Introduction
The deltorphins (Tyr-D-Ala-Phe-(Asp/Glu)-Val-Val-Gly-NH2) are the most selective natural opioid agonists for δ receptors. To understand the molecular basis of opioid bioactivity and selectivity, we synthesized eight deltorphin analogs with major conformational restraints imposed by cyclization. Linear peptides containing dibasic amino acid residues (Lys, Orn, Dab, Dap) in positions 2 and 4 were prepared and cyclization was achieved by a urea closure through reaction of bis(4-nitro-phenyl) carbonate with the side chain amino groups [1]. Molecular weights were confirmed using LSIMS mass spectrometry. Cyclic deltorphin analogs having 14–18-membered depsipetide ring structures were obtained. The peptides were tested in the guinea-pig ileum (GPI) and mouse vas deferens (MVD) assays [2] (Table 1). DEL1, DEL3 and DEL8 showed significantly enhanced potency relative to [Leu]Enk at the δ receptor and, in the case of DEL1 and DEL8, also high selectivity towards δ receptors, as determined in the MVD and GPI assays.
Table 1.
The analogs studied and their biological activities.
| Analog | GPI μ-selective IC50[nM] |
MVD δ-selective IC50[nM] |
||
|---|---|---|---|---|
| n | m | |||
| DEL1 | 4 | 2 | 65.4± 9.6 | 0.640 ± 0.043 |
| DEL2 | 3 | 2 | 460± 9 | 106 ± 7 |
| DEL3 | 4 | 1 | 25.4±2.0 | 0.483±0.065 |
| DEL4 | 3 | 1 | >10,000 | 27.1±3.1 |
| DEL5 | 4 | 4 | 888±99 | 11.8±1.6 |
| DEL6 | 3 | 4 | 1020±250 | 2.73±0.028 |
| DEL7 | 4 | 3 | >10,000 | 67.0±6.9 |
| DEL8 | 3 | 3 | 159±23 | 0.814±0.054 |
| [Leu5]enk | H-Tyr-Gly-Gly-Phe-Leu-OH | 246 ± 39 | 11.4 ± 1.1 | |
Bbn – backbone
Results and Discussion
NMR spectra were recorded in H2O/D2O (9:1), with addition of CD3COOD (DEL5-DEL8) at 305 K with peptide concentrations 3–9 mM, on a Varian Unity 500 Plus spectrometer with sodium 3-trimethylsilyltetradeuteriumpropionate, TSP, as internal standard. The assignment of proton chemical shifts was accomplished based on 2D proton spectra TOCSY (80 ms), NOESY (100 ms), ROESY (200 ms), and DQF-COSY. Due to poor solubility, spectra of DEL3 and DEL4 could not be collected. NMR data were processed using the XEASY [3] software. The 3JHNαH coupling constants were obtained from the DQF-COSY spectra. The structures of all but DEL3 and DEL4 were refined using 4.0 ns productive MD runs, utilizing the Time-Averaged Constraints procedure (TAV-MD) dedicated to structure determinations from NMR of small flexible peptides [4]. Prior to the productive MD, routine operations, including parameterization of new moieties [5], TAV-constrained model-building, energy minimization, thermalization, etc., were done. MD simulations were carried out using the AMBER, ver. 8.0 software [5]. Each set of conformations from the TAV-MD trajectories was clustered into 5 to 6 families of conformations. Analogs DEL3 and DEL4 were submitted to a similar 4.0 ns MD, not restrained by NMR data and with starting structures taken at random.
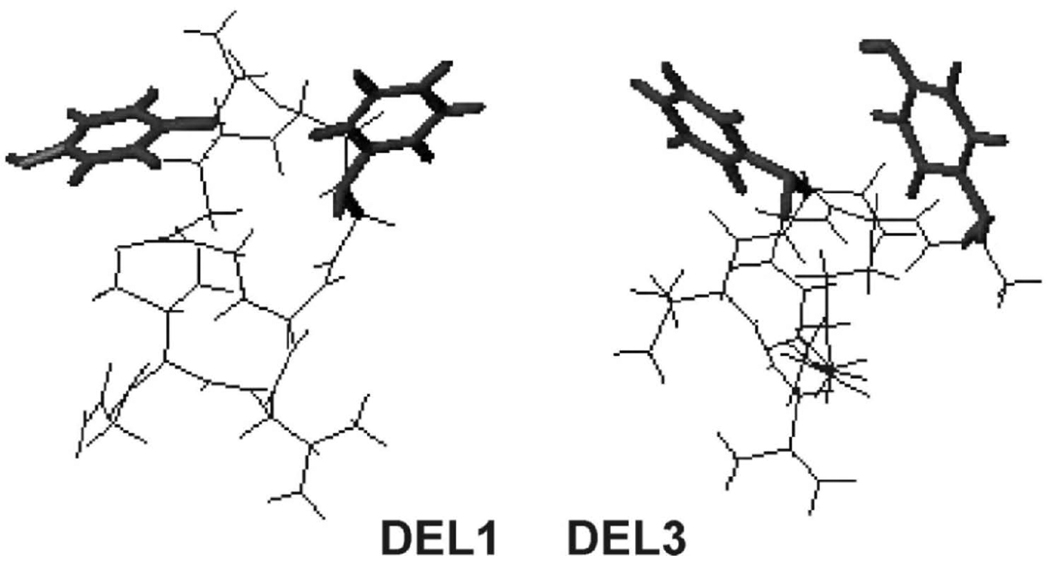
Both NMR-constrained and non-constrained MD results indicate that all studied analogs tend to take up well defined structures in solution. The highly potent (DEL1, DEL3 and DEL8) and δ-selective (DEL1 and DEL3) compounds share the following common features. 1) They have a moderate size 15–16-membered ring spanning residues 2 and 4. 2) The Tyr1 and Phe3 aromatic rings are preferably close to each other to enable a ring stacking interaction, e.g. they are on the average 5.8, 6.5 and 9.8 Å apart in DEL1, DEL3 and DEL8, respectively. 3) The structures of DEL1, DEL3 and DEL8 are compact, showing mean distances between the N- and the C-termini of 9.4, 6.7 and 9.9 Å, respectively.
Fig 1.
Refined conformations of DEL1 (NMR data-supported) and DEL3 (NMR not supported; see above) with Tyr1-Phe3 interacting aromatic rings exposed. The ring-ring centroid distances are equal to 5.8 and 6.5 Å, respectively.
Acknowledgments
Supported by Polish Ministry of Science grant 3T09A02328 (JI, AZ, EW), BW8000-5-0058-7 (JC, SR-M) and by grants from the CIHR and NIH (NNC and PWS).
References
- 1.Filip K, Oleszczuk M, Pawlak D, Wójcik J, Chung NN, Schiller PW, Izdebski J. J. Peptide Sci. 2003;9:649–657. doi: 10.1002/psc.510. [DOI] [PubMed] [Google Scholar]
- 2.DiMaio J, Schiller PW. Proc. Natl Acad. Sci. 1980;77:7162–7166. doi: 10.1073/pnas.77.12.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartles C, Xia T, Billeter M, Günter P, Wütrich K. J. Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 4.Pearlman DA. J. Biomol. NMR. 1994;4:1–16. doi: 10.1007/BF00178332. [DOI] [PubMed] [Google Scholar]
- 5.Case DA, Kollman PA, et al. AMBER 8. San Francisco: Univ of California; 2004. [Google Scholar]