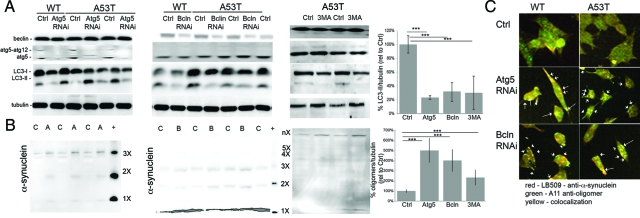
Blocking Autophagy Promotes the Accumulation of α-Syn Oligomers
To assess the effect of reduced autophagy in α-synuclein transfected cells, RNA interference against beclin or Atg5, or chemical inhibition by 3MA were used to block autophagy. Both genes have been identified previously as being critical for initiating autophagy (Figure 5, A and B). Atg5 RNAi reduced LC3-II by as much as 78.2% (± 2.7%) (Figure 5A; P < 0.001), whereas beclin RNAi reduced the levels of LC3-II by 70.3% (± 12.5%) (P < 0.001). Cells were also treated for 24 hours with 3-MA, an inhibitor of the PI3kinase/beclin pathway and we noted that there was a 72.4% (± 23%) decrease in LC3-II/tubulin levels (P < 0.01; Figure 5A). The control sequences (off-site target and vector alone) did not reduce LC3-II levels (data not shown). Following RNAi or 3MA treatment, there was a significant increase in oligomers/tubulin (Figure 5B; 486 ± 92% in Atg5 RNAi treated mutant cells, 405 ± 63% for beclin and 252 ± 62% for 3MA treated A53T cells, P < 0.01 for all), as well as a trend toward increased higher molecular-weight oligomeric forms, and an increase in aggregates that did not migrate out of the well (Figure 5B). The oligomers were evident under confocal microscopy as large, α-syn-positive aggregates that accumulated intracellularly and colabeled with the anti-oligomeric antibody, A11. The colabeled puncta (white arrows) were much more prominent in the RNAi treated A53T-transfected cells than the wild-type cells, or in the untreated cells (Figure 5C). The degree of green (A11) colocalization went from 6% in the untreated A53T cells to 38% in wild-type and 65% in A53T cells with Atg5 RNAi, and 33% and 58%, respectively, following beclin RNAi treatment.
Figure 5.
Inhibition of macroautophagy promotes the accumulation of aggregated and oligomeric α-synuclein. A: Following RNAi reduction of autophagy by targeting Atg5 or beclin for 48 hours, or addition of 10 mmol/L 3MA for 24 hours, there was a significant decrease in LC3-II/tubulin (20% to 30% of Ctrl levels for all treatments, ***P < 0.001) and decrease in beclin (only with beclin RNAi treatment). Densitometry is provided for LC3-II/tubulin levels following RNAi or 3MA treatment in the A53T cells. B: α-Synuclein oligomers were seen in the A53T cells following either Atg5 (A) or beclin (B) RNAi or 3MA treatment and levels substantially increased relative to the untreated group (C). In wild-type (WT) cells, there was an increase in a band corresponding to 4× α-synuclein in both the Atg5 and beclin RNAi treatments. A53T-expressing cells had increased levels of the 2× to 4× oligomers following beclin RNAi treatment when detected on a higher percentage gel. There was also a significant increase in higher molecular weight oligomers/aggregates at the gel interface. Densitometric analyses for A53T cells of oligomers/tubulin identify a significant increase in Atg5 and beclin RNAi, and 3MA-treated cells (***P < 0.001). C: There was increased puncta of α-synuclein (red) that was also oligomer-specific (red), as detected by the A11 antibody. Although this was evident in both WT and A53T cells, there were more α-synuclein aggregates (arrows) in the A53T cells. Scale bar = 5 μm. All values are expressed as mean ± SEM.
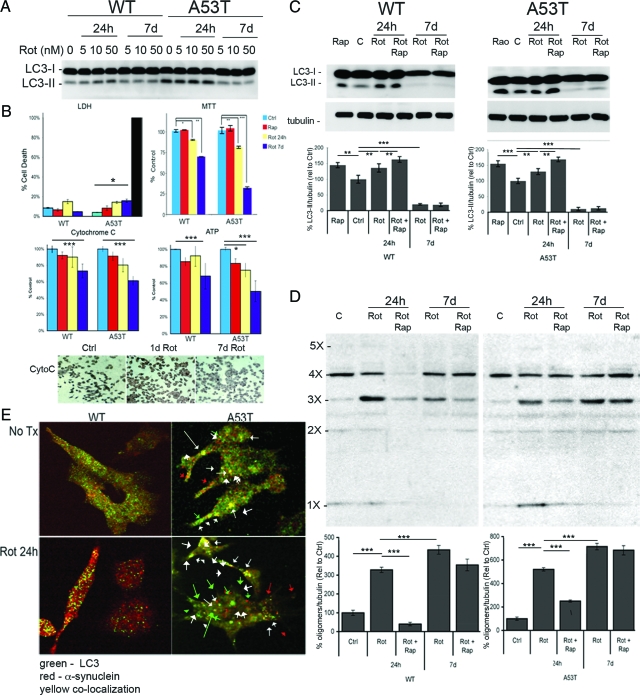
Oligomer α-Syn in Rotenone-Treated Cells and Clearance by Rapamycin
To establish if autophagy is required to maintain α-syn homeostasis and reduce the presence of α-synuclein oligomers and inclusions, oligomer formation was induced in M17 transfected cells by exposing the cells to the organic pesticide, rotenone. Varying concentrations and exposure times were tested, which showed that there was a substantial increase in autophagy response in the wild-type and A53T cells to rotenone after 24 hours and that the wild-type cells had significantly lower basal levels of LC3-II than A53T cells (Figure 6A). Chronic exposure (7 days) to rotenone at the concentrations used significantly reduced the level of LC3-II in the cells. Using an MTT assay, a significant decline in enzymatic activity was noted, although the number of cells was not significantly different between the vehicle and rotenone-treated groups (Figure 6B). The attenuation of metabolic activity was supported by the observation of a significant decrease in cytochrome c oxidase and ATP levels in these cells (Figure 6B). After 24 hours of rotenone treatment, a 16% decrease in cytochrome c oxidase levels and a 22.3% decrease in ATP was observed in the A53T cells compared with a 12.2% cytochrome c oxidase and 13.5% ATP loss in the wild-type cells. Following 7 days treatment, cytochrome c oxidase levels had fallen 24% in the wild-type cells and 38% in the A53T cells (P < 0.001), while ATP showed a similar reduction (32.5% and 50.5%, respectively, P < 0.001). Mutant cells treated for 7 days showed significant decline in cytochrome c oxidase, as indicated by reduced diaminobenzidine immunoreactivity. There was also a significant decrease in ATP levels in A53T cells following 24 hours of rotenone exposure (23.5% decrease, P < 0.05). Reduced metabolism in the cells was not due to cell death based on lactate dehydrogenase release, which was not significantly altered in the 24 hour treated wild-type or mutant cells. There was a moderate 18% increase in lactate dehydrogenase release in mutant cells exposed to rotenone over 7 days, but this was small compared with Triton X-100 treated cells (black bar) (P < 0.05; Figure 6B). While there was a mild decline in the wild-type cells, there was a larger relative decrease in MTT conversion in the mutant cells. Acute exposure did not appreciably hamper the autophagic response, as there was a robust increase in LC3-II. Following 7 days of rotenone treatment, however, the was a dramatic decline in activity, particularly in the A53T cells, resulting in only 5% to 10% of constitutive LC3-II levels, indicating that while early mitochondrial dysfunction may induce autophagy to stimulate organelle clearance, prolonged treatment results in decreased autophagy due to metabolic limitations. We did not note any loss of MTT conversion when autophagy was stimulated with rapamycin (Figure 6B).
Figure 6.
The organic pesticide rotenone impairs mitochondrial activity and prolonged exposure limits the efficacy of autophagy. A: Rotenone (5 to 50 nmol/L) induces autophagy in wild-type (WT) and (to a lesser degree) A53T α-synuclein-transfected cells within 24 hours, but chronic exposure (7 days) of rotenone, dramatically reduces the levels of LC3-II. B: At the same time, lactate dehydrogenase levels were only significantly different in 7-day treated A53T (P < 0.05) cells relative to controls, whereas cellular activity, as determined by MTT assay, was reduced by 10% in WT cells (P < 0.05) and 22% (P < 0.01) in the A53T cells after 24 hours. Following 7 days of treatment, the decrease was 32% (P < 0.01) and 70% (P < 0.001), respectively. Cytochrome C was also significantly reduced after 7 days of rotenone treatment in both cell lines, although the change was more apparent the mutant cells. Immunochemistry of cytochrome c oxidase activity in the mutant cells showed a clear decline in diaminobenzidine immunoreactivity after 7 days of rotenone treatment. This decline in metabolic levels was even greater at 7 days in the mutant cells as determined ATP levels. C: Whereas 50 nmol/L rotenone induced autophagy, as indicated by LC3-II levels, at a similar rate to 10 nmol/L rapamycin at 24 hours, prolonged exposure to this level of rotenone depressed the levels of LC3-II/tubulin. Densitometry results are shown for the samples (n = 6; **P < 0.01, ***P < 0.001). D: At 24 hours, rapamycin (added at the 20-hour point) was competent in removing most of the oligomeric α-synuclein in both the WT and A53T α-synuclein-transfected cells treated with 50 nmol/L rotenone. This clearance mechanism was not as effective after 7 days of exposure in the cell lines, though there was a minor reduction in 3× α-syn in the WT cells. Densitometry results (n = 6) are shown for levels of oligomers (*** P < 0.001). E: Confocal images of WT and A53T cells treated for 24 hours with 50 nmol/L rotenone and colabeled with LC3 (green) and α-synuclein (red). Although there was significant colocalization (yellow, white arrow) between LC3 and α-synuclein in punctate structures indicating AVs, there were also numerous green puncta of AVs without α-synuclein (green arrow) in the A53T cells, and red puncta (red arrow) that identify α-synuclein aggregates not being degraded via autophagy. All values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
At a concentration of 50 nmol/L rotenone, lower levels of mitochondrial activity were observed, but not cell death. At day 1, there was an increase in LC3-II/tubulin levels (65% and 75% in wild-type and A53T cells with rotenone and rapamycin), but at day 7, this level was significantly decreased (<20%) in both cell lines (Figure 6C). A sharp increase in α-syn oligomers was observed after 24 hours, and an even more dramatic increase in the level of α-syn oligomers (4.5× and 6.5× in wild-type and A53T cells, respectively) was seen at day 7 (Figure 6D). At day 1, this effect was reversible with rapamycin, where 4 hours treatment significantly reduced the α-syn “laddering” (dimers-pentamers) seen on immunoblots. Rapamycin may also act via inhibition of protein synthesis, thereby reducing the levels of new α-syn production, but following 7 days of rotenone treatment, we did not observe any appreciable clearance of α-syn oligomers. After 7 days of treatment, a significant decrease was observed, but this decrease was lower than the change at day 1 (80% reduction vs 20% reduction in wild-type cells, with 30% and <5% decline in mutant cells). When viewed by confocal microscopy, some of the α-syn aggregates that had formed colocalized with the vacuolar LC3-II staining (white arrow; Figure 6E). α-synuclein aggregates were also A11-positive (data not shown). We were also able to identify AVs (green only LC3-puncta; green arrow) that did not contain α-synuclein, as well as α-synuclein aggregates that were not engulfed by AVs (red only puncta; red arrow). The degree of colocalization of LC3 puncta with α-synuclein staining was 24% (green versus yellow), while α-synuclein to LC3 puncta (red versus yellow) was 38%, suggesting that there was a moderate preference for α-synuclein, particularly larger sized immunoreactive aggregates to be localized in AVs. From this, we surmise that macroautophagy is critical for the maintenance of α-synuclein levels, thereby reducing the levels that would result in oligomerization and aggregate formation. We conclude that metabolic health is essential to determine the efficacy of autophagic health and the ability to clear potentially neurotoxic α-synuclein oligomers.
Discussion
Protein misfolding and the accumulation of protein aggregates are hallmarks of several neurodegenerative diseases, including Alzheimer’s, Parkinson’s, fronto-temporal dementia, and Huntington’s disease.53,54 Intracellular inclusions are believed to disrupt normal neuronal activity, impeding cell function, and leaving the cell vulnerable to death.55,56,57 One event that is believed to contribute to this regressive event is reduced proteolysis associated with disease or aging.39,58,59,60 Both proteasomal and lysosomal activity are thought to be reduced with aging,60,61,62,63 with the net effect being decreased protein degradation leading to increased potential for protein misfolding and intracellular accumulation. Consequently, considerable efforts are underway to target degradative pathways therapeutically to reduce or remove intracellular inclusions.
Macroautophagy is an essential pathway for the clearance of aggregated proteins such as α-synuclein, though induction may be insufficient to clear all of the intracellular aggregates. In our analysis of DLB versus age-matched control brains, there was a substantial increase in autophagy (LC3-II and biomarkers of induction—beclin and p-P70S6 kinase) in the DLB patient brains. These results imply that while autophagy was stimulated in an attempt to clear aberrant α-syn inclusions, it was unable to remove all of the accumulated α-synuclein. These observations were recapitulated in the transgenic mice, where, in the A53T variant animals, there was an increase in the levels of beclin and LC3-II. As the α-synuclein transgenic mice do not develop overt aggregates, it is likely that the neurons in these mice are very efficient in maintaining protein homeostasis.31,32 There was a stronger autophagic response in mice with mutant variant α-syn compared with mice with similar levels of wild-type protein. Although gene-dosage effects of wild-type α-syn contributes to pathology in humans,64 there was no discernible difference in the level of autophagy markers when wild-type high-expressors were compared with low-expressors or nontransgenics suggesting that the mutant protein, which did elicit a response, is more pathogenic in mice. We also show that aging increased the level of beclin and LC3-II. Typically, LC3-II levels are very low due to efficient fusion of AVs to lysosomes. In our samples, there was an increase in LC3-II, as well as AVs. This may be a response to oligomer formation, or alternately, it is likely that autophagy may also be incomplete, or arrested, as has been demonstrated in Alzheimer’s disease,43 as well as in in vitro studies.49 Autophagy may be less efficient, and require a higher level of induction to maintain the neurons in the aged brain, a hypothesis supported by our data and that of others.65 In addition, we also observed that prolonged metabolic loss, as indicated by mitochondrial activity, could significantly decrease the level of macroautophagy, as indicated by LC3-II levels, with a dramatic increase in α-synuclein aggregation, which was not reversible when we attempted to reverse the cellular accumulation, suggesting that metabolic failure leads to an irreversible loss of autophagy, barring increased cellular respiration.
The absence or disruption of autophagy has been shown to lead to protein buildup and increased intracellular inclusions.31,32 As mutant α-synuclein cannot by degraded via lysosomal specific chaperone-mediated autophagy,28 reduced autophagy should result in accumulation of α-synuclein. We have shown that blocking the initiation of autophagy by targeting Atg5 or beclin expression through RNA interference clearly induced the accumulation of oligomeric α-synuclein in the mutant-expressing cells. Autophagy is critical for maintaining a balance in the level of α-synuclein, as we readily identified oligomeric forms of α-synuclein that comigrated with oligomers of recombinant α-syn in extracts of A53T-transfected cells following rotenone treatment. To a lesser degree, we also saw an increase of oligomeric α-synuclein in the wild-type cells following rotenone administration, suggesting that loss of one of the autophagic mechanisms (macro or chaperone-mediated autophagy) will result in increased synucleinopathy regardless of the isoform. Chemical induction of autophagy with rapamycin was successful in abrogating α-synuclein oligomers in cells acutely treated (24 hours) with 50 nmol/L rotenone. In cells treated for 24 hours with rotenone, an increase in LC3-II was observed, as well as an increase in the oligomeric laddering reminiscent of the DLB brains, suggesting that autophagy was stimulated by the toxic effects of rotenone, but was proteolytically insufficient to degrade all oligomers. Addition of rapamycin significantly reduced the levels of oligomeric α-synuclein in the acutely treated wild-type cells, indicating that it is possible to pharmacologically induce autophagy and limit accumulation of the pathogenic forms of the protein.
In addition to its role in misfolded protein clearance, autophagy also plays a central role in the clearance of defective mitochondria (mitophagy) in neurons. To perform this function, it is essential that there be sufficient cellular respiration and energy production (eg, ATP) to support the proteolytic process.66 It is also important for the cell that autophagy does not exacerbate cellular energy deficits by excessive clearance of mitochondria, such as may occur following chronic exposure to organic pesticides like rotenone that targets the mitochondrial complex I (NADH-ubiquinone oxidoreductase), or agents that promote energy depletion. Our data show that the levels of autophagy induced, and the efficiency of clearance of aggregated α-synuclein was dependent on the metabolic health of the cells as chronic, but not acute, exposure to rotenone led to a drop in metabolic activity that correlated with a steep reduction in LC3-II levels and a dramatic increase in oligomers. Overall, this implies that metabolic deficits significantly reduce the ability of autophagy to degrade α-syn. Clinically relevant correlates would include reduced mitochondrial function in PD patients with gene mutations linked to the mitochondria, such as DJ-1, PINK1, and parkin (67; as reviewed in68), or patients chronically exposed to organic pesticides, such as rotenone or paraquat, that have inhibited mitochondrial function and synucleinopathy.69,70,71
Overall, these studies provide a comprehensive analysis of the state of autophagy relative to α-synucleinopathy, and support the use of small molecule therapeutics to induce autophagy and promote protein clearance. This work indicates that the mammalian target of rapamycin pathway can be targeted with small molecules to induce autophagy or improve its clearance capacity, but rapamycin is not a good choice as there is limited, or no, brain penetrance. It may also be possible to target beclin/VPS34/PI3k to stimulate autophagy, as the signaling pathway was enhanced in response to the mutant variant protein. Finally, autophagy is critical for the prevention of toxic oligomer formation such as α-synuclein oligomers. While it is anticipated that therapeutic treatment of PD may optimize autophagy, it is important to establish the potential for success, particularly in aged individuals, or if intervention is proposed for patients with familial forms of PD, or those compromised by prolonged exposure to toxic agents such as organic pesticides.
Acknowledgments
We thank Dr. Anne Cataldo, Ms. Linda Hassinger, and Lyn Casey for their assistance with electron microscopy.
Footnotes
Address reprint requests to Karen Duff, Dept of Pathology, Columbia University Medical Center, 630 W168th St Rm 12-461, New York NY 10032. E-mail: ked2115@columbia.edu.
Supported by grants from the National Institute of Neurological Disorders and Stroke (NS050487, to L.C.), the Parkinson’s Disease Foundation (L.C.), and the Intramural Research Program of the National Institutes of Health, National Institute on Aging (Project Z01-AG000948, to M.R.C.).
Present affiliation of B.D. is Merritt Center for Neuromuscular Disorders, Columbia University, 630 W. 168th St, New York, NY 10032.
References
- Wakabayashi K, Matsumoto K, Takayama K, Yoshimoto M, Takahashi H. NACP, a presynaptic protein, immunoreactivity in Lewy bodies in Parkinson’s disease. Neurosci Lett. 1997;239:45–48. doi: 10.1016/s0304-3940(97)00891-4. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG. Lewy body diseases and multiple system atrophy as alpha-synucleinopathies. Mol Psychiatry. 1998;3:462–465. doi: 10.1038/sj.mp.4000458. [DOI] [PubMed] [Google Scholar]
- Irizarry MC, Growdon W, Gomez-Isla T, Newell K, George JM, Clayton DF, Hyman BT. Nigral and cortical Lewy bodies and dystrophic nigral neurites in Parkinson’s disease and cortical Lewy body disease contain alpha-synuclein immunoreactivity. J Neuropathol Exp Neurol. 1998;57:334–337. doi: 10.1097/00005072-199804000-00005. [DOI] [PubMed] [Google Scholar]
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- Goers J, Uversky VN, Fink AL. Polycation-induced oligomerization and accelerated fibrillation of human alpha-synuclein in vitro. Protein Sci. 2003;12:702–707. doi: 10.1110/ps.0230903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon R, Bar-Joseph I, Frosch MP, Walsh DM, Hamilton JA, Selkoe DJ. The formation of highly soluble oligomers of alpha-synuclein is regulated by fatty acids and enhanced in Parkinson’s disease. Neuron. 2003;37:583–595. doi: 10.1016/s0896-6273(03)00024-2. [DOI] [PubMed] [Google Scholar]
- Nielsen MS, Vorum H, Lindersson E, Jensen PH. Ca2+ binding to alpha-synuclein regulates ligand binding and oligomerization. J Biol Chem. 2001;276:22680–22684. doi: 10.1074/jbc.M101181200. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Lansbury PT., Jr Is there a cause-and-effect relationship between alpha-synuclein fibrillization and Parkinson’s disease? Nat Cell Biol. 2000;2:E115–E119. doi: 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. Alpha-synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275:34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- Outeiro TF, Putcha P, Tetzlaff JE, Spoelgen R, Koker M, Carvalho F, Hyman BT, McLean PJ. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS ONE. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Polymeropoulos MH. Intron-exon structure of ubiquitin c-terminal hydrolase-L1. DNA Res. 1998;5:397–400. doi: 10.1093/dnares/5.6.397. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, Squitieri F, Krieger E, Vanacore N, van Swieten JC, Brice A, van Duijn CM, Oostra B, Meco G, Heutink P. DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003;24:159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, Strain KJ, Maraganore DM. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, Al-Din A, Hillmer AM, Karsak M, Liss B, Woods CG, Behrens MI, Kubisch C. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco EV, Annesi G, Tarantino P, Rocca FE, Provenzano G, Civitelli D, Ciro Candiano IC, Annesi F, Carrideo S, Condino F, Nicoletti G, Messina D, Novellino F, Morelli M, Quattrone A. Glucocerebrosidase gene mutations are associated with Parkinson’s disease in southern Italy. Mov Disord. 2008;23:460–463. doi: 10.1002/mds.21892. [DOI] [PubMed] [Google Scholar]
- Mata IF, Samii A, Schneer SH, Roberts JW, Griffith A, Leis BC, Schellenberg GD, Sidransky E, Bird TD, Leverenz JB, Tsuang D, Zabetian CP. Glucocerebrosidase gene mutations: a risk factor for Lewy body disorders. Arch Neurol. 2008;65:379–382. doi: 10.1001/archneurol.2007.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LN, Ross BM, Wang Y, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, Waters C, Ford B, Frucht S, Ottman R, Marder K. Mutations in the glucocerebrosidase gene are associated with early-onset Parkinson disease. Neurology. 2007;69:1270–1277. doi: 10.1212/01.wnl.0000276989.17578.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharon-Peretz J, Badarny S, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson disease: phenotype-genotype correlation. Neurology. 2005;65:1460–1461. doi: 10.1212/01.wnl.0000176987.47875.28. [DOI] [PubMed] [Google Scholar]
- Clark LN, Kartsaklis LA, Gilbert RW, Dorado B, Ross BM, Kisselev S, Verbitsky M, Mejia-Santana H, Cote LJ, Andrews H, Vonsattel J-P, Fahn S, Mayeux R, Honig LS, Marder K: Association of glucocerebrosidase mutations with dementia with Lewy bodies. Arch Neurol 2009, 66:578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Gomes AV, Barnes JA, Dice JF. Selective degradation of annexins by chaperone-mediated autophagy. J Biol Chem. 2000;275:33329–33335. doi: 10.1074/jbc.M005655200. [DOI] [PubMed] [Google Scholar]
- Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. How do intracellular proteolytic systems change with age? Front Biosci. 1998;3:d25–d43. doi: 10.2741/a264. [DOI] [PubMed] [Google Scholar]
- Gordon PB, Hoyvik H, Seglen PO. Prelysosomal and lysosomal connections between autophagy and endocytosis. Biochem J. 1992;283:361–369. doi: 10.1042/bj2830361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, Jiang Y, Duff K, Uchiyama Y, Naslund J, Mathews PM, Cataldo AM, Nixon RA. Macroautophagy–a novel beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigelny IF, Bar-On P, Sharikov Y, Crews L, Hashimoto M, Miller MA, Keller SH, Platoshyn O, Yuan JX, Masliah E. Dynamics of alpha-synuclein aggregation and inhibition of pore-like oligomer development by beta-synuclein. Febs J. 2007;274:1862–1877. doi: 10.1111/j.1742-4658.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- Tsigelny IF, Crews L, Desplats P, Shaked GM, Sharikov Y, Mizuno H, Spencer B, Rockenstein E, Trejo M, Platoshyn O, Yuan JX, Masliah E. Mechanisms of hybrid oligomer formation in the pathogenesis of combined Alzheimer’s and Parkinson’s diseases. PLoS ONE. 2008;3:e3135. doi: 10.1371/journal.pone.0003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Vila M, Lincoln S, McCormack A, Picciano M, LaFrancois J, Yu X, Dickson D, Langston WJ, McGowan E, Farrer M, Hardy J, Duff K, Przedborski S, Di Monte DA. Lack of nigral pathology in transgenic mice expressing human alpha-synuclein driven by the tyrosine hydroxylase promoter. Neurobiol Dis. 2001;8:535–539. doi: 10.1006/nbdi.2001.0392. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Khoshaghideh F, Patel S, Lee SJ: Clearance of alpha-synuclein oligomeric intermediates via the lysosomal degradation pathaway. J Neurosci 2004, 24:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Ries V, Henchcliffe C, Kareva T, Rzhetskaya M, Bland R, During MJ, Kholodilov N, Burke RE. Oncoprotein Akt/PKB induces trophic effects in murine models of Parkinson’s disease. Proc Natl Acad Sci USA. 2006;103:18757–18762. doi: 10.1073/pnas.0606401103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Rockenstein E, Crews L, Masliah E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer’s and Parkinson’s diseases. Neuromolecular Med. 2003;4:21–36. doi: 10.1385/NMM:4:1-2:21. [DOI] [PubMed] [Google Scholar]
- Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, Olanow CW. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81:301–306. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Goedert M. The alpha-synucleinopathies: Parkinson’s disease, dementia with Lewy bodies, and multiple system atrophy. Ann NY Acad Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Sarkar S, Rubinsztein DC. Clearance of mutant aggregate-prone proteins by autophagy. Methods Mol Biol. 2008;445:195–211. doi: 10.1007/978-1-59745-157-4_13. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Difiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang L, Craddock J, Bruce-Keller AJ, Dasuri K, Nguyen A, Keller JN. Aging and dietary restriction effects on ubiquitination, sumoylation, and the proteasome in the heart. Mech Ageing Dev. 2008;129:515–521. doi: 10.1016/j.mad.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Sovak G, Cuervo AM. Protein degradation and aging. Exp Gerontol. 2005;40:622–633. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol. 2004;36:2376–2391. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. α-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. When lysosomes get old. Exp Gerontol. 2000;35:119–131. doi: 10.1016/s0531-5565(00)00075-9. [DOI] [PubMed] [Google Scholar]
- Plomp PJ, Gordon PB, Meijer AJ, Hoyvik H, Seglen PO. Energy dependence of different steps in the autophagic-lysosomal pathway. J Biol Chem. 1989;264:6699–6704. [PubMed] [Google Scholar]
- Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CM, Alfonso A, Steer C, Liu L, Przedborski S, Wolozin B. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich A, Flint Beal M. Parkinsonism genes: culprits and clues. J Neurochem. 2006;99:1062–1072. doi: 10.1111/j.1471-4159.2006.04102.x. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Fink AL. Pesticides directly accelerate the rate of alpha-synuclein fibril formation: a possible factor in Parkinson’s disease. FEBS Lett. 2001;500:105–108. doi: 10.1016/s0014-5793(01)02597-2. [DOI] [PubMed] [Google Scholar]
- Norris EH, Uryu K, Leight S, Giasson BI, Trojanowski JQ, Lee VM. Pesticide exposure exacerbates alpha-synucleinopathy in an A53T transgenic mouse model. Am J Pathol. 2007;170:658–666. doi: 10.2353/ajpath.2007.060359. [DOI] [PMC free article] [PubMed] [Google Scholar]