Summary
Blood coagulation is the result of a cascade of zymogen activation events, however, its initiation is allosteric. Factor VIIa circulates in a zymogen-like state and is allosterically activated by binding to tissue factor. Thrombin, the final protease generated in the blood coagulation cascade, has also been shown to exist in a low activity state in the absence of cofactors, and the structural features of this ‘slow’ form has been studied for many years. In this manuscript I will review the general features that render zymogens inactive and how proteolytic cleavage results in activation, but I will also show how this distinction is blurred by zymogens that have activity (protease-like zymogens) and proteases with low activity (zymogen-like proteases). This will then be applied in the analysis of slow thrombin to reveal how allosteric activation of thrombin simply reflects the conversion from a zymogen-like enzyme to an active serine protease.
Keywords: cofactor, regulation, serine protease, structure, thrombin, zymogen
How Serine Proteases Work
The mechanism by which serine proteases hydrolyse peptide bonds had been worked out in great detail by the mid-seventies [1]. Any discussion on what makes a zymogen inactive or what renders a protease zymogen-like requires a brief review of the steps and residues involved in proteolysis. All of the proteases involved in the blood coagulation cascade belong to the chymotrypsin family (S1 in the Merops Database, merops.sanger.ac.uk/index.htm) and share a two β-barrel domain structure, with the active site situated between the two domains. The classic representation of the conserved serine protease structure is shown in Fig. 1A, with the active site facing and substrates binding from left to right from N-to-C termini. For simplicity, a template numbering scheme is typically used for the catalytic domain of all chymotrypsin family members. The catalytic architecture is composed of a primary specificity pocket (S1 pocket, where the substrate P1 residue binds [2]), the catalytic triad of His57, Asp102 and Ser195, and the oxyanion hole formed by the main chain amides of Gly193 and Ser195 (Fig. 1B). Catalysis proceeds via several well defined steps: 1) docking of the P1 residue into the S1 pocket to align the scissile P1-P1′ peptide bond with the catalytic residues; 2) the Oγ of Ser195 initiates a nucleophilic attack on the carbonyl carbon of the P1 residue, potentiated by the transfer of the hydroxyl proton to His57 (delocalized by Asp102); 3) a tetrahedral transition state is formed and the negative charge on the P1 oxygen atom is stabilized by hydrogen bonding to the main chain amides of Gly193 and Ser195 (the oxyanion hole); 4) transfer of the proton from His57 to the N-terminus of the P1′ residue occurs with the collapse of the tretrahedral transition state to the trigonal acyl-enzyme intermediate, resulting in the breaking of the P1-P1′ peptide linkage; 5) the P’ region diffuses out of the active site and a single molecule of water diffuses in to hydrogen bond with His57; 6) a proton is withdrawn from the water as the oxygen atom attacks the carbonyl carbon of the acyl intermediate to form the second tetrahedral transition state, again stabilized by the oxyanion hole; 7) the tetrahedral transition state collapses with the transfer of the proton from His57 to Ser195; 8) the peptide containing the P1 residue diffuses out of the S1 pocket to complete the cycle. Thus, for a serine protease to be active it must have a preformed substrate binding pocket (S1 site of principal importance), and the precise positioning of residues that make up the catalytic triad and the oxyanion hole.
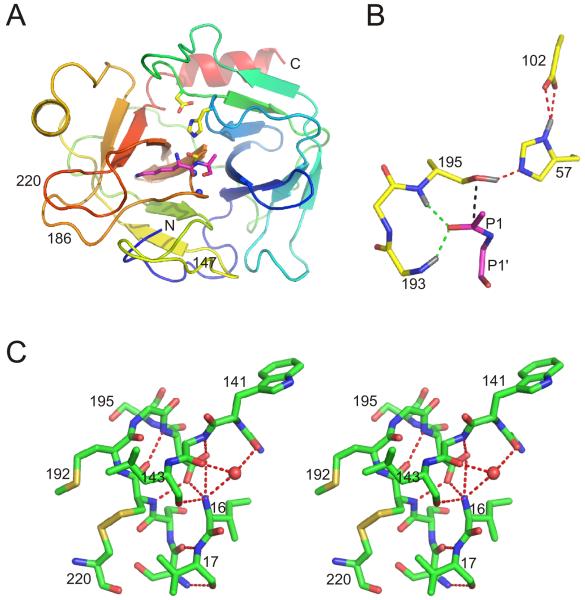
Fig. 1.
Serine protease structure. (A) Ribbon diagram of trypsin coloured from N (blue) to C (red) termini, oriented in the classic fashion, with the C-terminal β-barrel on the left and the N-terminal β-barrel on the right. The catalytic triad is shown as yellow sticks, and the N atoms of the oxyanion hole are shown as blue balls. The P1 Lys and P1′ Ala residues of BPTI are shown as magenta sticks to illustrate the principal substrate binding site. The 147, 186 and 220-loops are indicated. (B) A close up of the catalytic triad and oxyanion hole illustrates how the precise orientation of these residues with the scissile bond is required for catalysis (coloured as in (A)). The amide hydrogens of the oxyanion hole (and on His57 and Ser195) are shown in grey, and important hydrogen bonds are indicated by green and red dashed lines. The main chain residues of the substrate are shown, and the trajectory of the nucleophilic attack is indicated in black. (C) A stereo representation of the interactions between the N-terminus and adjacent residues for chymotrypsin illustrates the direct link with the activation domain.
Why Zymogens are Inactive
There are over twenty structures of zymogens of serine proteases deposited in the Protein Databank. The first two, chymotrypsinogen [3] and trypsinogen [4,5], were published in the 1970’s and showed interesting differences of potential functional importance. All residues of chymotrypsinogen were well defined in electron density, whereas in trypsinogen loops including residues 142-152, 184A-193, and 216-223 could not be modelled due to the presumed flexibility of these regions. Although these loops were defined in the chymotrypsinogen structure, they were in conformations quite different from that of the active enzyme (Fig. 2A). The catalytic triad was correctly formed in both structures (Fig. 2B), indicating that the proton transfer system is functional in zymogens. This is consistent with the ability of some zymogens to react with small substrates and inhibitors [6-8]. Although different in detail, both structures revealed an obscured S1 pocket and an incorrectly formed oxyanion hole (Fig. 2B). These two features are sufficient to account for the catalytic deficiency of zymogens toward peptide and natural substrates and inhibitors. Subsequent crystal structures of zymogen forms of chymotrypsin family serine proteases have helped refine the general features that account for inactivity, but also reveal many interesting variations. No two zymogen structures are identical, even when there are multiple copies of the same zymogen sharing the asymmetric unit, indicating that some of the defined structural features may be crystallization artefacts and suggesting an underlying flexibility of certain loops. It is clear that there is more than one way to confer inactivity upon a zymogen, but enough structural information exists to arrive at three general rules: 1) the S1 pocket is inaccessible or not formed; 2) Gly193 is either flexible or in a conformation that cannot stabilize the tetrahedral oxyanion; 3) three loops (excluding the activation loop itself) are either disordered or in a conformation significantly different from that of the active enzyme, 142-153, 184A-194, 215-225. These loops have been called the ‘activation domain’ [9], but here I will refer to them as ‘activation loops’ or ‘zymogen loops’.
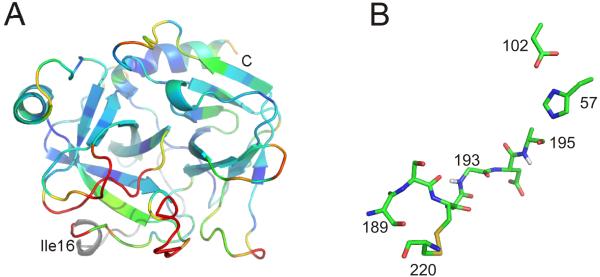
Fig. 2.
Zymogen structure. (A) A ribbon diagram of chymotrypsinogen (with the light chain in grey) is shown coloured according to Cα RMSD with chymotrypsin (blue to red for 0-2Å). The red region corresponds to the zymogen activation domain and is disordered in many other zymogen structures. (B) A close up of the catalytic region of chymotrypsinogen (with hydrogens of Gly193 and Ser195 in grey) shows an intact catalytic triad, but a non-catalytic oxyanion hole.
Prethrombin-2
There are two deposited structures of thrombin in a zymogen state, prethrombin-2 (pre2): human bound to the exosite I ligand hirugen [10]; and, bovine crystallized in two different conditions, but in the absence of any ligand [11]. These structures conform to what was already observed for trypsinogen and chymotrypsinogen, with the zymogen activation loops either fully disordered or ordered with high temperature factors in a conformation distinct from the active protease. As one might expect, it is the hirugen-bound human pre2 that possesses the ordered zymogen loops, and the apo bovine pre2 with disordered loops. In bovine pre2, the disordered regions are 141-146, 186A-188 and 217-224 (set at an occupancy value of 0.01 in the coordinate file), but resides on either side of these loops are clearly in different conformations to that of active thrombin. As in most other zymogen structures, the catalytic triad is correctly formed, but the S1 pocket and the oxyanion hole are not.
Zymogens Activation
Zymogens are activated by cleavage of the bond between residues 15 and 16; for serine proteases involved in hemostasis this bond is Arg-Ile (or Val). The new primary amine at the N-terminus of Ile16 becomes positively charged upon cleavage, and through a mechanism referred to as molecular sexuality [12], it inserts into a hydrophobic pocket formed by the zymogen activation loops (sometimes called the activation pocket), and is stabilized by a salt-bridge with Asp194. The contacts between the hydrophobic N-terminus and the zymogen activation loops are detailed in Fig. 1C, and are highly conserved among the chymotrypsin family members. The effect of the insertion of the new N-terminus on the overall structure or flexibility of the protein is limited to the zymogen activation regions described above (see Fig. 2A). The activating conformational changes are therefore quite local to Ile16, and one may therefore question whether they are sufficiently remote to constitute allostery. In any case, cleavage at residue 15 generally results in the ordering of the catalytic residues and the formation of the S1 pocket. How this is achieved is quite easy to envision. The hydrophobic side chains at 16 and 17 are driven out of the aqueous environment towards the cavity, while complementary electrostatics orient the side chain of Asp194 internally towards the pocket. As a consequence Cys191 is repositioned, allowing the main chain of Asp189 to make a double hydrogen bond to residue 17. This event effectively anchors the catalytic loop from 194 to 189, to rigidify the oxyanion hole and form the S1 pocket (side chain of 189 forms the base of the pocket). Cys220 is linked to the active site loop through a disulfide bond with Cys191, and its repositioning engenders van der Waals’ contacts for residues 220 and 221A with Val17. The final zymogen loop, 141-152, is stabilized by direct main chain hydrogen bonds to the new N-terminus (142 and 143) (see Fig. 1C).
Protease-like Zymogens and Zymogen-like Proteases
Most of the conformational changes I refer to above involve the stabilization of flexible regions critical for the catalytic function of the protease. This is because the zymogen loops in most structures are disordered or at least sample multiple conformations. It is therefore only a minor simplification to consider flexibility of the zymogen loops to be the general basis for zymogen inactivity. This conceptually decouples activation from cleavage of the Arg15-Ile16 peptide bond, since stabilization of these loops is all that is required. This is nicely illustrated by the activation of zymogens with strong ligands [13], high salt [14], and by staphylocoagulase [15]. In addition, there are zymogens that exhibit appreciable catalytic activity in the absence of other activator molecules (protease-like zymogens), and proteases that exist in a catalytically inert state in spite of being cleaved at the Arg15-Ile16 bond (zymogen-like proteases). The best examples of a protease-like zymogen and a zymogen-like protease are single-chain tissue plasminogen activator [16] and factor VIIa [17,18], respectively. The activity of the zymogen form of tPA has been shown to be in part due to the insertion of the side chain of Lys156 into the activation pocket [16,19], with its ammonium group forming an analogous salt-bridge to Asp194. However, it is still an effective enzyme when Lys156 is mutated, indicating that the zymogen activation loops are inherently more stable than for normal zymogens. Conversely, the activation loops are likely to be inherently more disordered in fVIIa, since its N-terminus is partially exposed unless the cofactor TF is bound. Due to space constraints I am not able to go into more detail about either of these fascinating and important enzymes, but the take-home message is that zymogenicity is related to the flexibility/stability of the activation loops.
‘Slow’ Thrombin is Zymogen-like
For about 30 years now researchers have been studying the effect of Na+ binding on the activity and structure of thrombin (for a thorough review see [20]). Thrombin is not unique among coagulation proteases in its ability to coordinate Na+, nor in expressing greater activity in its presence than in its absence (e.g. factors VIIa, Xa, IXa and APC [21]). However, Na+ is considered a cofactor for thrombin because its Kd at 37°C is reported to be similar to the concentration of Na+ in the blood [22], and so the two forms should be equally populated in vivo. Na+ binding leads to a general improvement in catalytic efficiency (10-times faster cleavage of S2238), including procoagulant substrates such as fibrinogen and PAR-1; it has therefore become known as the procoagulant ‘fast’ form. Conversely, Na+-free thrombin is a poor enzyme, except when bound to thrombomodulin (TM), which effectively alters the specificity from pro- to anticoagulant substrates; it is thus known as the ‘slow’ form [23]. This distinction appears to be supported by animal studies that show infused recombinant slow thrombin is indeed an anticoagulant, and that this effect is mediated by the protein C pathway [24]. Thrombin coordinates Na+ using the main chain oxygens of Arg221A and Lys224 and four water molecules [25] (Fig. 3A). These and adjacent residues of the 220-loop contact the 186-loop, the 147-loop and Val17. In other words, Na+ binding would appear to influence the stability of the entire zymogen activation domain. The ‘slow/fast’ nomenclature is specific to thrombin, and this perhaps explains why efforts to determine the molecular basis behind the low activity apo state have ignored the obvious link to zymogenicity. Isn’t it possible that, similar to fVIIa, full conversion from the zymogen form of thrombin to its active form requires the additional step of cofactor binding (either Na+ or TM)? To answer this question it is necessary to determine the structure of ‘slow’ thrombin.
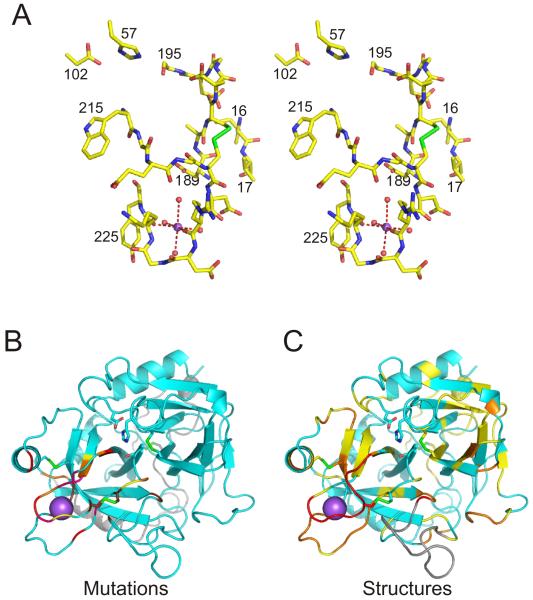
Fig. 3.
Thrombin structure and Na+ binding site. (A) A stereo view of the catalytic region and the Na+ binding site of thrombin (Na+ purple ball and coordinated waters red balls). (B) A ribbon diagram of thrombin, with catalytic residues and disulfide bonds as sticks and Na+ as large purple ball. The light chain of thrombin is coloured gray and the heavy chain is coloured cyan. Residues whose mutation to Ala results in a 0.5-0.75 log unit loss of Na+ affinity are in yellow, 0.75-1 in orange, 1-1.5 in red, and above 1.5 in magenta. Other mutations with large effects on Na+ binding are shown in chocolate. (C) Same representation as (B), but residues are coloured according to the degree to which the Cα position of slow thrombin deviates from the fast form. This is expressed in terms of a signal to noise ratio, as defined previously [20], with yellow from 2-4, orange from 4-10 and red above 10.
Structural Features of Slow Thrombin
Based on the assumption that the slow form was a distinct, well defined conformational state of thrombin, researchers have attempted for many years to determine its structure by X-ray crystallography. Zhang and Tulinksky [25] correctly identified the coordination site of Na+ by growing crystals in its presence and subsequently replacing it with the more electron-dense Rb+. As a natural corollary, they soaked out Na+ and determined the structure of what they hoped would be thrombin in the slow state. Although Na+ was missing from the structure, the conformation was identical to the Na+-bound state. They concluded that crystal contacts may stabilize the fast state regardless of the occupancy of the Na+ binding site. Furthermore, the crystals were grown in the presence of the exosite I ligand hirugen, and it is known that exosite I binding has the same conformational effect as Na+ binding (i.e. stabilization of the fast conformation) [26]. So it was clear that obtaining a crystal structure of the slow form would require non-liganded thrombin and a modicum of good luck.
Mutagenesis studies were initially more successful in defining the regions involved in Na+ binding, and the associated conformational change. I recently published a review of these studies, classifying the importance of the residues according to their effect on Na+ affinity [20]. Those residues of medium to extreme importance (0.5 log effects and above) are shown in Fig. 3B. With few exceptions, the residues involved in the cofactor activation of thrombin belong to the zymogen activation loops. This is perhaps unsurprising when considering the location of the Na+ binding site (Fig. 3B), but for exosite I ligands this truly is a long-range allosteric effect.
Today the PDB contains several depositions of thrombin structures with ‘slow’ in the titles. I recently conducted a thorough analysis of the structures [20] and concluded that the two copies of Na+-free thrombin in 1MH0 [27] and in 1SGI [28] (same crystal, but different resolution) are indistinguishable from acknowledged fast forms, whereas a second class represented by 2AFQ [29] (wild-type), 2GP9 [30] (D102Q), and 1RD3 [31] (E217K) were structurally distinct from fast thrombin. When the regions that differ significantly (Cα RMSD signal to noise ratio above 2) in these structures are plotted (Fig. 3C), the correspondence with the mutagenesis data (Fig. 3B) and the zymogen structures (Fig. 2A) is strikingly clear. The altered conformation of the zymogen loops of the slow thrombins has the same functional effect seen for zymogens; namely, the blocking of the S1 pocket and the malformation of the oxyanion hole, in spite of having its N-terminal Ile16 stably incorporated in the activation pocket.
Conclusions
To some it may seem odd for a 2009 State-of-the-Art article to review work that was done in the seventies, but the goal of this review is to place past and ongoing work on protease allostery (thrombin in particular) into the correct historical and scientific context. There are several examples of serine proteases in haemostasis that require cofactor binding to achieve full activation. In some cases this is due to solely to co-localization, while in others there is an accompanying allosteric activation event. To some degree, all coagulation proteases from fVIIa through to thrombin can be activated toward small substrates through either coordination of cations or macromolecular cofactor binding. Thus the apo forms of these proteases have not completed the conversion to their fully active states, and are therefore still zymogen-like to some degree. The issue of physiological relevance is decided by whether these low-activity states exist in and around the blood clot, or whether they are merely artefacts of contrived in vitro conditions.
Acknowledgements
I thank the Scientific Committee of ISTH 2009 for the invitation to contribute this manuscript. Funding for my work on thrombin is provided by the National Institutes of Health (HL68629), the British Heart Foundation (UK) and the Medical Research Council (UK).
References
- 1.Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–58. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- 2.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27(2):157–62. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 3.Denton J, Lane DA, Thunberg L, Slater AM, Lindahl U. Binding of platelet factor 4 to heparin oligosaccharides. Biochem J. 1983;209(2):455–60. doi: 10.1042/bj2090455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kossiakoff AA, Chambers JL, Kay LM, Stroud RM. Structure of bovine trypsinogen at 1.9 A resolution. Biochemistry. 1977;16(4):654–64. doi: 10.1021/bi00623a016. [DOI] [PubMed] [Google Scholar]
- 5.Fehlhammer H, Bode W, Huber R. Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin. J Mol Biol. 1977;111(4):415–38. doi: 10.1016/s0022-2836(77)80062-4. [DOI] [PubMed] [Google Scholar]
- 6.Morgan PH, Robinson NC, Walsh KA, Neurath H. Inactivation of bovine trypsinogen and chymotrypsinogen by diisopropylphosphorofluoridate. Proc Natl Acad Sci U S A. 1972;69(11):3312–6. doi: 10.1073/pnas.69.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan PH, Walsh KA, Neurath H. Inactivation of trypsinogen by methane sulfonyl fluoride. FEBS Lett. 1974;41(1):108–10. doi: 10.1016/0014-5793(74)80965-8. [DOI] [PubMed] [Google Scholar]
- 8.Lonsdale-Eccles JD, Neurath H, Walsh KA. Probes of the mechanism of zymogen catalysis. Biochemistry. 1978;17(14):2805–9. doi: 10.1021/bi00607a016. [DOI] [PubMed] [Google Scholar]
- 9.Huber R, Bode W. Structural basis of the activation and action of trypsin. Accounts of Chemical Research. 1978;11(3):114–22. [Google Scholar]
- 10.Vijayalakshmi J, Padmanabhan KP, Mann KG, Tulinsky A. The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3(12):2254–71. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malkowski MG, Martin PD, Guzik JC, Edwards BF. The co-crystal structure of unliganded bovine alpha-thrombin and prethrombin-2: movement of the Tyr-Pro-Pro-Trp segment and active site residues upon ligand binding. Protein Sci. 1997;6(7):1438–48. doi: 10.1002/pro.5560060708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bode W, Huber R. Induction of the bovine trypsinogen-trypsin transition by peptides sequentially similar to the N-terminus of trypsin. FEBS Lett. 1976;68(2):231–6. doi: 10.1016/0014-5793(76)80443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode W. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. II. The binding of the pancreatic trypsin inhibitor and of isoleucine-valine and of sequentially related peptides to trypsinogen and to p-guanidinobenzoate-trypsinogen. J Mol Biol. 1979;127(4):357–74. doi: 10.1016/0022-2836(79)90227-4. [DOI] [PubMed] [Google Scholar]
- 14.Londsdale-Eccles JD, Kerr MA, Walsh KA, Neurath H. Catalysis by zymogens: increased reactivity at high ionic strength. FEBS Lett. 1979;100(1):157–60. doi: 10.1016/0014-5793(79)81154-0. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425(6957):535–9. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 16.Renatus M, Engh RA, Stubbs MT, Huber R, Fischer S, Kohnert U, Bode W. Lysine 156 promotes the anomalous proenzyme activity of tPA: X-ray crystal structure of single-chain human tPA. EMBO J. 1997;16(16):4797–805. doi: 10.1093/emboj/16.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruf W, Dickinson CD. Allosteric regulation of the cofactor-dependent serine protease coagulation factor VIIa. Trends Cardiovasc Med. 1998;8(8):350–6. doi: 10.1016/s1050-1738(98)00031-0. [DOI] [PubMed] [Google Scholar]
- 18.Olsen OH, Persson E. Cofactor-induced and mutational activity enhancement of coagulation factor VIIa. Cell Mol Life Sci. 2008;65(6):953–63. doi: 10.1007/s00018-007-7480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachias K, Madison EL. Converting tissue-type plasminogen activator into a zymogen. J Biol Chem. 1996;271(46):28749–52. doi: 10.1074/jbc.271.46.28749. [DOI] [PubMed] [Google Scholar]
- 20.Huntington JA. How Na(+) activates thrombin - a review of the functional and structural data. Biol Chem. 2008;389(8):1025–35. doi: 10.1515/BC.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page MJ, Di Cera E. Is Na+ a coagulation factor? Thromb Haemost. 2006;95(6):920–1. doi: 10.1160/TH06-05-0239. [DOI] [PubMed] [Google Scholar]
- 22.Di Cera E. Thrombin. Mol Aspects Med. 2008;29(4):203–54. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells CM, Di Cera E. Thrombin is a Na(+)-activated enzyme. Biochemistry. 1992;31(47):11721–30. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 24.Gruber A, Marzec UM, Bush L, Di Cera E, Fernandez JA, Berny MA, Tucker EI, McCarty OJ, Griffin JH, Hanson SR. Relative antithrombotic and antihemostatic effects of protein C activator versus low-molecular-weight heparin in primates. Blood. 2007;109(9):3733–40. doi: 10.1182/blood-2006-07-035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang E, Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys Chem. 1997;63(23):185–200. doi: 10.1016/s0301-4622(96)02227-2. [DOI] [PubMed] [Google Scholar]
- 26.Ayala Y, Di Cera E. Molecular recognition by thrombin. Role of the slow-->fast transition, site-specific ion binding energetics and thermodynamic mapping of structural components. J Mol Biol. 1994;235(2):733–46. doi: 10.1006/jmbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 27.Pineda AO, Savvides S, Waksman G, Di Cera E. Crystal structure of the anticoagulant slow form of thrombin. J Biol Chem. 2002;277(43):40177–80. doi: 10.1074/jbc.C200465200. [DOI] [PubMed] [Google Scholar]
- 28.Pineda AO, Carrell CJ, Bush LA, Prasad S, Caccia S, Chen ZW, Mathews FS, Di Cera E. Molecular dissection of Na+ binding to thrombin. J Biol Chem. 2004;279(30):31842–53. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 29.Johnson DJ, Adams TE, Li W, Huntington JA. Crystal structure of wild-type human thrombin in the Na+-free state. Biochem J. 2005;392(Pt 1):21–8. doi: 10.1042/BJ20051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pineda AO, Chen ZW, Bah A, Garvey LC, Mathews FS, Di Cera E. Crystal structure of thrombin in a self-inhibited conformation. J Biol Chem. 2006;281(43):32922–8. doi: 10.1074/jbc.M605530200. [DOI] [PubMed] [Google Scholar]
- 31.Carter WJ, Myles T, Gibbs CS, Leung LL, Huntington JA. Crystal structure of anticoagulant thrombin variant E217K provides insights into thrombin allostery. J Biol Chem. 2004;279(25):26387–94. doi: 10.1074/jbc.M402364200. [DOI] [PubMed] [Google Scholar]