Abstract
RGS7, RGS11, and their binding partner Gβ5 are localized to the dendritic tips of retinal ON bipolar cells (ON-BPC), where mGluR6 responds to glutamate released from photoreceptor terminals by activation of the RGS7/RGS11 substrate, Gαo. To determine their functions in retinal signaling, we investigated cell-specific expression patterns of RGS7 and RGS11 by immunostaining, and measured light responses by electroretinography in mice with targeted disruptions of the genes encoding them. RGS7 staining is present in dendritic tips of all rod ON-BPC, but missing in those for subsets of cone ON-BPC, whereas the converse was true for RGS11 staining. Genetic disruption of either RGS7 or RGS11 produced delays in the ON-BPC-derived electroretinogram b-wave, but no changes in the photoreceptor-derived a-wave. Homozygous RGS7 mutant mice had delays in rod-driven b-waves, whereas RGS11 mutant mice had delays in rod-driven, and especially in cone-driven b-waves. The b-wave delays were further enhanced in mice homozygous for both RGS7 and RGS11 gene disruptions. Thus, RGS7 and RGS11 act in parallel to regulate the kinetics of ON bipolar cell responses, with differential impacts on the rod and cone pathways.
Introduction
The R7 regulator of G-protein signaling (RGS) proteins RGS6, RGS7, RGS9, and RGS11 are largely restricted to neurons (Watson et al., 1994; Gold et al., 1997; Snow et al., 1998; Thomas et al., 1998; Zhang et al., 2000; Krishna et al., 2002), where they are thought to act as negative regulators of G-protein signaling through acceleration of GTP hydrolysis by Gi/o class G-proteins (for review, see Ross and Wilkie, 2000; Cowan et al., 2001). Whereas specific functions have been attributed to photoreceptor- and brain-specific isoforms of RGS9 (Rahman et al., 1999, 2003; Zhang et al., 1999; Chen et al., 2000; Lyubarsky et al., 2001; Cabrera-Vera et al., 2004; Nishiguchi et al., 2004; Kovoor et al., 2005; Krispel et al., 2006), the physiological functions of other R7 RGS proteins are not known.
Retinal ON bipolar cells respond to glutamate from photoreceptor terminals through the activation of mGluR6 (Nawy and Copenhagen, 1987; Nakajima et al., 1993; Masu et al., 1995), which activates Gαo. Light results in decreased glutamate release, leading to inactivation of mGluR6 and Gαo (Nawy, 1999; Dhingra et al., 2000, 2002; Vardi et al., 2000), opening of cation-selective channels of unknown molecular identity (Shiells et al., 1981; Slaughter and Miller, 1981; Shiells and Falk, 1990; Nawy and Jahr, 1991; Yamashita and Wässle, 1991), and membrane depolarization. These responses occur on a subsecond time scale, suggesting that some mechanism is required to accelerate GTP hydrolysis by Gαo, which in the absence of a GTPase accelerating protein (GAP) requires tens of seconds to hydrolyze GTP.
The GTPase accelerating proteins, or GAPs, that act on Gαo in the mGluR6 pathway have not yet been identified. The variant of RGS20 known as Ret-RGS1 (Dhingra et al., 2004) is present in ON bipolar cells, and has been shown to be capable of acting as a GAP for Gαo in vitro. Likewise, RGS11 and RGS7 have been reported to be present in the dendrites of ON bipolar cells, where they colocalize with mGluR6 and their obligate partner subunit, Gβ5 (Morgans et al., 2007); however, the differential distribution of RGS11 and RGS7 in different types of bipolar cells was not investigated. Gβ5 knock-out mice (Rao et al., 2007) have virtually no retinal bipolar cell-derived b-wave response in their electroretinogram (ERG). This result suggests an important role for R7 RGS proteins in signal transduction within ON bipolar cells, because their light-induced depolarization is the source of the b-wave. However, the photoreceptor synaptic connections to bipolar cells are abnormal in Gβ5 knock-out mice, so it remains unclear whether in wild-type mice R7 RGS proteins are important for signaling in ON bipolar cells after synapses have developed.
To examine the potential roles of RGS7 and RGS11, we have used immunofluorescence to study in detail their distributions in retinal neurons, and recorded ERG responses of mice with genetic deficiencies in RGS7, RGS11, or both.
Materials and Methods
Mutant mice.
RGS7 (011655-UNC) and RGS11 (011653-UNC) knock-out/mutant mice were generated by Lexicon Genetics, and obtained from Mutant Mouse Regional Resource Centers (University of North Carolina, Chapel Hill, NC). The superscript tm1Lex refers to each mutant allele. The RGS11 mutant allele (RGS11tm1Lex) is an effective null allele and produces no detectable RGS11 protein. The RGS7 mutant allele (RGS7tm1Lex) is a targeted deletion of exon 10, which produces a low amount of a truncated protein that reacts with RGS7 antibodies. RGS7(+/tm1Lex)RGS11(+/tm1Lex) mice were obtained by crossing RGS7(tm1Lex/tm1Lex)RGS11(+/+) mice with RGS7(+/+)RGS11(tm1Lex/tm1Lex) mice in our laboratory. RGS7(+/tm1Lex)RGS11(tm1Lex/tm1Lex) were further produced by crossing of RGS7(+/tm1Lex)RGS11(+/tm1Lex) mice with RGS7(+/+)RGS11(tm1Lex/tm1Lex) mice. We have attempted to obtain RGS7(tm1Lex/tm1Lex)RGS11(tm1Lex/tm1Lex) double homozygotes using the breeding paradigm of intercrossing of RGS7(+/tm1Lex)RGS11(+/tm1Lex) double heterozygote mice but with limited success (1 of 77 offspring). Thus, the majority of the RGS7(tm1Lex/tm1Lex)RGS11(tm1Lex/tm1Lex) double homozygotes were produced by intercrossing of RGS7(+/tm1Lex)RGS11(tm1Lex/tm1Lex) mice.
Genotyping.
Mouse tails (1–2 mm long) were digested with 750 μl of proteinase K (0.5 mg/ml) in lysis buffer (50 mm Tris, 50 mm EDTA and 0.5% SDS) at 55°C overnight. Genomic DNA was extracted with 750 μl of 25:24:1 of phenol:chloroform:isoamyl alcohol (Invitrogen). The extracted DNA (∼500 μl) was precipitated with 500 μl of isopropanol, washed again with 70% ethanol, air dried, and resuspended in 100–150 μl of Tris-EDTA. Genotyping for the wild-type allele and mutant allele of mRGS7 and mRGS11 were performed using the strategy recommended by Lexicon Genetics. The following pairs of primers were used for PCR: 1285–96 and 1285–74 for wild-type mRGS7 allele; Neo3a and 1285–74 for mRGS7 mutant allele; 1286-upper and 1286-lower for wild-type RGS11 allele; Neo3a/1286–57 for mRGS11 mutant allele. Details of the sequences are as follows: 1285–96, 5′CCTACACCAGAAACCAAGCC3′; 1285–74, 5′GACAGTCAGTGCTCAAACCC3); 1286-upper, 5′AGTTAAGGGCATTGGAGACCGT3′; 1286-lower, 5′CCAAAGAAACCGAAAGTGTGTTAGGG3′;1286–57, CTTCCAATATCCACCCTAGC; Neo3a, 5′GCAGCGCATCGCCTTCTATC3.
PCRs were performed on Robocycler (Stratagene) using Taq polymerase (Invitrogen, catalog #10342–020) in 20 μl reaction volume with the following components: 1× Taq polymerase buffer, 1.5 mm MaCl2, 0.05 mm of each dNTP, 0.5 μm of each primer, 2 μl of genomic DNA, and 0.05 u/μl Taq polymerase. The amplification protocol is: 1 cycle of 94°C for 2 min, 30 cycles of 94°C for 20 s, 55°C for 30 s, 68°C for 1 min 10 s, and 1 cycle of 68°C for 10 min. The wild-type RGS7 allele has a PCR product of 290 bp, the RGS7 mutant allele has a PCR product of 379 bp, the RGS11 wild-type allele has a PCR product of 203 bp, and the RGS11 mutant allele has a product of 753 bp.
Mice were used as adults 6–8 weeks of age from either C57BL/6J strain (for some immunostaining experiments), or from a mixed 129S5/SvEvBrd–C57BL/6J (129-C57) background (for comparing mice with targeted RGS mutations and wild-type or heterozygous littermates). Mice with the Rgs7tm1Lex and Rgs11tm1Lex alleles had a similar mixed 129S5/SvEvBrd–C57BL/6J (129-C57) background. ERGs were recorded from homozygous RGS7 mutants (Rgs7tm1Lex/Rgs7tm1Lex), homozygous RGS11 mutants (Rgs11tm1Lex/Rgs11tm1Lex), or double homozygotes (Rgs7tm1Lex/Rgs7tm1Lex/Rgs11tm1Lex/Rgs11tm1Lex) and compared with their wild-type or heterozygous littermates or, for the double homozygotes, to mixed 129-C57 background mice wild-type or heterozygous at both RGS7 and RGS11 loci [for details, see, Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory; World Wide Web URL: http://www.informatics.jax.org.]
The mice were reared and housed in a room with a 12 h light (< 40 lux)/12 h dark cycle. All animal procedures conformed to US Public Health Service and Institute for Laboratory Animal Research guidelines and were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
ERG recording.
Animals were dark-adapted overnight in a ventilated light-tight box. The next day, they were prepared for recording under red illumination [light-emitting diode (LED), λ > 620 nm]. Pupils were dilated to 3 mm in diameter with topical atropine (0.5%), tropicamide (1%), and phenylephrine (2.5%) before anesthesia and at least 1 h before recording ERGs. A drop of proparacaine hydrochloride (0.5%) was used for corneal anesthesia. The mice were initially anesthetized with an intraperitoneal injection of ketamine (70 mg/kg) and xylazine (7 mg/kg; both drugs from Vedco). A single dose of maintenance anesthesia, ketamine (56 mg/kg) and xylazine (5.6 mg/kg), was administered after ∼45 min via a subcutaneous needle fixed to the flank.
Rectal temperature was maintained between 36 and 37°C with an electrically heated blanket (CWE). The animal was housed in an aluminum Faraday cage to insulate the recorded signals from external static electrical fields. The animal's head was held steady to reduce noise originating from respiratory and other movements, using an aluminum head holder with a hole for the upper incisors to fix the upper jaw. This fixation ensured that the jaw remained open throughout the recording. Moist room air was pumped through a clear polyvinylchloride (PVC) pipe kept close to the open mouth. The head holder also served as the ground. All animals were recovered after the ERG recording session.
Recording sessions lasted up to 2 h. ERGs were recorded differentially between Dawson/Trick/Litzkow nylon/silver electrode fiber electrodes (Dawson et al., 1979) moistened with normal saline and placed on the two eyes. Eyes were covered with contact lenses that were pressure molded from 0.19 mm clear ACLAR film (Ted Pella) for the stimulated eye and 0.7 mm opaque PVC for the nonstimulated eye [for details, see the study by Sagdullaev et al. (2004)]. Both lenses were placed over a cover of 1.2% methylcellulose in 1.2% saline. The signals were amplified (DC to 500 Hz), digitized at 2 KHz, and sent to the computer for averaging, display and storage, and subsequent analysis.
A custom-made LED [λmax, 462 nm; −5.8–1.9 log scotopic Troland seconds (sc td s)]-based stimulator provided the light stimuli. The maximum stimulus pulse width was <5 ms. The light was collected in a cone internally coated with white paint and sent through a fiber optic cable (0.5 inch diameter, 36 inches long; Edmund Optics) into the Faraday cage. A diffuser and a stainless-steel cone at the distal end of the fiber-optic cable kept very close to the stimulated eye provided a Ganzfeld stimulus. ERGs were recorded using brief full field flashes from darkness, or in the presence of a steady rod suppressing backgrounds of 1.8 log sc td (λmax, 462 nm). The half time of the pulse width was taken as time 0 for the ERG recording. Flash intensity was varied by doubling the pulse widths of the stimuli and by choosing banks of preattenuated LEDs.
ERG records were only used for analysis if the presence of a stable background was verified for 15–20 min. (Bui and Fortune, 2004). The intervals between flashes were adjusted so that the response returned to baseline before another stimulus was presented. For the lowest intensity stimulus the interflash interval was 2 s, and for the highest intensities it was kept at 10 s. Low amplitude responses, such as the scotopic threshold response (STR), were averaged over many trials (40) and higher amplitude responses over fewer trials (10–20). A digital 60 Hz notch filter was applied off-line. Another stimulus paradigm for isolation of cone-driven signals is described in appropriate sections of Results.
Light calibration.
A calibrated photometer (IL1700, International Light Research) with a filter corrected for human scotopic vision was used to measure stimulus intensities. Our use of this approach was justified because a previous study on murine ERG a-waves had shown that the spectral sensitivity of the mouse rods is very similar to the Commission Internationale de I'Eclairage scotopic spectral efficiency (Lyubarsky et al., 1999). The luminance was recorded as sc cd m−2. To estimate the energy of the flash stimuli, a 1 KHz train of current pulses, each having a 75.77 ms duration, was used to activate the different LED settings within the stimulator. Assuming that the luminance of the LEDs was constant for the different pulse widths used, we calculated the luminance–time product for each of the single flash stimuli used and expressed them in units of sc cd s m−2. The luminance values and the luminance–time product were multiplied with the mouse dilated pupillary area measured in square millimeters to get retinal illuminance and retinal illuminance-time product values in units of scotopic Trolands and scotopic Troland seconds respectively. Based on the study of Saszik et al., 2002, a flash of 1 sc td s should produce 121.6 photoisomerizations per rod in the mouse retina, and this factor was used to estimate the photoisomerizations indicated in the figures. An alternative value of 181 photoisomerizations (sc td s)−1 has also been reported based on measured bleaching rates (Lyubarsky et al., 2004), which would shift the indicated number of photoisomerizations by a factor of 1.5.
Data processing.
The oscillatory potentials (OPs) were extracted off-line by bandpass filtering in the frequency domain between 55 and 250 Hz using a Butterworth second order filter implemented in Matlab (v. 7.01, The Math Works). The input data were processed in both the forward and reverse directions to eliminate phase distortion. Sigmaplot 9.0 (Systat Software) was used to graph the data and for statistical analysis.
Intravitreal injections.
Injections were performed using a trinocular stereo dissecting microscope (6.6× magnification) under dim red illumination (>620 nm) to avoid light-adapting the rods. Pharmacological agents were delivered via a 26 gauge steel needle with a conical style noncoring point fixed on a 10 μl Hamilton microsyringe (Hamilton Company) and inserted at a 45° angle into a small pilot hole 0.5 mm behind the limbus (created by a 30 gauge needle). A ∼1 μl volume of balanced salt solution, pH 7.4, containing GABA was slowly injected over a 1 min interval to achieve an estimated final concentration of ∼35 mm in a vitreal volume of 20 μl (Saszik et al., 2002). After injection, the ERG was monitored until the drug effects on the waveform were stable (∼40–45 min) before collecting the recordings shown.
Tissue preparation.
Light-adapted adult mice were used for immunohistochemical analysis. Animals were killed by carbon-dioxide inhalation and the eyes were rapidly excised from the head. The corneas were slit open, the lens was expressed, and the eyes were immersed in 4% formaldehyde in 0.1 m cacodylate buffer, pH 7.4, for 5 min at 4°C. The eyes were rinsed in PBS, pH 7.4, cryoprotected in 30% sucrose in PBS overnight at 4°C, embedded in OCT embedding medium (Tissue-Tek), and fast frozen on dry ice. Frozen sections through the retinal layers were taken along the vertical meridian of the eyecup at a thickness of 10–12 μm and collected onto Superfrost/Plus microscope slides (Fisherbrand, Fisher Scientific). Sections were stored at −20°C until use.
Retinal wholemounts were prepared after excising the eye and removing the cornea and lens (as described in the previous paragraph). The neural retina was rapidly isolated free of the retinal pigmented epithelium in 1× PBS (4°C, pH 7.4) and then immediately fixed lightly in 4% paraformaldehyde in 0.1 m cacodylate buffer, pH 7.4, for 5 min at 4°C. The vitreous humor was removed, and relaxing cuts were made in the retinal margin to allow the retina to flatten. The retina was rinsed in 1× PBS and immunolabeled free-floating.
Antibodies and antisera.
Details of primary antibodies used in this study are presented in Table 1. Secondary antisera were raised in goat or donkey and were specific for either mouse, rabbit, or sheep immunoglubulins and conjugated to fluorescent dyes: Alexa Fluor-488, Alexa Fluor-555, and Alexa Fluor-647 (dilution: 1:200–1:500; Invitrogen).
Table 1.
Primary antibodies and antisera
| Antigen | Host | Dilution | Source | Catalog or reference # | Reference |
|---|---|---|---|---|---|
| RGS7 | Rabbit | 1:500–1:1000 | Bethyl Laboratories /T. G. Wensel | R4612 | Morgans et al., 2007 |
| RGS11 | Rabbit | 1:500–1:1000 | Bethyl Laboratories /T. G. Wensel | R4969 | Chen et al., 2003 |
| Gβ5 | Goat | 1:500–1:1000 | Bethyl Laboratories /T. G. Wensel | G4719 | Morgans et al., 2007 |
| Gβ5 | Rabbit | 1:500–1:1000 | Bethyl Laboratories /T. G. Wensel | R5214 | Watson et al., 1994a |
| mGluR6 | Rabbit | 1:500 | Antibodies | 61–181 | Mojumder et al., 2007 |
| mGluR6 | Sheep | 1:100 | C. W. Morgans, OHSU, Beavertown, OR | Morgans et al., 2006 | |
| PKC α | Mouse | 1:500–1:1000 | Upstate Biotechnology | 05–54 clone M4 | |
| Gαo | Mouse | 1:1000 | Millipore Bioscience Research Reagents | Mab 3073 | |
| Chx-10 | Sheep | 1:50 | Exalpha Biologicals | X1180P | Mojumder et al., 2007 |
| Ribeye (ctbp2/ribeye) | Mouse | 1:5000 | BD Biosciences Pharmingen | 612044 | Morgans et al., 2007 |
OHSU, Oregon Health & Science University.
aThe antibodies were produced by Bethyl Laboratories in rabbit host from the inoculating antigen described by Watson et al. (1994).
Immunolabeling.
Immunofluorescent labeling of frozen sections was performed using protocols similar to those used previously (Sherry et al., 2006; Mojumder et al., 2007, 2008). Sections were thawed, fixed in 4% formaldehyde for 10–15 min at room temperature to improve section adherence to the slide, rinsed, treated with 1–2% NaBH4 to reduce autofluorescence, and rinsed again. Nonspecific labeling was blocked using 10% normal goat serum or normal donkey serum, 5% bovine serum albumin, 0.5–1% fish gelatin, and 0.1% Triton X-100 in 1× PBS (blocker). Excess blocker was removed and the sections were incubated in a combination of primary antibodies diluted in blocker for 24 h at 4°C. The sections were rinsed in 1× PBS (three times for 10 min), incubated in the blocking solution for 30 min, and then incubated with a combination of secondary antibodies for 30–45 min at room temperature. Sections were rinsed, coverslipped in a fade-retardant mounting medium (Prolong Gold, Invitrogen), and examined in the microscope.
Retinal wholemounts were immunolabeled free-floating. Wholemounts were treated with 1–2% NaBH4 for 1–2 min, rinsed in deionized water followed by 1× PBS, and incubated in blocker solution for 1 h at room temperature to block nonspecific labeling. Retinas were incubated in primary antibody for 3 d at 4°C. They were then were rinsed in 1× PBS for 2 h at room temperature and incubated free-floating in secondary antibody at room temperature for 1 h. Retinas were rinsed in 1× PBS for 2 h at room temperature, flattened onto microscope slides with the ganglion cell side up, coverslipped with Prolong Gold, and examined in the confocal microscope.
To confirm the specificity of immunolabeling methods, sections were processed in the absence of primary antibodies or by substituting normal animal serum corresponding to the host species of the primary antibody. These treatments eliminated labeling, as expected. All antibodies and antisera were diluted in the appropriate blocker solution.
In some of our experiments, we used two primary antibodies from rabbit for double labeling using modification of a protocol described previously (Hunyady et al., 1996; Shindler and Roth, 1996; Wang et al., 1999). In this experiment, a high-density labeling of a target protein for one immunofluorescent channel was achieved through tyramide signal amplification (TSA) using the catalytic activity of horseradish peroxidase (HRP), whereas the second target was then detected by sequential use of conventional secondary immunofluorescence labeling described above. Covalent coupling of highly reactive, short-lived tyramide radicals to residues (principally the phenol moiety of protein tyrosine residues) in the vicinity of the HRP–target interaction site, yields stable signal resistant to loss by diffusion.
Tyramide stock solution was prepared by dissolving one vial of the solid Alexa-488 labeled-tyramide provided by the manufacturer (Invitrogen) in 150 μl of DMSO. All endogenous peroxidase activity was quenched by incubating each slide in a peroxidase-quenching buffer (1× PBS + 1–3% H2O2) for 60 min at room temperature. The specimen was incubated with 1% blocking reagent for 60 min at room temperature. The section was then incubated with primary antibody diluted in 1% blocking reagent overnight. To find out the optimal primary antibody concentration, serial dilutions of primary antibody were used in a preliminary experiment. To ensure that there was no nonspecific staining in the retina and that the peroxidase-quenching buffer worked successfully, a control slide with blocking solution but without primary antibody was processed in parallel. The section was rinsed with 1× PBS three times for 10 min each. A working solution of the HRP conjugate was prepared by diluting the stock solution 1:100 in 1% blocking solution. HRP conjugate working solution (15 μl) was applied to each section and incubated for 30 min at room temperature. The section was then rinsed with 1× PBS three times for 10 min each. Tyramide working solution was prepared by diluting the tyramide stock solution 1:200 in 0.0015% H2O2 just before labeling. Tyramide working solution (15 μl) was applied to each section and incubated for 10 min at room temperature. The sections were then rinsed with 1× PBS three times for 10 min each. The sections were subsequently incubated overnight with the other primary antibody diluted in blocker using standard antibody concentrations described in Table 1. The sections were rinsed in 1× PBS (three times for 10 min), incubated in the blocking solution for 30 min, and then incubated with a secondary antibodies (of different emission spectra from that fluorescent labeled tyramide) for 30–45 min at room temperature. Another control consisting of primary plus secondary antibody incubation but no TSA was used to ensure that the use of a fluorescent secondary antibody targeting the low-abundance primary antibody in the tissues would be below the level of detection by the confocal microscope. Sections were rinsed, coverslipped in a fade-retardant mounting medium (Prolong Gold, Invitrogen), and examined in the confocal microscope.
Imaging.
Confocal images were acquired using a confocal laser-scanning microscope (LSM) with a krypton–argon laser (LSM 510; Zeiss). Images were captured sequentially for each channel using either 40× (numerical aperture = 1.30) or 63× (numerical aperture = 1.40) oil-immersion objective lenses. Stacks of serial optical sections were collected at a step size of 0.3–0.6 μm. Each image shown is either a maximum projection of an image stack, or a single, representative optical section processed with Zeiss LSM personal computer software and prepared using Adobe Photoshop 6.0 software (Adobe Systems).
Results
Overlapping, but distinguishable, distributions of RGS7 and RGS11 in the dendritic tips of ON bipolar cells in the outer plexiform layer
As reported previously (Morgans et al., 2007), we found that antibodies for all three proteins, RGS7, RGS11, and Gβ5, displayed strong punctate staining in the outer plexiform layer (OPL). Moreover, the staining coincided with staining for mGluR6 (Fig. 1), the metabotropic glutamate receptor specific for dendritic tips of ON bipolar cells (Vardi and Morigiwa, 1997), and was found at the tips of cells staining for Gαo (Fig. 2 ), the G-protein of the mGluR6 cascade, consistent with colocalization with the mGluR6 transduction machinery in the dendritic tips of ON bipolar cells. However, in contrast to previous reports, we found that the distributions of these three GAP proteins were not identical, and varied between rod and cone ON bipolar cells.
Figure 1.
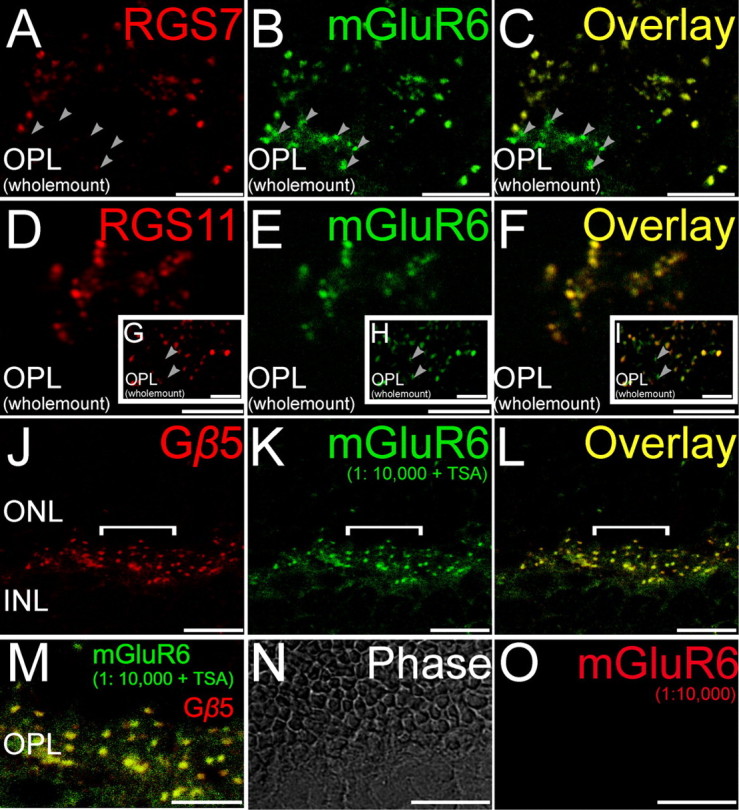
Double staining for GAP proteins and mGluR6. Double labeling using antibodies directed against RGS7, RGS11, and Gβ5 (red) and the ON bipolar cell marker mGluR6 (green). A–C, (Wholemount), Labeling for RGS7 colocalized with labeling for mGluR6. Some closely clustered dendritic tips likely representing ON cone bipolar cells were immunopositive for mGluR6 but not RGS7 (arrowheads). D–F (Wholemount), Labeling for RGS11 in the mouse retina colocalized extensively with labeling for mGluR6 especially at tightly clustered ON bipolar cell dendritic tips representing putative ON cone bipolar cells. G–I (Wholemount insets), Some isolated dendritic tips, likely representing rod bipolar cells, were immunopositive for mGluR6 but not RGS11 (arrowheads). J–L (Radial section), Labeling for Gβ5 in the mouse retina colocalized extensively with labeling for mGluR6. M (Radial section), The overlay of the region indicated by the white horizontal line in J–L is shown here in an expanded scale to show the colocalization of Gβ5 and mGluR6. N, O (Radial section), Tyramide signal amplification did not produce any visible immunolabeling at the dilution of 1:10,000 for the red channel, eliminating the possibility of cross reaction of the secondary antibodies detected in the red channel with the primary detected in the green channel. Scale bars: A–I, M–O, 5 μm; J–L, 10 μm. ONL, Outer nuclear layer; INL, inner nuclear layer.
Figure 2.
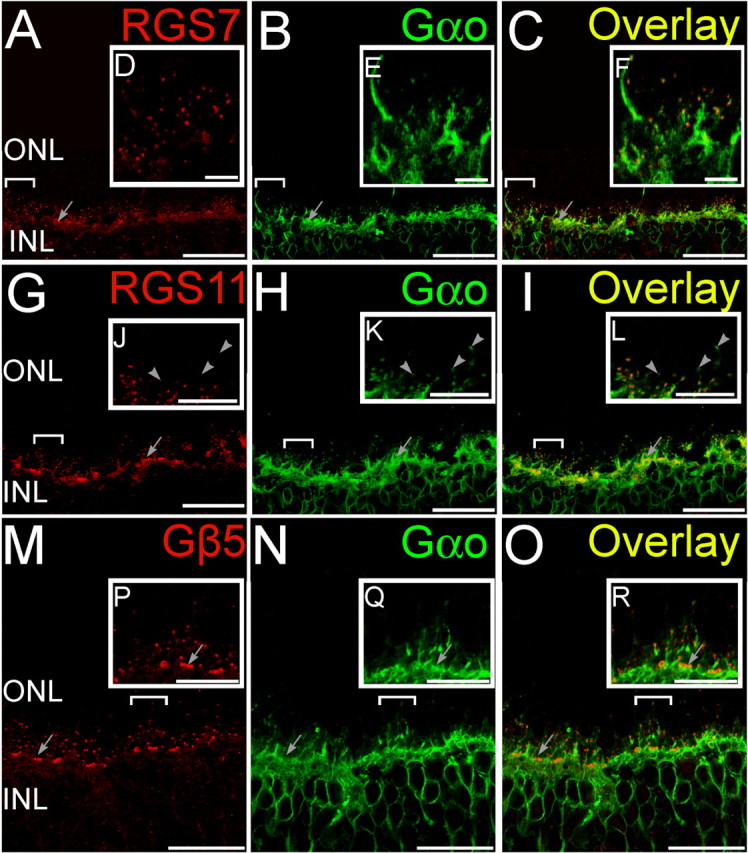
Double staining for GAP proteins and Gαo. Double labeling using antibodies directed against RGS7, RGS11, and Gβ5 (red) and Gαo (green). A–C (Radial section), Whereas Gαo staining was widely distributed in ON bipolar cells, the punctuate staining for RGS7 (A) in the OPL colocalized with labeling for Gαo (B) at the dendritic tips of ON bipolar cells as seen from the overlay (C). Clusters of RGS7 staining (arrows) indicate their presence in cone bipolar cells. D–F (Insets of bracketed regions in A–C magnified), The punctuate staining for RGS11 (D) in the OPL colocalized with labeling for Gαo (E) at the dendritic tips of ON bipolar cells as seen from the overlay (F). G–I (Radial section), Clusters of RGS11 staining (arrows) indicate their presence in cone bipolar cells. J–L (Insets of bracketed regions in G–I magnified), Some rod-bipolar cell dendritic tips (arrowheads) were devoid of detectable staining for RGS11. M–O (Radial section), The punctuate staining for Gβ5 (M) in the OPL colocalized with labeling for Gαo (N) at the dendritic tips of ON bipolar cells as seen from the overlay (O). P–R (Insets of bracketed regions in M–O magnified), Clusters of Gβ5 staining (arrows) indicate their presence in cone bipolar cells. Scale bars: A–C, G–I, and M–O, 20 μm; D–F, J–L, and P–R, 5 μm. ONL, Outer nuclear layer; INL, inner nuclear layer.
Staining for Gβ5 was uniform between rod and cone ON bipolar cells (Figs. 1, 3, 4), and we were unable to identify any clear cases in which cells stained for mGluR6 but not Gβ5 or for Gαo but not Gβ5. These results are consistent with the idea that Gβ5 is present in all dendritic tips of ON bipolar cells, likely associated with either RGS7 or RGS11, or both. In contrast, RGS7 staining was more intense in rod bipolar cells than in cone ON bipolar cells (Figs. 1–4). To obtain a semiquantitative measure of staining intensities, fluorescence intensity profiles for RGS7, RGS11, or Gβ5 were measured for ON cone bipolar cell dendritic tips (identified by clustering and appearance of multiple peaks, for example, gray trace, Fig. 3J) and for single peaks corresponding to rod bipolar cell dendritic tips. The average ratio of cone to rod tip peak intensity profile over 70 pairs of adjacent rod and cone ON bipolar cells, compared pairwise to ensure identical conditions for staining and imaging, was found to be significantly less than unity (Fig. 3K) for RGS7, indicating more intense signal for RGS7 in rod bipolar cells than in ON cone bipolar cells. However, the same ratios for RGS11 or Gβ5 were not significantly different from unity, indicating that these two proteins showed similar staining intensities in the dendritic tips of both ON cone and rod bipolar cells.
Figure 3.
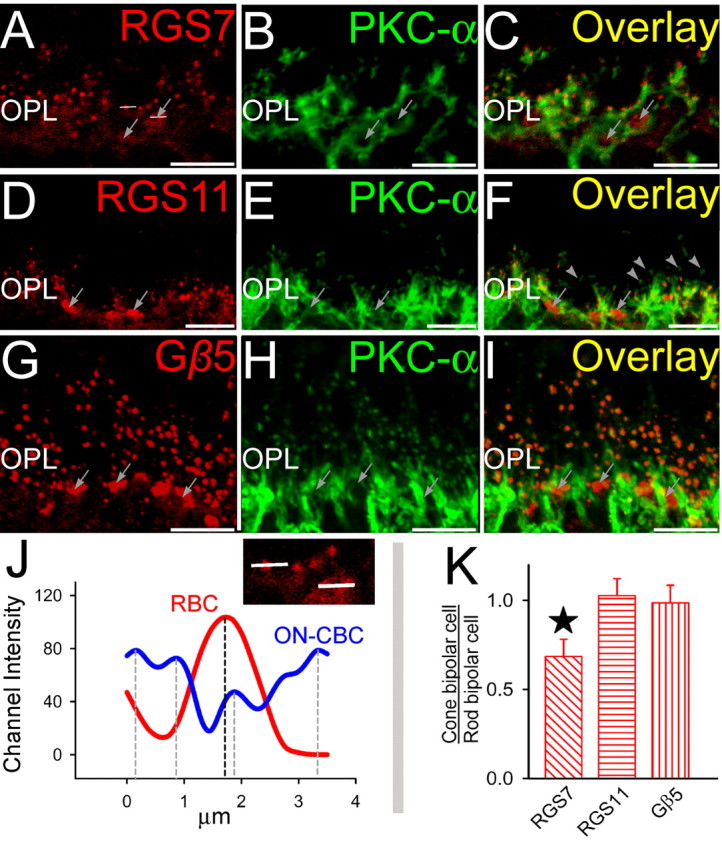
Double staining for GAP proteins and rod bipolar cell marker PKCα. Double labeling using antibodies directed against RGS7, RGS11, and Gβ5 (red) and PKC-α (green). A–C (Radial section), The punctuate staining for RGS7 (A) in the OPL colocalized with labeling for PKC-α (B) at the dendritic tips of rod bipolar cells as seen from the overlay (C). Clusters of RGS7 staining (arrows) indicate their presence in cone bipolar cells. The horizontal lines are representative channel intensity graphed in figure J. D–F (Radial section), The punctuate staining for RGS11 (D) in the OPL colocalized with labeling for PKC-α (E) at the dendritic tips of rod bipolar cells as seen from the overlay (F). Clusters of RGS11 staining (arrows) indicate their presence in cone bipolar cells. Some rod-bipolar cell dendritic tips were devoid of detectable staining for RGS11. G–I (Radial section), The punctuate staining for Gβ5 (G) in the OPL colocalized with labeling for PKC-α (H) at the dendritic tips of rod bipolar cells as seen from the overlay (I). Clusters of Gβ5 staining (arrows) indicate their presence in cone bipolar cells. J, Typical channel intensity profiles for rod and cone bipolar cell dendritic tips. Intensities were measured along white lines in A across a rod dendritic tip and a cluster of cone bipolar cell dendritic tips. Rod bipolar cells had a discrete single peak, whereas cone-bipolars (being closely packed) showed multiple peaks. The peak channel intensity profiles are indicated by dashed vertical lines. K, Average peak channel intensity profiles (across 70 channel intensity profile values for each antigen) for cone bipolar tips as a ratio of peak intensities for rod bipolar cells (across 70 channel intensity profile values) for RGS7, RGS11, and Gβ5. Statistically significant difference is indicated by the star above the vertical bar (paired t test; p > 0.01). Scale bar, 20 μm. ONL, Outer nuclear layer; INL, inner nuclear layer.
Figure 4.
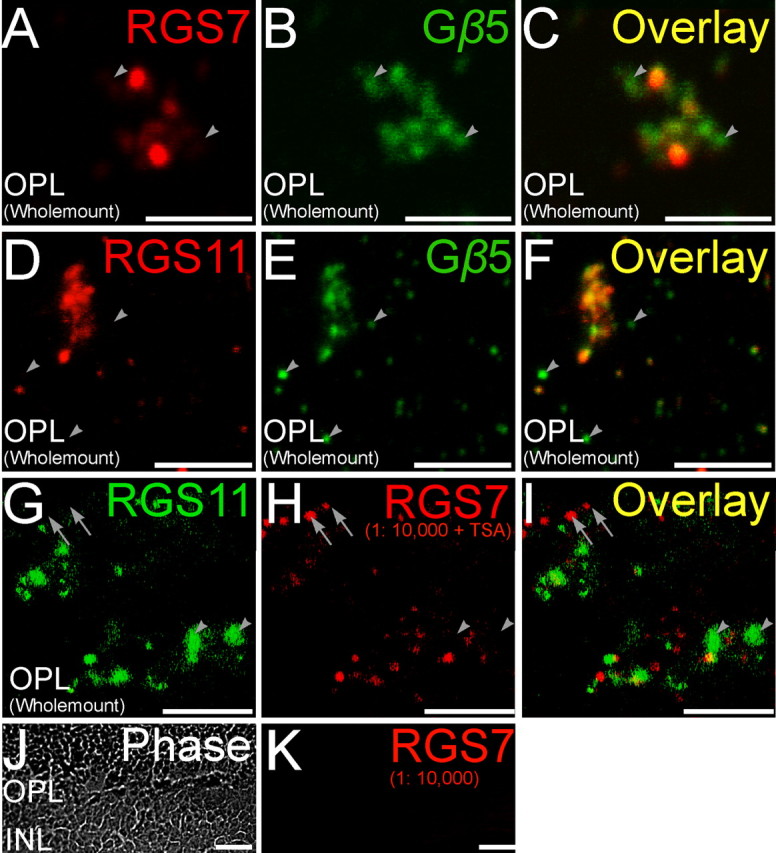
Double staining for RGS proteins and Gβ5. Double labeling using antibodies directed against RGS7 and RGS11 (red) and Gβ5 (green). A–C (Wholemount), A magnified wholemount view of the OPL shows ON cone bipolar cells in tight clusters. Some of the Gβ5 immunopositive puncta did not have significant labeling for RGS7 (arrowheads). D–F (Wholemount), RGS11 immunopositive puncta in the OPL showed good colocalization with closely spaced clusters of Gβ5 immunopositive puncta belonging to ON cone bipolar cells. However, isolated Gβ5 immunopositive puncta likely belonging to rod bipolar cells did not have detectable levels of RGS11 immunofluorescence (arrowheads). G–I, Noncoincident immunofluorescence for RGS7 or RGS11 in the OPL (wholemount). Double labeling using antibodies directed against RGS11 (green) and RGS7 (red). Staining for RGS7 and RGS11 did not overlap for some dendritic tips (arrowhead, RGS11+, RGS7−; arrow, RGS7+, RGS11−). Scale bars: A–I, 5 μm. J, K (Radial section), Tyramide signal amplification did not produce any visible immunolabeling at the dilution of 1:10,000 for the red channel, eliminating the possibility of cross reaction of the secondary antibodies in the red channel with the primary in the green channel. Scale bars: J, K, 10 μm.
Although many cone ON bipolar cells stained for RGS7, staining was undetectable in a subset of cone ON bipolar cell dendritic tips, identified by their clustering pattern and staining for mGluR6 (Fig. 1), Gαo (Fig. 2), or Gβ5 (Fig. 4), or by lack of staining for PKCα (a marker for rod bipolar cells, but not cone ON bipolar cells, Fig. 3). A reverse pattern was observed for RGS11. Although labeled dendritic tips were fairly uniform in intensity of RGS11 staining (Figs. 1–4), some dendritic tips that stained for mGlurR6 (Fig. 1) or Gαo (Fig. 2) did not stain for RGS11, and these appeared by morphology to be rod bipolar cells. This hypothesis was confirmed by double staining for rod bipolar cell marker PKCα, which revealed that some, but not all, rod bipolar cell dendritic tips lack detectable RGS11 staining (Fig. 3). If these dendritic tips belong to different bipolar cells, then this observation suggests that at the biochemical level there may be more than one class of rod bipolar cells.
These results lead to the conclusions that some rod bipolar cells dendritic tips lack RGS11, and some ON cone bipolar cell dendritic tips lack RGS7, but that all ON bipolar cells contain Gβ5. Double staining would be expected to reveal ON cone bipolar cells staining for Gβ5 but not RGS7, and some rod bipolar cells staining for Gβ5 but not RGS11. These predictions are borne out by the results (Fig. 4). The conclusion of overlapping, but not identical, distributions of RGS11 and RGS7 is further confirmed by double staining for these two RGS proteins (Fig. 4G–I). Although some cells stain for both RGS11 and RGS7, some cone ON bipolar dendritic tips lack detectable RGS7, and some rod bipolar dendritic tips lack RGS11.
RGS7 and RGS11 mutant mice
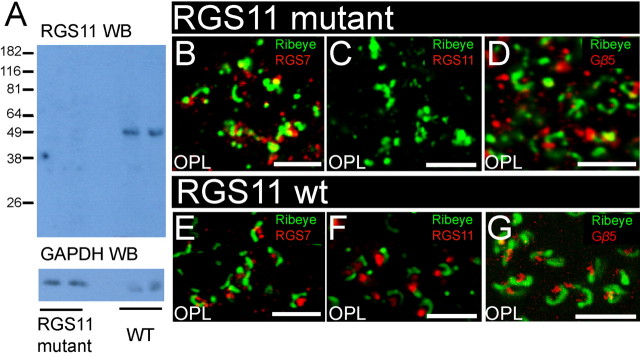
To determine the functional roles of RGS7 and RGS11 in retinal signaling, we obtained mice with targeted disruptions of the RGS7 or RGS11 genes and bred them to homozygosity at each locus. The RGS7 mutation consists of a targeted disruption of a single exon of RGS7, and this mutation apparently leads to formation of a truncated protein which reacts with the RGS7 antibodies (Fig. 5A). The amount of the truncated gene product appears to be much less than that of full-length RGS7 in the wild-type retinas. Immunofluorescence in the mutant animals confirmed greatly decreased staining for RGS7 in the RGS7 mutant animals (Fig. 5B), with diminished staining detectable in dendritic tips of rod bipolar cells. These results suggest that the RGS7 mutation is likely to function as a hypomorphic allele, and functional consequences observed by electroretinography (see below) support this idea.
Figure 5.
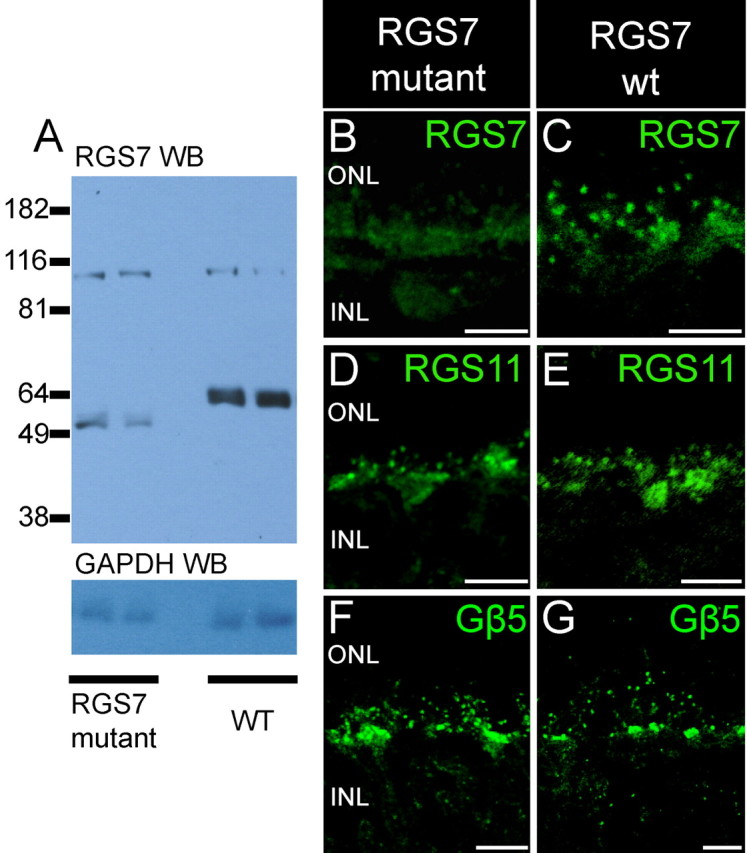
Absence of full-length protein in RGS7 mutant mice. A, Top panel, Western blot analysis of retinal lysates (1/30 of a retina) from two RGS7 mutant mice and two wild-type mice using affinity purified anti-bovine RGS7 antibody R4163. The anti-RGS7 antibody detected a band at molecular weight of ∼ 58 kDa in the lanes containing retinal lysates from wild-type mice. In the lanes containing retinal lysates from RGS7 mutant mice, the anti-RGS7 antibody detected a band at a position lower than that of the wild-type RGS7 band. The signal intensity from the mutant RGS7 protein was much lower than that of the wild-type full-length RGS7 protein. Bottom panel, Western blot analysis of the same blot with GAPDH antibody to show that comparable amounts of proteins were loaded. B, C, Diminished RGS7 staining in RGS7 mutant mice. The brightest punctate staining for RGS7 observed in rod bipolar cell dendritic tips in wild-type mice (C) is missing in mice homozygous for the RGS7 gene disruption (B) and is greatly diminished in the clusters of cone ON bipolar cell dendritic tips. Staining for RGS11 (D, E) and staining for Gβ5 (F, G) are not noticeably different in wild-type and RGS7 mutant mice. Scale bars: B–G, 10 μm.
Figure 6A shows that by immunoblotting no RGS11 could be detected in mice homozygous for the targeted disruption of the RGS11 gene. There was no detectable RGS11 staining in RGS11 knock-out mice compared with the wild-type control (Fig. 6C,F). RGS7 gene disruption did not appear to affect the expression or distribution of RGS11 (Fig. 5D,E) and Gβ5 (Fig. 5F,G). Likewise, by immunostaining, there was no noticeable effect of the RGS11 knock-out on the expression and distribution of RGS7 (Fig. 6B,E) or Gβ5 (Fig. 6D,G). In addition to verifying altered R7 RGS protein levels in the mutant retinas, these results serve to confirm the specificity of the immunofluorescence signals obtained with the antibodies and conditions used in our studies of wild-type mice.
Figure 6.
Absence of protein in RGS11 mutant mice. A, Top panel, Western blot analysis of retinal lysates (1/15 of a retina) from two RGS11 mutant mice and two wild-type mice using anti-mouse RGS11 serum R4970. Whereas the RGS11 antiserum detected a single band (∼50 kDa) with the expected size of RGS11, no band at that position was detected in the lanes containing retinal lysates from RGS11 mutant mice. Bottom panel, Western blot analysis of the same blot with GAPDH antibody to show that comparable amounts of proteins were loaded. B–G, Loss of immunostaining for RGS11, but not other antigens, in RGS11 mutant mice. The bright staining for RGS11 (red) postsynaptic to ribeye staining (green) in the outer plexiform layer (C) is eliminated in mice homozygous for the targeted disruption of the RGS11 gene (F). In contrast, staining for RGS7 (red, B, E) and staining for Gβ5 (D, G) were unaffected by disruption of the RGS11 gene. Scale bars: B–G, 10 μm.
Delayed scotopic b-wave ERG responses in mutant mice
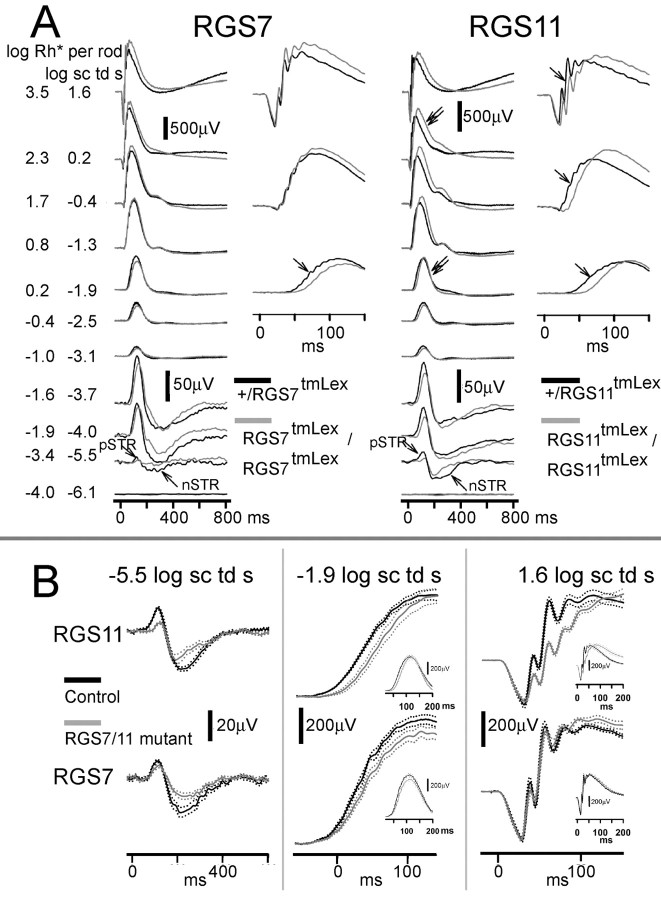
To examine the contribution of RGS7 and RGS11 to the light-evoked retinal responses, we recorded ERGs from RGS7 and RGS11 mutant mice and compared the recordings to their heterozygous or wild-type litter mate controls. Figure 7 shows representative waveforms for RGS7 mutant animals compared with their heterozygous controls for a range of stimulus intensities for the fully dark-adapted condition. Very low stimulus intensities (for example, −5.5 and −4 log sc td s) produced the negative and positive scotopic threshold responses (n-STR and p-STR) which are known to originate from light-evoked responses of amacrine or ganglion cells distal to bipolar cells in rodents (Saszik et al., 2002; Bui and Fortune, 2004; Mojumder et al., 2008). RGS7 mutation caused an attenuation of the n-STR (−5.5 log sc td s). For higher scotopic intensities (for example, −1.9 log sc td s) that produced an ERG response driven primarily by light-evoked activity of the rod-rod bipolar cell circuit, there was a delay in the leading edge of the b-wave response and a slight decrease in amplitude in this RGS7 mutant. There was no change in the trailing edge of the b-wave response. For higher intensities (e.g., 1.6 log sc td s) that produced a mixed rod plus cone driven ERG response, there was no change observed in the photoreceptor-derived leading edge of the a-wave response or the rod plus ON cone bipolar cell driven b-wave. Although, this particular animal showed a delayed trailing edge of the b-wave response, the averaged data (see below) showed no significant difference from controls in the trailing edge.
Figure 7.
Effects of RGS7 and RGS11 gene disruptions on ERG flash responses. A, ERG responses for representative subjects to brief flashes of increasing stimulus energies, from bottom to top, for the fully dark-adapted condition. Gray traces (left), RGS7 mutants; gray traces (right), RGS11mutants; black traces, heterozygous littermate controls. pSTR, Positive scotopic threshold response; nSTR, negative scotopic threshold response. Unlabeled single arrows indicate a delay in the leading edge of the b-wave. Double arrows indicate a delay in the trailing edge of the b-wave that was found to be significant for the RGS11 mutants on average at higher energies. B, Average ERG responses on expanded time scales. Solid traces show average responses for mutant mice (gray) and wild-type (black) littermates, with dotted traces showing ±1 SEM. Top panel (left), ERG responses for the RGS11 mutants for very low intensities (−5.5 log sc td s) show attenuation for both the positive and negative scotopic threshold responses (p-STR and n-STR). Middle, Rod bipolar cell driven b-wave for scotopic intensities (−1.9 log sc td s) was delayed. Middle inset, The trailing edge of the b-wave response was unchanged. Right, The mixed rod plus cone driven b-wave for higher stimulus intensities (1.6 log sc td s) showed a delayed leading edge, but there was no significant change in the leading edge of the a-wave response. Right inset, Trailing edge of the b-wave was delayed in RGS11 mutant animals. Bottom panel (left), ERG responses for the RGS7 mutants for very low intensities (−5.5 log sc td s) show that the n-STR was attenuated. Middle, The rod bipolar cell driven b-wave for scotopic intensities (−1.9 log sc td s) was delayed and slightly decreased in amplitude. Middle inset, The trailing edge of the b-wave response was unchanged. Right, There was no significant change in the b-wave for the mixed rod plus cone ERGs for higher stimulus intensities (1.9 log sc td s) or the leading edge of the a-wave. Right inset, Trailing edge of the b-wave was not significantly delayed in RGS7 mutant animals (n = 4 for RGS7 mutants and their littermate controls; n = 9 for RGS11 and n = 6 for RGS11 littermate controls).
The RGS11 knock-out showed ERG changes that were distinct from those in the RGS7 mutants. For low stimulus intensities (−5.5 log sc td s), both the n- and p-STRs were attenuated. Higher scotopic intensities (for example, −1.9 log sc td s) showed a delay in the leading edge of the rod-bipolar cell-derived leading edge of the b-wave response, but no change in the trailing edge of the b-wave response. Higher intensities (e.g., 1.6 log sc td s) showed a delay in both the leading and trailing edges of the b-wave response. However, there was no change in the leading edge of the photoreceptor-derived a-wave, indicating that the delays observed in the b-wave were most likely caused by changes in signaling downstream of the photoreceptor phototransduction cascades.
The averaged ERG waveforms (average ± SEM) for the RGS7 mutants compared with wild-type or heterozygous controls (Fig. 7A, left) show that the reduction in the n-STR (−5.5 log sc td s) and delay in the scotopic b-wave (−1.9 log sc td s) were significant. However, ERG changes for the mixed rod-cone ERGs were not significant, indicating that the RGS7 mutation primarily affected the light-evoked responses from the rod bipolar cells and the third-order responses in the scotopic circuit (i.e., downstream to rod−rod bipolar cells). ERG changes for the RGS11 knock-out (Fig. 7A, right), however, produced an attenuation of both the n- and p-STRs and a delay in the leading edge of the scotopic b-wave, indicating that RGS11 mutation affected the light-evoked responses from the rod bipolar cells and the third-order responses in the scotopic circuit (i.e., downstream to rod− rod bipolar cells). There was delay in the leading and trailing edges of mixed rod plus cone-derived b-waves in the RGS11 mutants; we verified (see below) that the fully dark adapted cone-derived signals contributed to this delay. The leading edges of the photoreceptor-derived a-waves were unchanged in RGS11 mutants, similar to RGS7 mutants, indicating the effects of RGS11 mutation on the ERG most likely occur downstream from the photoreceptors.
To investigate whether the delayed mixed rod plus cone-derived b-wave seen at higher stimulus intensities could have contributions from the fully dark-adapted cone circuit, we used a cone-isolation paradigm. In this paradigm, the rod response was saturated by a 250 ms intense flash (3.0 log sc td). Preliminary experiments using high intensity flashes (1.9 log sc td s) showed that there was no measurable ERG response at this transient background, indicating that this transient flash suppressed both rod and cone responses. Cone responses are known to recover faster than rod responses from the effects of a saturating flash of light (Baylor, 1987; Lyubarsky et al., 1999). Studies in rodents have found that the ERG responses probed between 1200 and 5000 ms after a rod saturating flash in rodents (Pennesi et al., 2003; Mojumder et al., 2008) are predominantly cone-derived. We used a more conservative time of 750 ms after the flash response to probe for a cone-only response in the fully dark-adapted condition. We used this earlier interstimulus interval because our preliminary observations indicated that at these early times the cone-derived responses lacked significant oscillatory potentials on the leading edge of the b-wave, which could confound interpretation of the delay in the leading edge of the b-wave. As seen in Figure 8A, the probe flash response produced a b-wave response lacking prominent a-waves as expected from a cone-driven response.
Figure 8.
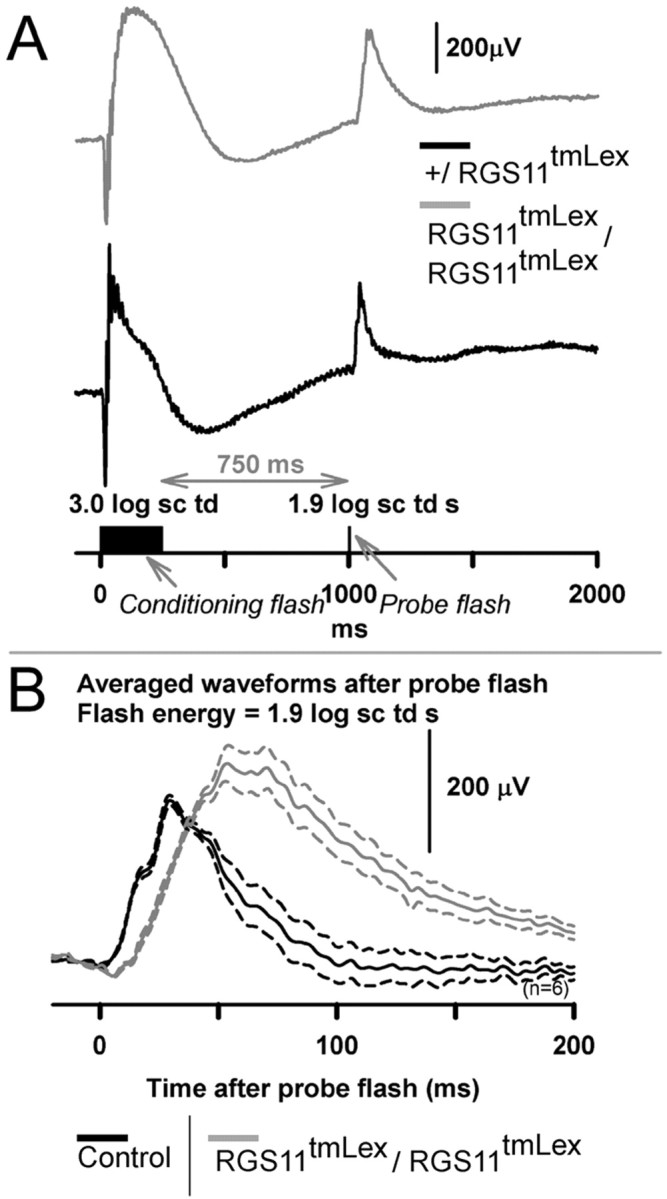
Effects of RGS11 mutation on cone isolated, dark-adapted ERG. A, Schematic showing the protocol for deriving the cone-driven dark-adapted ERG. A high energy conditioning flash (3.0 log sc td, 250 ms duration) for the fully dark adapted condition was used to suppress rods fully and cones partially. Test flashes (1.9 log sc td s) presented at 750 ms from the termination of the conditioning flash probed for cone-only responses. Gray trace, RGS11 mutant; black trace, heterozygous littermate control. The responses for the conditioning flash were subtracted from paired flash response to yield responses for the test flash only. B, Averaged high-energy test-flash (1.9 log sc td s) responses (n = 6, each for RGS11 mutant: gray traces; littermate control: black) show a delayed leading and trailing edge of the b-wave for the RGS11 mutants. The RGS11 mutant b-wave amplitudes were greater than controls.
Figure 8B shows that the leading and trailing edges for the average b-wave response for the cone-isolated b-wave for the RGS11 mutant animal were substantially delayed compared with its control. This result indicates that RGS11 in the cone-circuit contributes to the shaping of both the leading and trailing edges of the b-wave for the mixed rod plus cone-derived fully dark-adapted ERG.
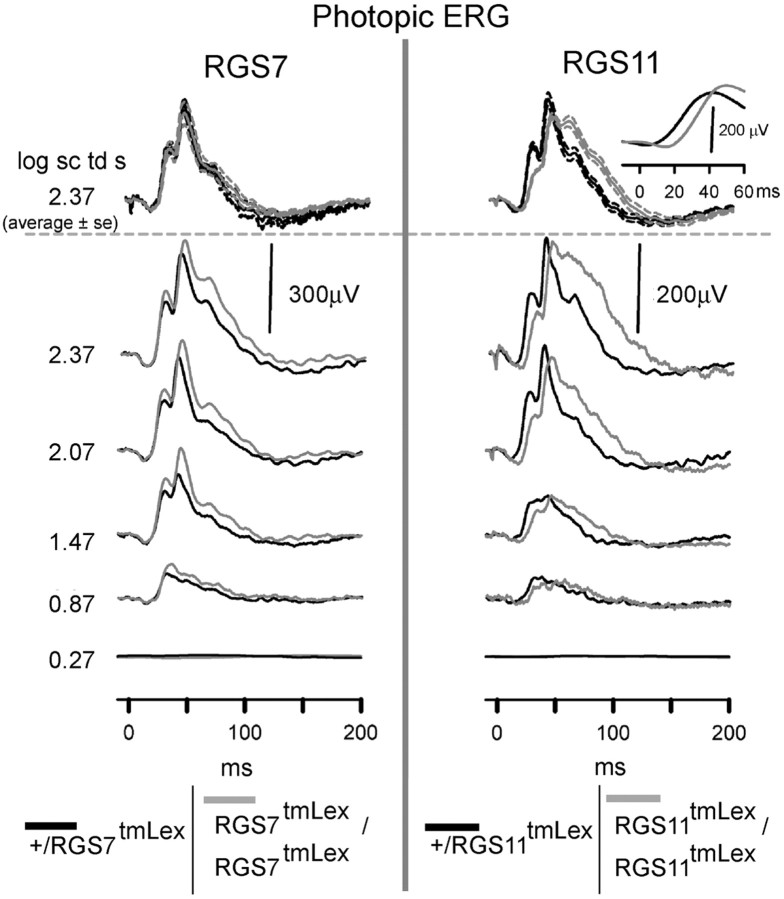
RGS11 mutation but not RGS7 mutation produces a delay in the photopic b-wave
Figure 9, bottom panel, shows representative ERG waveforms for RGS7 (left) and RGS11 (right) mutant animals compared with their heterozygous controls recorded for a background of 1.8 log sc td, which was enough to suppress all rod activity (Krishna et al., 2002; Mojumder et al., 2008). RGS11 mutation produced a delay in both the leading and trailing edges of the b-wave response for all stimulus intensities, very similar to the fully dark-adapted cone-isolated response. The averaged (±SEM) ERG waveforms for a high intensity flash (2.3 log sc td s) show that the delays were significant for the RGS11 mutant animals. This delay can be more prominently observed (right inset) after filtering the waveforms of oscillatory potentials (Butterworth high-pass filter 55 Hz). The RGS7 animals did not show significant differences from the controls under photopic conditions.
Figure 9.
RGS11 but not RGS7 mutation delays the b-wave of the photopic ERG. ERG responses for representative subjects to a range of increasing stimulus energies, from bottom to top, recorded for a rod suppressing background of 1.8 log sc td. The top most traces represent averaged ERG response for a high-energy flash of 2.3 log sc td s (n = 4 for RGS7 mutants and their littermate controls; n = 9 for RGS11and n = 6 for RGS11 littermate controls). Gray traces (left), RGS7 mutants; gray traces (right), RGS11mutants; black traces, heterozygous littermate control; dotted traces, ±SEM. Inset top right corner shows the averaged b-wave traces after filtering OPs.
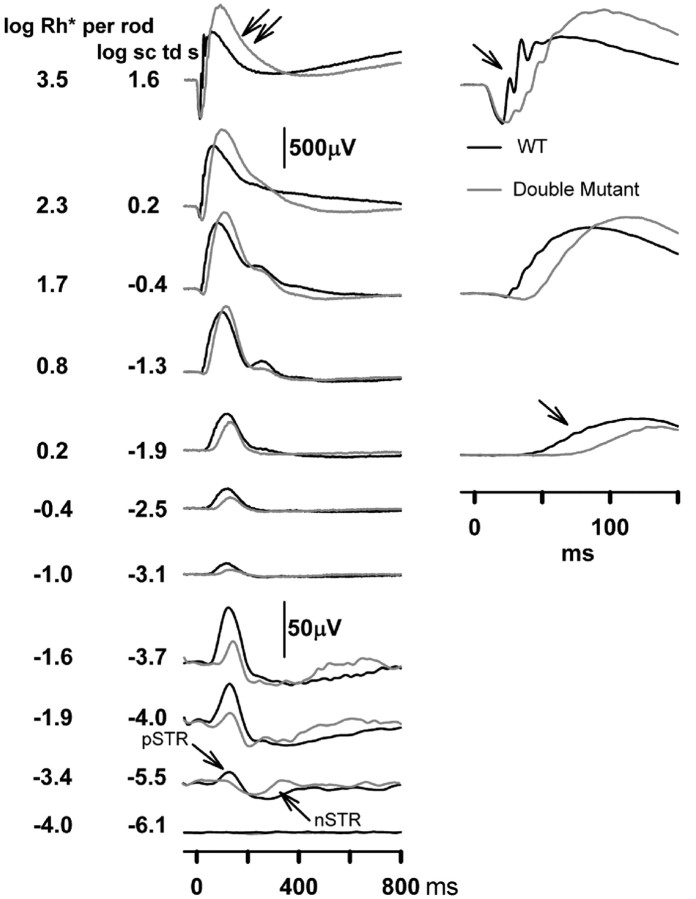
Simultaneous disruption of RGS7 and RGS11 genes produces an additive effect on ERG responses
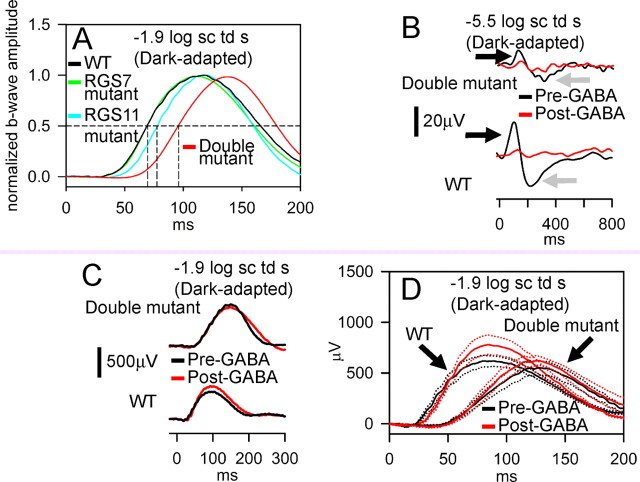
Mice were bred to homozygosity for the disrupted alleles at both the RGS7 and RGS11 loci, and their ERG responses recorded. The results (Figs. 10, 11) show clearly that both proteins are important for the kinetics of the bipolar cell responses. The most sensitive responses, the n-STR and p-STR, were both attenuated even more severely in the double-mutants than in either of the single mutants (Fig. 10). Over a wide range of intensities, the b-wave responses were considerably more delayed. At a stimulus energy of −1.9 log sc td s (Fig. 11A), normalized averaged responses show a striking phenotype: a substantially greater delay in the rising edge of the b-wave responses in the double mutant than in either the RGS11 mutant or RGS7 mutant mice. Thus, both RGS7 and RGS11 contribute to shaping the b-wave responses, and they appear to act in parallel branches of the signal transduction cascade, either in the same cell or, as suggested by the immunostaining results, perhaps in different sets of bipolar cells.
Figure 10.
Effects of RGS7+RGS11 gene disruptions on ERG flash responses. ERG responses for representative subjects to brief flashes of increasing stimulus energies, from bottom to top, for the fully dark-adapted condition. Gray traces, RGS7 and RGS11 double mutants; black traces, WT littermate control. pSTR, Positive scotopic threshold response; nSTR, negative scotopic threshold response. Unlabeled single arrows indicate a delay in the leading edge of the b-wave. Double arrows indicate a delay in the trailing edge of the b-wave.
Figure 11.
Additive effects of RGS7 and RGS11 gene disruptions on time course of the dark-adapted ERG responses arising from bipolar cells. A, Averaged ERG responses for WT (includes animals either homozygous WT or heterozygous for either RGS7 or RGS11, black, n = 14), RGS7 mutant (green, n = 4), RGS11 mutant (cyan, n = 9) and RGS7+RGS11 double mutant (red, n = 4) normalized to the maximal ERG amplitude for −1.9 log sc td s flash. Horizontal dashed line, Half-max amplitude; vertical dashed lines, time for half-max b-wave for the different traces (for WT and RGS7 mutant, 69.5 ms; RGS11 mutant, 77.5 ms; RGS7+RGS11 mutant, 96 ms). B, Dark-adapted ERG response recorded from representative subjects for a low energy flash of −5.5 log sc td s showing abolition of p- and n-STRs (black and gray arrows respectively) following intravitreal injection of GABA (red traces) for both RGS7+RGS11 double mutants (double mutant, black trace, top) and WT (wild type, black trace, bottom). C, Dark-adapted ERG response recorded from representative subjects for a flash energy of −1.9 log sc td s. Intravitreal injection of GABA (red traces) did not significantly alter the time course of the leading edge on the b-wave for either RGS7+RGS11 double mutants (black trace, top) or WT (black trace, bottom). D, Averaged dark-adapted ERG responses recorded for a flash energy of −1.9 log sc td s. WT, Wild type; double mutant, RGS7+RGS11 double mutants; red traces, after intravitreal injection of GABA; black traces, before intravitreal injection of GABA. Dotted traces indicate ±1 SEM. Number of animals: WT = 3; double mutants = 6.
Although rod and cone ON-bipolar cell responses dominate the b-wave (Robson and Frishman, 1995; Naarendorp et al., 2001; Saszik et al., 2002; Mojumder et al., 2008), there have been suggestions that voltage changes in inner retinal neurons may contribute to it in rabbit (Dong and Hare, 2000, 2002, 2003). To test the possibility that the changes observed in b-waves of the mutant mice are caused by changes in other neurons of the inner retina, we used intravitreal injection of GABA to block signals from neurons of the proximal inner retina. Intravitreal injection of GABA is known isolate the light-evoked potentials from the rod-bipolar cells and to confer linearity on the amplitude-intensity function of the murine dark-adapted b-wave for low-intensities, by suppressing proximal inner-retinal potentials such as the negative and positive scotopic threshold responses (Saszik et al., 2002). As shown in Figure 11B–D, GABA injection had little effect on the phenotypical differences between the RGS7/11 double mutants and wild-type mice. Although the scotopic threshold responses (n-STR and p-STR) were suppressed in animals of both genotypes (Fig. 11B), confirming the effective suppression of inner retinal signaling by GABA, the delay in the rising phase of the b-wave resulting from disruption of the R7 RGS protein genes was still prominent (Fig. 11C,D). Thus, it can be concluded with confidence that delays in ON-bipolar cell responses are largely responsible for the altered b-waves observed in the mutant mice, and that these alterations do not depend on signaling by the proximal inner retina.
Discussion
There are three major conclusions that emerge from the results presented in this study. The first is that RGS7 and RGS11 have distinct, albeit closely related and perhaps partially redundant, functions in the retina. This conclusion emerges from the observation that they are found in different cell types, including, in some cases, different dendritic tips of rod bipolar cells and cone ON bipolar cells. This conclusion is further supported by the observation that both scotopic threshold responses and b-wave responses are clearly different in mice with genetic disruptions of both genes compared with mice with a disruption of either gene by itself. The increased effects observed in double mutants is consistent with the hypothesis suggested by the differential immunostaining that there are some rod BPC dendrites whose signaling is regulated by RGS7 and not RGS11, and others in which both contribute to regulating response kinetics. The ERG b-wave is an average over all of these, so if in retinas lacking only one RGS protein some cells are delayed and others not, but all are delayed in the double mutants, an increased delay in the average would be expected. Additionally, in those dendrites normally expressing both, removing two RGS proteins instead of one may just result in a lower total concentration of RGS proteins, and thus slower kinetics. Additional support for different but overlapping functions of RGS7 and RGS11 comes from the observation that changes in b-wave responses are largely confined to rod-driven responses in RGS7 mutants, whereas they are most striking in responses involving cone inputs in RGS11 mutants.
The second major conclusion is that, as hypothesized at the outset of this work, RGS7 and RGS11 regulate the kinetics of the b-wave responses mediated by inactivation of Gαo. The simplest explanation for this effect is that in wild-type mice RGS7 and RGS11 directly bind to Gαo activated by the complex of glutamate with mGluR6 and accelerate its GTP hydrolysis. Although both proteins have been shown to have this activity in vitro (Hooks and Harden, 2004), we have no direct evidence that this is their precise function in retinal ON bipolar cells. As an example of an alternative hypothesis, it could be that one or both of these RGS proteins regulates the response to a completely different G-protein-coupled receptor (GPCR) in bipolar cells, whose activation in turn regulates bipolar cell response kinetics by effects on levels of cyclic nucleotides, calcium, or phosphoinositides. The viability of such alternative hypotheses is sustained by the observation that most of the Gαo in bipolar cells is not localized to the dendritic tips, unlike mGluR6, RGS7, RGS11, and Gβ5. We cannot say with certainty that the effects of removing RGS7 and RGS11 are attributable to their presence in bipolar cell dendritic tips, although their striking colocalization with mGluR6 makes this the most economical interpretation.
The persistence of the effects of RGS7/RGS11 disruption after blocking inner-retinal activity proximal to bipolar cells with GABA excludes the possibility that other mechanisms, such as feedback from amacrine cells, involving these two RGS proteins, contribute substantially to the observed changes in the b-wave response (excluding the oscillatory potentials). It is also clear that the effects are exerted in the bipolar cells and not the photoreceptors, as photoreceptor responses, represented by the a-wave, are unaltered in mice with RGS7 and RGS11 defects.
A third major conclusion is that RGS7 and RGS11 are not required for light responses of ON bipolar cells on a subsecond time scale. Although the responses are delayed when these proteins are lacking, the delays are on the order of tens of milliseconds rather than seconds, as observed, for example, in photoreceptor responses of RGS9 knock-out mice (Chen et al., 2000). Given the slow GTPase kinetics for isolated Gαo, it seems very likely that a third protein, and possibly others as well, in ON bipolar cells, is capable of serving as a GAP for Gαo. It would be interesting to explore the possibility that ret-RGS1 (Dhingra et al., 2004) plays this role.
Finally, we note that the experiments described here are best suited for identifying defects in signaling by photoreceptors or ON bipolar cells, because these contribute the a- and b-waves which dominate the ERG records, and thus our major findings concern the roles of RGS7 and RGS11 in ON bipolar cell responses. There may be even more severe defects in signaling by inner retinal neurons that we are unable to detect because of their lack of strong effects on the ERG. The same consideration applies to the roles of these R7 RGS proteins in the brain, where RGS7 may be the most abundant RGS protein. The Lexicon data avail-able on-line (http://www.informatics.jax.org/searches/accession_report.cgi?id=MGI:3528964#Additional_Info) indicates some behavioral abnormalities in the RGS7 mutants, but we have not yet determined either the extent or the mechanisms of such abnormalities. Future studies of RGS7 and RGS11 in the brain will likely implicate them in GPCR signaling pathways there, in addition to their roles in the retina.
Footnotes
This work was supported by National Institutes of Health Grant R01-EY11900 (T.G.W.) and Welch Foundation Grant Q0035. We thank Dr. Saumil Patel (University of Texas Medical School at Houston) for his valuable advice on making the custom-made LED-based stimulator used for electroretinography.
References
- Baylor DA. Photoreceptor signals and vision. Proctor Lecture. Invest Ophthalmol Vis Sci. 1987;28:34–49. [PubMed] [Google Scholar]
- Bui BV, Fortune B. Ganglion cell contributions to the rat full-field electroretinogram. J Physiol. 2004;555:153–173. doi: 10.1113/jphysiol.2003.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Hernandez S, Earls LR, Medkova M, Sundgren-Andersson AK, Surmeier DJ, Hamm HE. RGS9-2 modulates D2 dopamine receptor-mediated Ca2+ channel inhibition in rat striatal cholinergic interneurons. Proc Natl Acad Sci U S A. 2004;101:16339–16344. doi: 10.1073/pnas.0407416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CW, He W, Wensel TG. RGS proteins: lessons from the RGS9 subfamily. Prog Nucleic Acid Res Mol Biol. 2001;65:341–359. doi: 10.1016/s0079-6603(00)65009-2. [DOI] [PubMed] [Google Scholar]
- Dawson WW, Trick GL, Litzkow CA. Improved electrode for electroretinography. Invest Ophthalmol Vis Sci. 1979;18:988–991. [PubMed] [Google Scholar]
- Dhingra A, Lyubarsky A, Jiang M, Pugh EN, Jr, Birnbaumer L, Sterling P, Vardi N. The light response of ON bipolar neurons requires Gαo. J Neurosci. 2000;20:9053–9058. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Jiang M, Wang TL, Lyubarsky A, Savchenko A, Bar-Yehuda T, Sterling P, Birnbaumer L, Vardi N. Light response of retinal ON bipolar cells requires a specific splice variant of Gαo. J Neurosci. 2002;22:4878–4884. doi: 10.1523/JNEUROSCI.22-12-04878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Faurobert E, Dascal N, Sterling P, Vardi N. A retinal-specific regulator of G-protein signaling interacts with Gαo and accelerates an expressed metabotropic glutamate receptor 6 cascade. J Neurosci. 2004;24:5684–5693. doi: 10.1523/JNEUROSCI.0492-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CJ, Hare WA. Contribution to the kinetics and amplitude of the electroretinogram b-wave by third-order retinal neurons in the rabbit retina. Vision Res. 2000;40:579–589. doi: 10.1016/s0042-6989(99)00203-5. [DOI] [PubMed] [Google Scholar]
- Dong CJ, Hare WA. GABAc feedback pathway modulates the amplitude and kinetics of ERG b-wave in a mammalian retina in vivo. Vision Res. 2002;42:1081–1087. doi: 10.1016/s0042-6989(02)00032-9. [DOI] [PubMed] [Google Scholar]
- Dong CJ, Hare WA. Temporal modulation of scotopic visual signals by A17 amacrine cells in mammalian retina in vivo. J Neurophysiol. 2003;89:2159–2166. doi: 10.1152/jn.01008.2002. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks SB, Harden TK. Purification and in vitro functional analysis of R7 subfamily RGS proteins in complex with Gbeta5. Methods Enzymol. 2004;390:163–177. doi: 10.1016/S0076-6879(04)90011-0. [DOI] [PubMed] [Google Scholar]
- Hunyady B, Krempels K, Harta G, Mezey E. Immunohistochemical signal amplification by catalyzed reporter deposition and its application in double immunostaining. J Histochem Cytochem. 1996;44:1353–1362. doi: 10.1177/44.12.8985127. [DOI] [PubMed] [Google Scholar]
- Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, Axelrod JD, Cheyette BN, Simon MI, Lester HA, Schwarz J. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna VR, Alexander KR, Peachey NS. Temporal properties of the mouse cone electroretinogram. J Neurophysiol. 2002;87:42–48. doi: 10.1152/jn.00489.2001. [DOI] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Lyubarsky AL, Falsini B, Pennesi ME, Valentini P, Pugh EN., Jr UV- and midwave-sensitive cone-driven retinal responses of the mouse: a possible phenotype for coexpression of cone photopigments. J Neurosci. 1999;19:442–455. doi: 10.1523/JNEUROSCI.19-01-00442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky AL, Naarendorp F, Zhang X, Wensel T, Simon MI, Pugh EN., Jr RGS9-1 is required for normal inactivation of mouse cone phototransduction. Mol Vis. 2001;7:71–78. [PubMed] [Google Scholar]
- Lyubarsky AL, Daniele LL, Pugh EN., Jr From candelas to photoisomerizations in the mouse eye by rhodopsin bleaching in situ and the light-rearing dependence of the major components of the mouse ERG. Vision Res. 2004;44:3235–3251. doi: 10.1016/j.visres.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y, Shigemoto R, Takadac M, Nakamurag K, Nakaog K, Katsukig M, Nakanishia S. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–765. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- Mojumder DK, Frishman LJ, Otteson DC, Sherry DM. Voltage-gated sodium channel alpha-subunits Na (v) 1.1, Na (v) 1.2, and Na (v) 1.6 in the distal mammalian retina. Mol Vis. 2007;13:2163–2182. [PubMed] [Google Scholar]
- Mojumder DK, Sherry DM, Frishman LJ. Contribution of voltage-gated sodium channels to the b-wave of the mammalian flash electroretinogram. J Physiol. 2008;586:2551–2580. doi: 10.1113/jphysiol.2008.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Ren G, Akileswaran L. Localization of nyctalopin in the mammalian retina. Eur J Neurosci. 2006;23:1163–1171. doi: 10.1111/j.1460-9568.2006.04647.x. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Wensel TG, Brown RL, Perez-Leon JA, Bearnot B, Duvoisin RM. Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells. Eur J Neurosci. 2007;26:2899–2905. doi: 10.1111/j.1460-9568.2007.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarendorp F, Sato Y, Cajdric A, Hubbard NP. Absolute and relative sensitivity of the scotopic system of rat: electroretinography and behavior. Vis Neurosci. 2001;18:641–656. doi: 10.1017/s0952523801184142. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Iwakabe H, Akazawa C, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for L-2-amino-4-phosphonobutyrate. J Biol Chem. 1993;268:11868–11873. [PubMed] [Google Scholar]
- Nawy S. The metabotropic receptor mGluR6 may signal through Go, but not phosphodiesterase, in retinal bipolar cells. J Neurosci. 1999;19:2938–2944. doi: 10.1523/JNEUROSCI.19-08-02938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy S, Copenhagen DR. Multiple classes of glutamate receptor on depolarizing bipolar cells in retina. Nature. 1987;325:56–58. doi: 10.1038/325056a0. [DOI] [PubMed] [Google Scholar]
- Nawy S, Jahr CE. cGMP-gated conductance in retinal bipolar cells is suppressed by the photoreceptor transmitter. Neuron. 1991;7:677–683. doi: 10.1016/0896-6273(91)90380-i. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- Pennesi ME, Howes KA, Baehr W, Wu SM. Guanylate cyclase-activating protein (GCAP) 1 rescues cone recovery kinetics in GCAP1/GCAP2 knockout mice. Proc Natl Acad Sci U S A. 2003;100:6783–6788. doi: 10.1073/pnas.1130102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Gold SJ, Potenza MN, Cowan CW, Ni YG, He W, Wensel TG, Nestler EJ. Cloning and characterization of RGS9-2: a striatal-enriched alternatively spliced product of the RGS9 gene. J Neurosci. 1999;19:2016–2026. doi: 10.1523/JNEUROSCI.19-06-02016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, Kovoor A, Chen CK, DiLeone RJ, Schwarz SC, Selley DE, Sim-Selley LJ, Barrot M, Luedtke RR, Self D, Neve RL, Lester HA, Simon MI, Nestler EJ. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- Rao A, Dallman R, Henderson S, Chen CK. Gbeta5 is required for normal light responses and morphology of retinal ON-bipolar cells. J Neurosci. 2007;27:14199–14204. doi: 10.1523/JNEUROSCI.4934-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson JG, Frishman LJ. Response linearity and kinetics of the cat retina: the bipolar cell component of the dark-adapted electroretinogram. Vis Neurosci. 1995;12:837–850. doi: 10.1017/s0952523800009408. [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Sagdullaev BT, DeMarco PJ, McCall MA. Improved contact lens electrode for corneal ERG recordings in mice. Doc Ophthalmol. 2004;108:181–184. doi: 10.1007/s10633-004-5734-1. [DOI] [PubMed] [Google Scholar]
- Saszik SM, Robson JG, Frishman LJ. The scotopic threshold response of the dark-adapted electroretinogram of the mouse. J Physiol. 2002;543:899–916. doi: 10.1113/jphysiol.2002.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry DM, Mitchell R, Standifer KM, du Plessis B. Distribution of plasma membrane-associated syntaxins 1 through 4 indicates distinct trafficking functions in the synaptic layers of the mouse retina. BMC Neurosci. 2006;7:54. doi: 10.1186/1471-2202-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiells RA, Falk G. Glutamate receptors of rod bipolar cells are linked to a cyclic GMP cascade via a G-protein. Proc Biol Sci. 1990;242:91–94. doi: 10.1098/rspb.1990.0109. [DOI] [PubMed] [Google Scholar]
- Shiells RA, Falk G, Naghshineh S. Action of glutamate and aspartate analogues on rod horizontal and bipolar cells. Nature. 1981;294:592–594. doi: 10.1038/294592a0. [DOI] [PubMed] [Google Scholar]
- Shindler KS, Roth KA. Double immunofluorescent staining using two unconjugated primary antisera raised in the same species. J Histochem Cytochem. 1996;44:1331–1335. doi: 10.1177/44.11.8918908. [DOI] [PubMed] [Google Scholar]
- Slaughter MM, Miller RF. 2-amino-4-phosphonobutyric acid: a new pharmacological tool for retina research. Science. 1981;211:182–185. doi: 10.1126/science.6255566. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci U S A. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA, Danielson PE, Sutcliffe JG. RGS9: a regulator of G-protein signalling with specific expression in rat and mouse striatum. J Neurosci Res. 1998;52:118–124. doi: 10.1002/(SICI)1097-4547(19980401)52:1<118::AID-JNR11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Vardi N, Morigiwa K. ON cone bipolar cells in rat express the metabotropic receptor mGluR6. Vis Neurosci. 1997;14:789–794. doi: 10.1017/s0952523800012736. [DOI] [PubMed] [Google Scholar]
- Vardi N, Duvoisin R, Wu G, Sterling P. Localization of mGluR6 to dendrites of ON bipolar cells in primate retina. J Comp Neurol. 2000;423:402–412. doi: 10.1002/1096-9861(20000731)423:3<402::aid-cne4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Wang G, Achim CL, Hamilton RL, Wiley CA, Soontornniyomkij V. Tyramide signal amplification method in multiple-label immunofluorescence confocal microscopy. Methods. 1999;18:459–464. doi: 10.1006/meth.1999.0813. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Katz A, Simon MI. A fifth member of the mammalian G-protein beta-subunit family. Expression in brain and activation of the beta 2 isotype of phospholipase C. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- Yamashita M, Wässle H. Responses of rod bipolar cells isolated from the rat retina to the glutamate agonist 2-amino-4-phosphonobutyric acid (APB) J Neurosci. 1991;11:2372–2382. doi: 10.1523/JNEUROSCI.11-08-02372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Lai Z, Simonds WF. Differential expression of the G protein beta(5) gene: analysis of mouse brain, peripheral tissues, and cultured cell lines. J Neurochem. 2000;75:393–403. doi: 10.1046/j.1471-4159.2000.0750393.x. [DOI] [PubMed] [Google Scholar]
- Zhang K, Howes KA, He W, Bronson JD, Pettenati MJ, Chen C, Palczewski K, Wensel TG, Baehr W. Structure, alternative splicing, and expression of the human RGS9 gene. Gene. 1999;240:23–34. doi: 10.1016/s0378-1119(99)00393-5. [DOI] [PubMed] [Google Scholar]