Abstract
The oxidative folding pathways of two four-disulfide proteins of the ribonuclease family, ONC † and RNase A, which have similar three-dimensional folds but only 30 % sequence homology, are compared. In this study, a mechanism for the oxidative folding pathway of ONC is proposed. In particular, the kinetic roles and thermodynamic characteristics of key intermediates along the oxidative folding pathway, specifically, the structured intermediates, I1, I2 and I3, previously identified as des-[19–68, 30–75], des-[30–75], and des-[19–68], respectively, are discussed. In addition, the effects of temperature on the oxidative folding pathway have been examined. Differences in the folding mechanism between ONC and RNase A are attributed to the differences in their amino acid sequences and related inter-residue interactions, including differences in hydrophobic interactions. Compared to RNase A, ONC utilizes more efficient interactions along the oxidative folding pathway to adopt its native fold more rapidly.
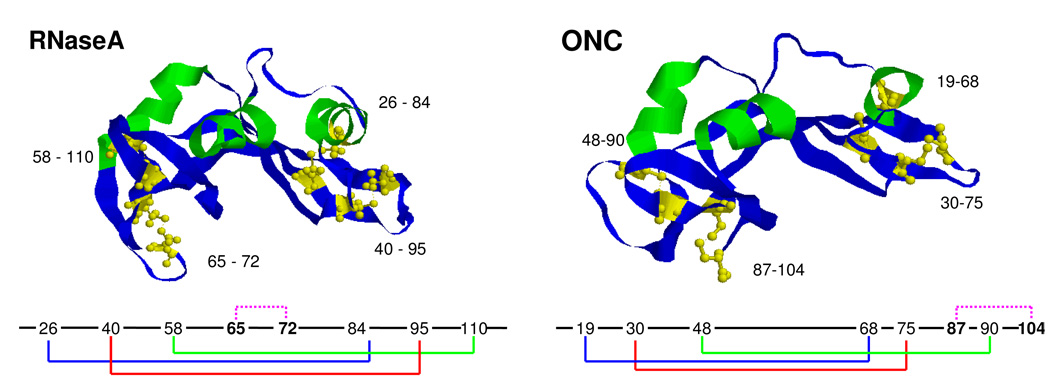
By examining the oxidative folding pathways of two homologs of the ribonuclease family, ONC in this paper, compared with our earlier work on RNase A (1, 2), insight is gained about oxidative folding of proteins. Of special interest in comparing these two four-disulfide proteins with the same tertiary fold (Fig. 1), but with only 30% sequence homology (3), is that three of the four native disulfide bonds ([19–68], [30–75], and [48–90] of ONC, and [26–84], [40–95], and [58–110] of RNase A) are in structurally homologous positions. The fourth native disulfide bond ([65–72] of RNase A), which closes a loop in the interior of the backbone, is replaced by the [87–104] disulfide bond of ONC, which closes the C-terminal region of the protein (see Fig. 1). Further, since the [65–72] disulfide bond of RNase A is the first one to form in the oxidative folding pathway (4), ONC must fold by another oxidative pathway because it lacks this homologous disulfide bond. Clearly, the differences in the amino acid sequences of these two structurally homologous proteins must account for anticipated differences in their oxidative folding pathways. Therefore, the goal of this investigation was to characterize the oxidative folding pathway of ONC, by identifying the kinetic mechanism by which ONC attains the correct disulfide bonding, coupled with the attainment of the native-like structure, thereby providing an understanding of the physical-chemical properties of two members of the ribonuclease family.
Figure 1.
Crystral structures of RNase A, 124 residues (PDB code: RSA1), and ONC, 104 residues (PDB code: 1ONC). Disulfide bonds are shown as ball and stick models. The disulfide bond connectivity is shown below the corresponding crystal structure.
An additional interest in ONC is the discovery that ONC is cytotoxic to various cancerous cell lines under in vivo conditions (3). As a result, ONC has undergone clinical trials to treat unresectable malignant mesothelioma (UMM). The clinical trials have just been completed, and a new drug application is being submitted (5). Current research has focused on understanding the critical steps in the cytotoxic action of ONC on cancerous cells (6, 7). The efficacy of the cytotoxicity of ONC is due to its selective uptake into cancerous cells and its resistance to proteolysis in regulatory pathways after undergoing endocytosis (3). One drawback to ONC is that it is not as effective as a catalyst compared to the catalytic activity of other riboncleases such as RNase A. Efforts have been made to increase the catalytic efficiency of ONC. However, the catalytic efficiency of RNase A has not been matched (8, 9). Further work to understand the physico-chemical properties of members of the ribonuclease family would aid continuing work to utilize ribonucleases as chemotherapeutics.
In our laboratory, RNase A has been used as a model system to study the protein folding problem. In particular, RNase A has been used to develop methods to study the oxidative folding of disulfide-bond containing proteins to determine how they attain both the correct disulfide linkages and the biologically-active structure from a reduced form having no disulfide bonds. By application of these methods to RNase A, two types of intermediate species along the oxidative folding pathway have been identified and characterized: unstructured ensembles (nS) and structured species (nS*). The oxidative folding mechanism of RNase A has been established in terms of these types of species (1, 2, 10, 11). In the early stages of oxidative folding, a pre-equilibrium between unstructured ensembles with zero, one, two, three, and four disulfide bonds is established. Each of these ensembles, except the fully-reduced form, contains native and non-native disulfide bonds. The rate-determining step in the mechanism was identified (1) as a redox-independent SH/S-S reshuffling step from an unstructured ensemble (3S) to two structured species (2), des-[40–95] and des-[65–72], each designated as 3S*, which oxidize rapidly to the native form. The structured 3S* intermediates have been analyzed by NMR (12, 13). Further work is being carried out to identify the residues responsible for the stabilization of these structured species (R.F. Gahl, R.E. Oswald, and H.A. Scheraga, work in progress).
Methods developed to study the oxidative-folding process in RNase A have been applied for the separation and purification of key disulfide-bond species in the oxidative folding of ONC. In a previous study (14), three structured intermediate species, des-[19–68, 30–75], des-[30–75], and des-[19–68] of ONC were identified. In the present study, their thermodynamic stabilities are characterized with the use of CD spectroscopy, and their roles in the kinetic pathways are identified. In addition, the temperature-dependence of the folding mechanism is also studied. By analyzing the effect of temperature on the various folding stages, the nature of the interactions in key species that drive the folding are identified. The information gained from these studies provides further understanding about the physico-chemical properties of the ribonuclease fold.
Materials and Methods
Materials
Wild type onconase was prepared from cDNA kindly provided by R. J. Youle (15) and purified as described in earlier studies (14, 16). DTTox was purified by RP-HPLC to 99.999% as confirmed by RP-HPLC and Ellman analysis (17). All other highest-quality chemicals were purchased from Sigma, Fisher, and Anatrace, and used without any further purification.
Preparation of reduced onconase
Reduced onconase, R-ONC, was prepared by adding lyophilized ONC to 4 mL of a buffer (0.1 M Tris, 1 mM EDTA, pH 8) containing 6 M GdnHCl and 100 mM DTTred (to an ONC concentration of 2 mg/mL) under humidified argon. After 2 – 4 hours, the reduction was stopped by reducing the pH to 3 using 100 µL of glacial acetic acid and immediately desalting the mixture on a G25 size-exclusion column equilibrated with 50mM acetic acid at pH 3 to avoid air oxidation. As soon as R-ONC eluted, it was frozen and stored at −70 °C. After desalting, water and acetic acid were removed by lyophilization, and R-ONC was stored as aliquots in 3 mM acetic acid at a concentration of 2.8 mg/mL at −70 °C. These aliquots were diluted to 17 µM (0.20 mg/mL) for oxidative folding experiments.
Two methods were used to measure the concentration of R-ONC: UV absorption at 280 nm, and thiol content with Ellman’s reagent (17). To determine the concentration of R-ONC by UV absorption, 7400 ± 600 M−1·cm−1 was used as the molar absorptivity, ε280, at 280 nm, determined by fitting the linear region of an absorbance-at-280 nm vs. concentration curve to the Beer-Lambert equation. To determine the concentration of R-ONC by Ellman’s analysis, a 15 µL aliquot of R-ONC stored in 3 mM acetic acid was added to 435 µL of a buffer (0.1M Tris, pH 8.3) that was previously purged of oxygen with humidified argon. A 50 µL aliquot of 15 mM DTNB solution in a pH 8.3, 0.1M Tris buffer was added to the 450 µL solution containing R-ONC. The thiol content of R-ONC was measured from the absorbance of TNB at 412 nm, using 13600 M−1·cm−1 (17) to determine the concentration of free TNB in the solution, which corresponds to the thiol content. The TNB concentration was divided by 8 (R-ONC has 8 cysteines) to obtain the concentration of R-ONC. Each method gave the same concentration of R-ONC within experimental error (~5 %).
Preparation of solutions for oxidative folding
The oxidative folding of ONC was studied under an anaerobic environment in which the redox potential of the solution was controlled by the relative concentrations of DTTox and DTTred. Tests were performed to verify (i) that there were only trace amounts of dissolved oxygen that would not affect the rate of oxidative folding over the course of the experiment, and (ii) that the method used to purge oxygen from the solution did not affect the concentration of the oxidizing agents or of R-ONC during the oxidative folding process. Oxidative folding solutions, including DTTox, were purged with humidified argon before R-ONC, or in some cases DTTred to control the redox conditions, were added to start the folding process. Also, as a control, DTTred was added to a purged protein-free solution to make sure that oxygen had been removed by the purging process. Under all of the conditions used to study the oxidative folding of ONC, the extent of DTTred oxidation by oxygen was no more than 5 % for the duration of the oxidative folding experiments.
The mechanism for thiolate oxidation by DTTox is outlined in ref. (10). For every two thiolates that are oxidized by one molecule of DTTox, one molecule of DTTred is produced. DTTred can also react with disulfide bonds and this is taken into account in analyzing the kinetics, mentioned later. Therefore, it is important to make sure that no dissolved oxygen reacts not only with the thiolate ions of oxidative folding intermediates but also with the DTTred that is produced; i.e., that the thiolate content remains constant throughout the duration of the oxidative folding experiment.
Oxidative folding was also studied under a range of temperatures. To insure that the concentration of species in the folding solutions did not change with temperature, the concentration of a solution containing 5 mM DTTox (0.1 M Tris, pH 8.0) was monitored over time at each temperature at 330 nm using 22.3 M−1·cm−1 for the molar absorptivity of DTTox, determined by plotting absorbance at 330 nm vs. concentration according to the Beer-Lambert equation. In addition, the humidified argon had to be equilibrated at the temperature of the reaction before it was used to purge the solution of oxygen. The concentration of DTTox did not change by more than 5 % over the duration of time used to monitor oxidative folding.
The pH of Tris buffer is temperature-dependent (18). For studies at different temperatures, this effect had to be taken into account in order to eliminate pH-dependent effects on the oxidative folding pathway. Our oxidation studies were performed at pH 8.0, which had to be kept constant at the different temperatures used. The temperature dependence of the pH of the buffer was measured at temperatures ranging from 15 to 57 °C. The slope (ΔpH/ΔT) of the pH vs. Temperature curve was determined to be −0.026 ± 0.0012 and is comparable to other values in the literature, −0.028 (18, 19). To adjust for this effect at different temperatures, the pH’s of buffers containing 0.1M Tris were adjusted to compensate for this variation from ambient temperature, 22 °C, to a given temperature to perform oxidative folding, at pH 8.0 at 15 – 57 °C, e.g., the pH of a buffer for a 15 °C or 37 °C experiment was adjusted to 7.7 or 8.3, respectively, at 25 °C.
Oxidative Folding of ONC and SCX-HPLC analysis
The range of redox conditions used to elucidate the oxidative folding pathway of ONC, starting with R-ONC, were 25 – 115 mM DTTox, 0 – 66 µM DTTred in the presence of 25 mM DTTox, at 25 °C. For studies at temperatures varying from 15 ° to 57 °C, the redox condition used was 25 mM DTTox and 16 µM DTTred. The concentration of R-ONC was 17 µM in all experiments. Precipitation was observed in experiments at concentrations higher than 17 µM (unpublished results). To ensure that no precipitation occurred during the folding experiments, a UV-Vis spectrum was taken at the beginning and end of the experiments. There was no change between each of the spectra. In addition, constant thiolate content for the duration of the oxidative folding experiment was verified by measuring the thiolate content using Ellman’s reagent at different times during the folding process.
After a redox solution was purged of oxygen by humidified argon, R-ONC, or in some cases DTTred, was added to the folding mixture under a certain redox condition and folding was allowed to proceed. The reaction mixture was kept under humidified argon for the duration of the folding reaction. At certain times, an aliquot was removed and added to a blocking buffer (2M Tris, pH 8.6, 20 mM EDTA), which contained enough AEMTS to constitute a 100-fold molar excess with respect to the free thiol content in the mixture. The blocking buffer that contained the AEMTS was diluted 10-fold upon addition of an aliquot of the oxidative folding mixture. The blocking of the free thiols by AEMTS was allowed to proceed for 2 min. The blocking reaction was stopped by the addition of glacial acetic acid. The aliquots were then frozen and maintained at −70 °C and could be stored at this temperature. Before the aliquots could be analyzed, the frozen solution was thawed and the buffer salts and unreacted AEMTS were removed by desalting on a G25 size- exclusion column equilibrated with 0.2% acetic acid using absorbance at 280 nm to monitor the eluted species.
Complete blocking was verified by a negative Ellman’s test. There was no absorbance at 412 nm indicating that all of the free thiolates had been blocked by AEMTS. The effectiveness of desalting with a G25 column was verified by comparing an AEMTS-blocked aliquot desalted with the G25 column and an aliquot desalted by RP-HPLC, the latter being a more effective but more time-consuming procedure with which to desalt samples. Desalting by RP-HPLC was performed on a water/acetonitrile buffer system with 0.09% TFA using a 25 cm × 4.6 mm SULPELCO Discovery® BIO Wide Pore C18, 5 mm particle size column and absorbance at 210 nm to monitor the eluting species, i.e., to demonstrate that salt had been removed. After the sample was injected with 100% water/ 0 % acetonitrile running through the column and the buffer salts, DTTox, AEMTS-blocked DTTred, and unreacted AEMTS eluted off the column, a constant flow of 20% water/ 80 % acetonitrile was passed through the column to elute and collect the intermediate disulfide-bonded species of ONC. The acetonitrile, water, and TFA were removed by lyophilization before reconstituting the protein species in 3 mM acetic acid to be analyzed by SCX-HPLC. Independent chromatograms of each of these protein-containing aliquots from both methods, analyzed by SCX-HPLC, were identical. Use of size-exclusion G25 columns was sufficient to analyze aliquots taken at different times during oxidative folding experiments.
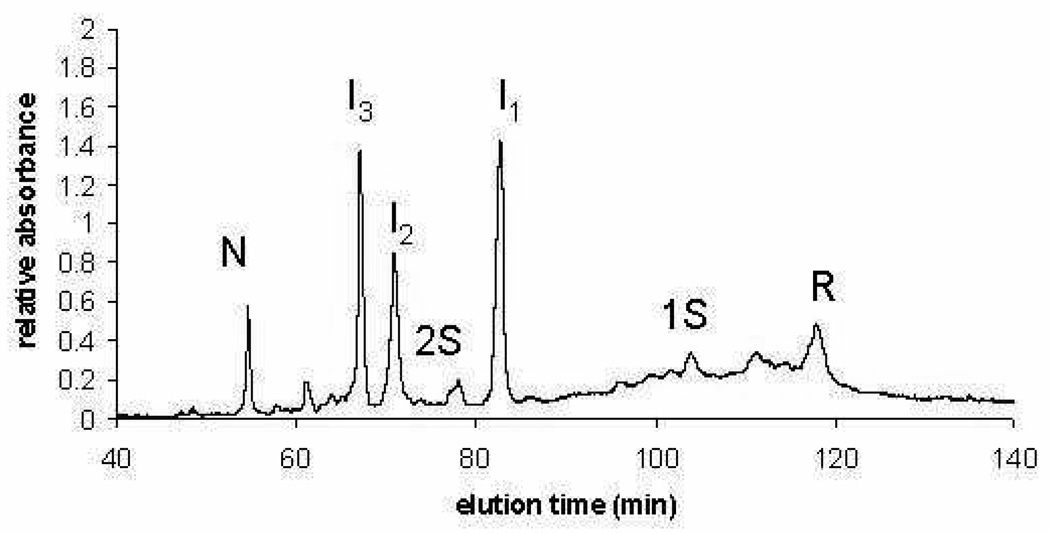
After an aliquot from an oxidative folding mixture was blocked with AEMTS and properly desalted using a size-exclusion G25 column, it was loaded directly onto an SCX-HPLC for separation and analysis using a TOSOH Biosciences TSKgel™ column SP-5PW 7.5 cm × 7.5 mm, analytical strong cation-exchange (SCX) column. The running buffer was 25 mM HEPES 2 mM EDTA pH 8. The gradient used to separate the various AEMTS-blocked species was 0–480 mM NaCl in 120 min. The running buffer of the size-exclusion G25 column, 3 mM acetic acid, is compatible with the SCX-HPLC column and buffer system. The amount of each species was estimated quantitatively by monitoring the absorbance at 280 nm, and the relative amounts of each species was computed by integrating the area of each corresponding peak. The observed intermediate species along the folding pathway under conditions reported in the Figure Legend are indicated in Fig. 2.
Figure 2.
Example of an SCX-HPLC analysis of the distribution of AEMTS-blocked intermediate species from the oxidative folding of ONC after 90 min in 25 mM DTTox and 66 µM DTTred at 25 °C and pH 8.0. The species were separated on a TOSOH Biosciences TSKgel™ SCX column, SP-5PW 7.5 cm × 7.5 mm, using pH 8, 25mM HEPES, 3mM EDTA as a running buffer and an NaCl gradient for separation. I1, I2 and I3 have been identified as des-[19–68, 30–75], des-[30–75], and des-[19–68], respectively (14).
For one experiment, it was necessary to determine the products of the direct oxidation of unblocked I1, i.e., des-[19–68, 30–75], to N. In order to prepare a sufficient amount of unblocked I1 from R-ONC, the oxidation of R-ONC was carried out with 25 mM DTTox, at 25 °C, pH 8.0, for 15 min. The subsequent oxidation of R-ONC was stopped by the addition of acetic acid. The products were separated by RP-HPLC as described above, the difference being that a gradient of 30 – 45 % acetonitrile over 60 min, instead of a constant mixture of 20% water/ 80% acetonitrile, was used to separate the unblocked species in the reaction mixture.
After unblocked I1 was collected, water, TFA and acetonitrile were removed by lyophilization. Unblocked I1 was reconstituted in 50 mM acetic acid buffer and its concentration (10 µM) was determined by Ellman’s reagent. Using the procedure described above, the oxidation of I1 to N was then examined.
Analysis of the Kinetics of Oxidative Folding
In order to determine the mechanism of oxidative folding of ONC, the observed redox-independent rate constants for the various steps in a kinetic scheme were determined under the redox conditions mentioned in the previous section. By examining various possible mechanisms, the following one was found to provide the best fit to the experimental variation of the concentrations of each species with time:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
This mechanism is describable by the following system of differential equations:
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
The concentrations of DTTox and DTTred, for use in equation 9–equation 15, were obtained from the following equations:
| (16) |
| (17) |
The system of differential equations, 9 – 15, was solved numerically by using the 4th order Runge-Kutta method (20), requiring that the computed time-dependent curves for the evolution of each species provided a best fit to the experimental data. Thus, the best-fit redox-independent observed rate constants were determined by a simplex algorithm in conjunction with a modified Monte-Carlo method to randomize the data sets before fitting (21). Random noise was generated by using a Box-Muller algorithm (22), which provides a Gaussian distribution of height 1 and standard deviation 1. Noise was included in the experimental values by adding a number chosen randomly from the generated Gaussian error distribution, and multiplied by the experimentally-determined standard deviation of that experimental value. By applying this procedure to each experimental value and thereby providing a Runge-Kutta solution for each value, 50 copies of each rate constant were obtained. An arithmetical average of each rate constant and its standard deviation resulted form this procedure. By analyzing these additional data sets, the accuracy and precision of the k’s were improved.
To verify that the rate constants produced by this fitting procedure uniquely describe the time-dependence of each of the species, a new set of “experimental” data was generated from a new set of k’s, computed by changing each of the k’s of column 2 of Table 1 by ± 30 %. The Runge-Kutta procedure was then applied to this new set of “experimental” data to compute a new set of k’s (column 3 of Table 1). Both sets, shown in Table 1, are in agreement within standard deviations of the experimentally determined set, given that this test was performed for only one redox condition whereas the original set was obtained by combining the data from all the redox conditions examined.
Table 1.
Observed rate constants for each step in the oxidative folding mechanism (eq. 1–eq. 8) determined at pH 8.0, 25 °C
| Redox-independent rate constants | k’s (M−1 min−1)a | k’s (M−1 min−1)b |
|---|---|---|
| k1 | 14.0 ± 0.14 | 18.0 ± 0.75 |
| k2 | 2770 ± 51 | 2600 ± 357 |
| k3 | 0.19 ± 0.01 | 0.20 ± 0.01 |
| k4 | 14 ± 4.3 | 20 ± 5 |
| k5 | 0.62 ± 0.01 | 0.63 ± 0.02 |
| k6 | 45 ± 7.5 | 50 ± 7 |
| k7 | 1.00 ± 0.07 | 1.0 ± 0.2 |
| k8 | 1.28 ± 0.02 | 1.25 ± 0.07 |
| k9 | 0.39 ± 0.03 | 0.41 ± 0.04 |
| k10 | 0.59 ± 0.01 | 0.65 ± 0.04 |
| k11 | 0.28 ± 0.01 | 0.22 ± 0.06 |
computed from the original experimental data
computed from the “experimental” data, created by varying the original k’s to test their uniqueness.
As noted by Thannhauser and co-workers (23), the intermediate species can have different absorbtivities at 280 nm because the intermediates have varying degrees of structure that would affect the measured absorptivity. This effect was corrected for by use of a plot of area under a peak vs. concentration of a particular species, measured by NTSB analysis (24).
CD spectra of the Structured Intermediates, I1, I2, I3 and N
The extent of native-like structure for the structured intermediates, I1, I2, and I3 was determined by CD spectroscopy with an AVIV Biomedical (model 202-01) spectropolarimeter and compared to the CD spectrum of N. The mean residue ellipticity (MRE) was measured in the far-UV region, 190 to 260 nm. Samples were prepared by populating the intermediates by oxidative folding to maximize the quantity of the intermediates (25 mM DTTox, 16 µM DTTred, 25 °C, pH 8.0, 90 min). Aliquots of the folding mixture were blocked with AEMTS and separated by SCX-HPLC as described in the section, “Oxidative Folding of ONC and SCX-HPLC analysis”. Each of the blocked intermediates was collected as they eluted off the column, and the pH of the collected fraction was adjusted to 3 with glacial acetic acid. The SCX-HPLC buffer salts and NaCl from each intermediate were removed by RP-HPLC as described in the section “Oxidative Folding of ONC and SCX-HPLC analysis.” Water, acetonitrile, and TFA were removed by lyophylization, and the intermediates were each reconstituted in 50mM acetic acid to be analyzed by CD spectroscopy at pH 3.0. This pH was chosen to avoid removal of the blocking groups during the CD measurements because the disulfide bond between the protein moiety and the blocking group is more stable at a low pH (25). The CD spectrum of 50 mM acetic acid was subtracted from the CD spectra of the protein solutions. To convert the observed circular dichroism signal from mdeg to MRE, the concentration of each of the solutions had to be measured accurately. The concentration of each solution was determined by thiolate analysis utilizing NTSB and sulfite-reduction of disulfides, as described in ref. (24). The disulfide content of each intermediate was obtained from the additional thiols resulting from sulfite reduction of its disulfide bonds (24). Each of the intermediates has a different number of thiols produced by reduction of intramolecular disulfide bonds and of the disulfide bonds produced by covalent attachment of the thiols to the blocking group, -SCH2CH2NH3+; I1, has 12 free thiols (4 from the two S-S bonds, 4 from the free thiols blocked with AEMTS, and 4 from the cystamine group of the released blocking group, -SCH2CH2NH3+), I2 and I3 each has 10 free thiols (6 from the three S-S bonds, 2 from the free thiols blocked with AEMTS, and 2 from the cystamine group of the released blocking group, -SCH2CH2NH3+), and N has 8 free thiols (8 from the 4 S-S bonds). After CD spectra were obtained for all of the blocked intermediates and N-ONC, they were checked by SCX-HPLC to insure that the disulfide bonds of each of the species did not undergo possible removal of the blocking group with attendant SH/S-S reshuffling. This SCX-HPLC method to analyze AEMTS-blocked species is sufficient to test for these side reactions because it can separate species that have the same number of disulfide bonds, I2 and I3, and species that contain different numbers of disulfide bonds, which would have a different number of blocking groups,-SCH2CH2NH3+, that give rise to different cationic affinities.
Thermal Stability of AEMTS-blocked Intermediates I1, I2, I3 and N
Thermal denaturation of AEMTS-blocked I1, I2, I3 and N was monitored by CD spectroscopy, and samples were prepared as described in the previous section. The percent of folding was monitored by CD at 198 nm, the wavelength at which there was the greatest difference between the folded and unfolded spectra. The temperature range studied was from 25 ° to 97.5 °C. The temperature was incremented 5 °C in the range corresponding to folded and unfolded species, 2.5 °C as the folding transition approached, and 1 °C during the folding transitions. Each solution was equilibrated to maintain the temperature within 0.1 °C for 2 min before collecting data for 60 seconds. The corresponding error in the CD (0.8 mdeg) was observed to be maintained for 5 minutes of temperature equilibration and 5 minutes of data collection. Heating and cooling curves were obtained to show reversibility of the denaturation process. After each thermal unfolding experiment, the sample was checked by SCX-HPLC to insure that the disulfide bonds of the species did not reshuffle.
The thermal denaturation curves were fit to a two-state model, and thermodynamic parameters were extracted by the following equations:
| (18) |
| (19) |
| (20) |
Results
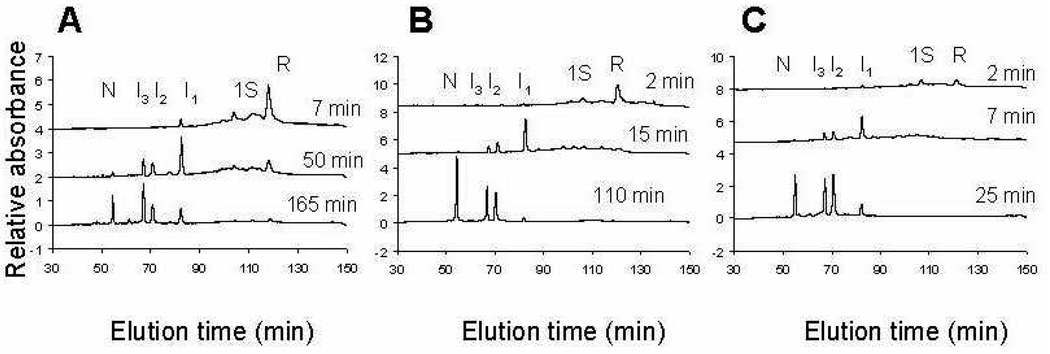
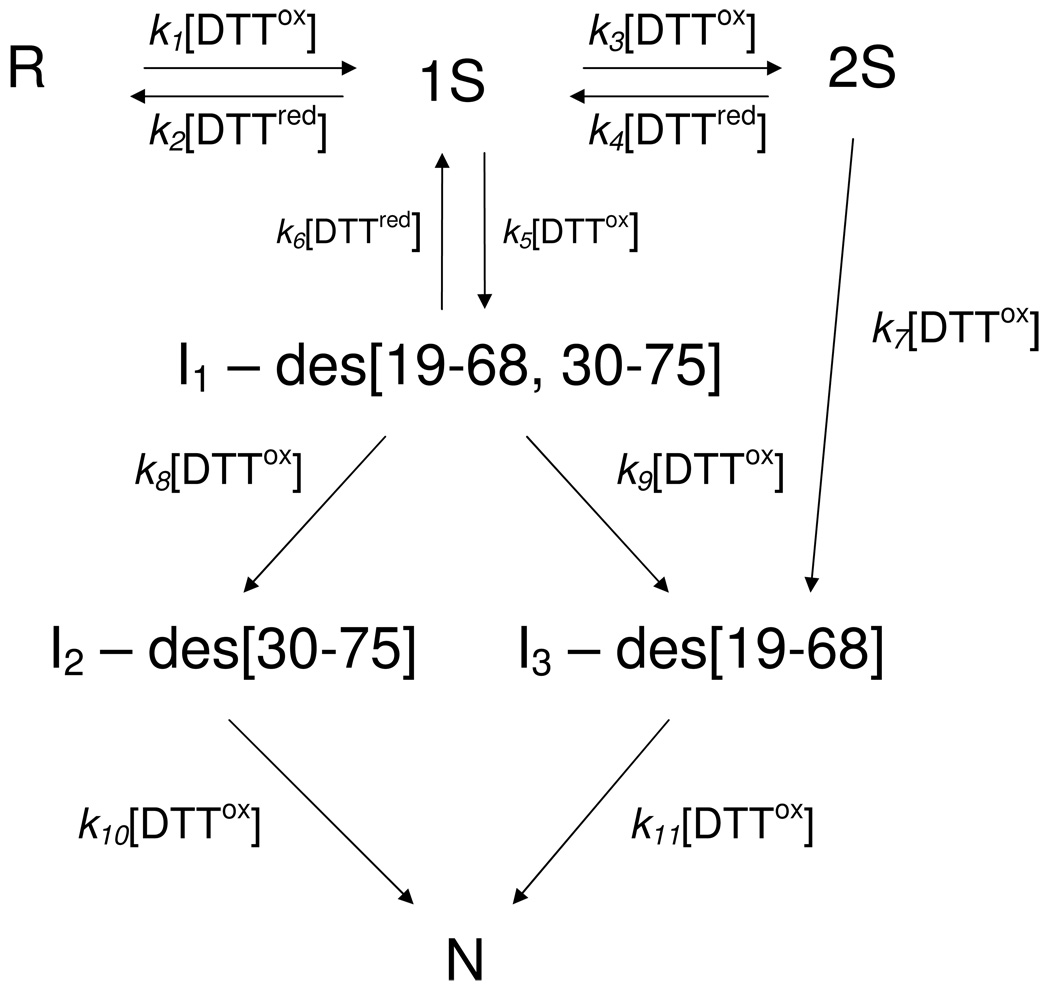
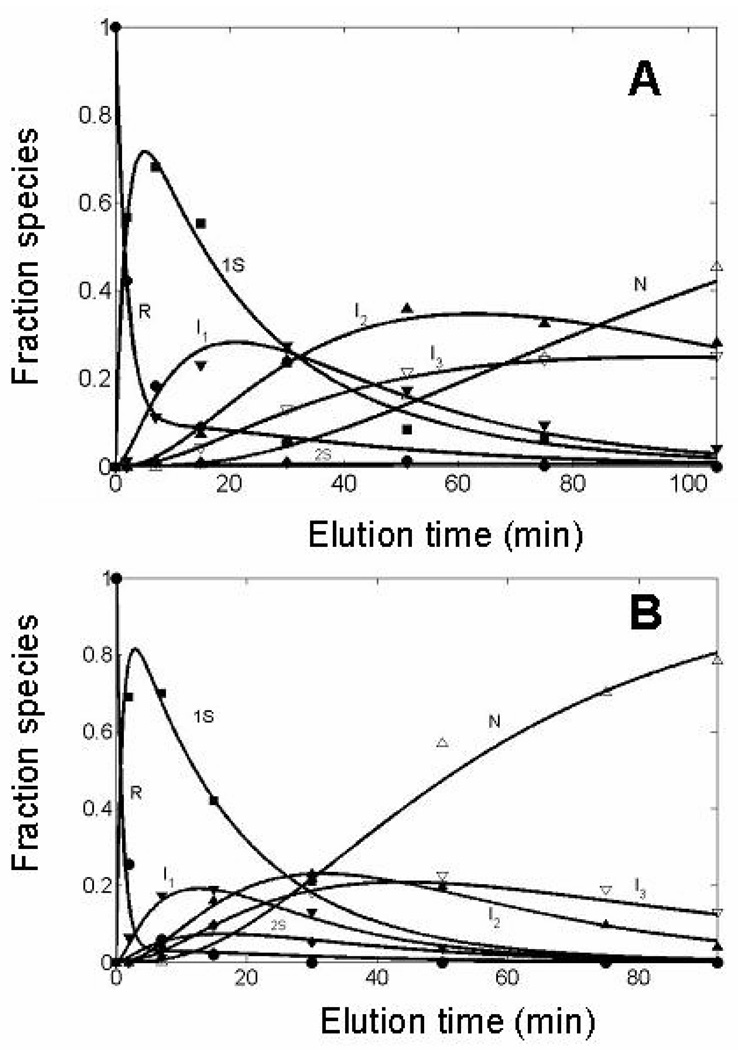
The separation of intermediate blocked species at different reaction times and different elution times is shown for three redox conditions in panels A, B, and C of Fig. 3. The relative populations of each of the intermediate species in chromatograms of this type were obtained quantitatively by integrating the area under curves produced from each of the intermediates. It was observed that the elution times of the intermediates are very reproducible, and the assignment of a consistent baseline for integration of peak area could be obtained by superimposing each of the corresponding chromatograms. Analysis of the relative changes in populations for each intermediate species, by Runge-Kutta solutions for the system of equation 9–equation 15, for oxidative folding reactions under a variety of redox conditions, led to the oxidative folding mechanism of eqn. 1–eqn. 8, shown in Fig. 4, which provided the best fit to the experimental data. The time-dependence of the various species are shown in Fig. 5 for two oxidizing conditions, 25 mM DTTox and 70 mM DTTox, respectively. The observed rate constants that correspond to each of the steps in the mechanism over the entire redox range are presented in column 2 of Table 1.
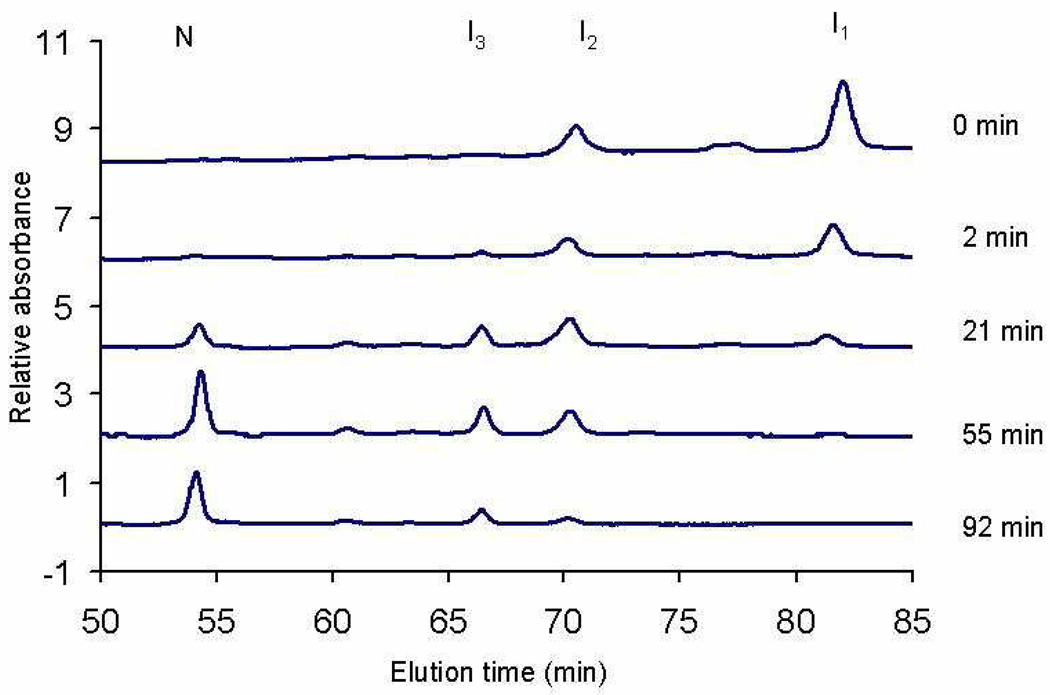
Figure 3.
Different distributions of intermediate species under different conditions. Panel A: 25 mM DTTox and 66 µM DTTred, 25 °C and pH 8.0 at 7, 50, and 165 min into the oxidative folding process. Panel B: 25 mM DTTox without DTTred, 25 °C and pH 8.0 at 2, 15, and 110 min into the oxidative folding process. Panel C: 70 mM DTTox without DTTred, 25 °C and pH 8.0 at 2, 7 and 25 min into the oxidative folding process.
Figure 4.
Oxidative folding mechanism for ONC. No 3S or 4S ensembles were detected under all of the oxidative folding conditions. The only three-disulfide-bond species are I2 and I3, and the only four-disulfide-bond species is N.
Figure 5.
Changes in the distribution of species (R-ONC, 1S, 2S, I1, I2, I3, N-ONC) over time, fit to the oxidative folding mechanism shown in Fig. 4. The condition analyzed in Panel A is 25 mM DTTox, 25 °C and pH 8.0 and that in Panel B is 70 mM DTTox, 25 °C and pH 8.0.
The oxidative folding of ONC is much faster than that of RNase A. According to previous work (2), 60 % of RNase A is recovered in 400 min in the presence of 100 mM DTTox and 16 µM RNase A. However, 80 % of ONC is recovered in 95 min in the presence of 70 mM DTTox and 17 µM ONC. The following analysis of the kinetic and thermodynamic role of various structured and unstructured species involved in the oxidative folding of ONC provides insight into the more efficient folding of ONC.
Kinetic Mechanism – Unstructured Ensembles
The 1S and 2S peaks in the chromatogram in Fig. 2 were identified as having one and two-disulfide species by mass spectrometry, and as unstructured (14) because they did not survive a reduction pulse before blocking with AEMTS. By the same criterion, I1, I2, I3 and N are structured.
As shown in Fig. 3, the first step in the oxidative folding of R-ONC by DTTox is the formation of an unstructured ensemble of species containing one disulfide bond (1S). This ensemble elutes right before R-ONC on the chromatogram and does not consist of a single peak. Rather, the number of peaks corresponds to different 1S species eluting at different times, 95 – 118 min, as seen in panels A, B, and C in Figure 3. In the mechanism in Fig. 4, there is a pre-equilibrium between R-ONC and the 1S ensemble, between the 1S and 2S ensembles, and between the 1S ensemble and I1. The observed rate constants for oxidation and reduction of the 1S and 2S ensembles do not correspond to those of a single disulfide species; each member of the ensemble contributes to the observed rate constant. To take account of the possible number of species in each ensemble to obtain the intrinsic rate constant for each species in the ensemble, the observed rate constants are modified by inclusion of appropriate statistical factors, as shown in Table 2. For the unstructured ensembles, 1S and 2S, the magnitude of these modified rate constants are different, namely 0.5 and 0.013 for the oxidation to form 1S and 2S, respectively, and 2770 and 7 for the reduction of 1S and 2S, respectively. The different modified rate constants reflect the different intrinsic reactivities of the members of each of the unstructured ensembles. However, according to ref. 2, the modified rate constants of RNase A are of similar magnitude for the oxidation and reduction of the 1S, 2S, 3S and 4S ensembles, which indicates that the free energy barriers of each step are the same. Therefore, while there is still no native-like structure in these ensembles, the different free energy barriers in ONC reflect different interactions in the unfolded state of ONC compared to RNase A.
Table 2.
Modified rate constantsa involved in unstructured ensembles at 25 °C, pH 8.0.
| kfb | kfave,c | kintrad | krb | krave,c | |
|---|---|---|---|---|---|
| k1 | 14 | 0.5 | 70.9 | - | - |
| k2 | - | - | - | 2770 | 2770 |
| k3 | 0.19 | 0.013 | 1.84 | - | - |
| k4 | - | - | - | 14 | 7 |
In units of M−1min−1
kf and kr refer to observed rate constants for oxidation and reduction steps, respectively. k1 and k3 correspond to oxidations of R and 1S, respectively, while k2 and k4 correspond to the reductions of 1S and 2S, respectively.
kfave and krave are modified for statistical factors that arise from the oxidation or reduction of each member of the respective ensembles to give the average rate among all of the members of the ensembles. For k1, there are 28 different ways to form the 1S ensemble from R, and therefore k1 is divided by 28. For k3, there are 15 ways to form the 2S ensemble from any member in the 1S ensemble, and therefore k3 is divided by 15. In a similar manner for the rate constants corresponding to reductions, k2 is not corrected because there is only one way to form R from any member of the 1S ensemble. k4 is divided by 2 because there are two ways of forming a 1S species from any member of the 2S ensemble.
Another quantity that helps characterize the interactions in these unstructured ensembles is kintra. kintra is a redox independent rate constant that reflects the relative rate of intramolecular SH/S-S reshuffling. The reshuffling process in the oxidation of two cysteines to form a disulfide bond is characterized by the following steps:
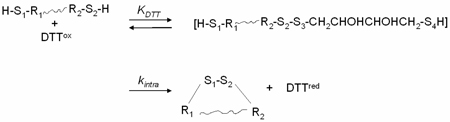 |
(21) |
First, H-S1-R1R2-S2-H forms an unstable mixed-disulfide with DTTox to a degree determined by KDTT. The value of KDTT has been derived earlier (26) and is 7.05 × 10−3M−1· S4 could react with S3 to regain a molecule of DTTox, or S1 could attack S2 with a rate constant kintra to release a molecule of DTTred and leave S1 and S2 in a disulfide bond; i.e. S4 of the dithiothreitol moiety could release H-S1-R1R2-S2-H and regain a molecule of free DTTox, or S1 can attack S2 at a rate of kintra to release a molecule of DTTred and leave S1 and S2 oxidized. kintra can be computed with the equation:
| (22) |
where kfave is an intrinsic rate constant, obtained from the observed k’s of Table 1 for oxidation reactions, modified by statistical factors, similarly for krave. Values for kintra are shown for the 1S and 2S ensembles in Table 2.
In the context of the oxidative folding pathway, the observed oxidation or reduction rate constants, kf or kr, can be multiplied by the concentrations of DTTox and DTTred, respectively, used in these experiments and compared to the rate of intramolecular reshuffling reactions. In all reactions, the rate of reshuffling (in terms of kintra) for either species is 102 – 103 times larger, than the average rate of oxidation or reduction. This indicates that the individual species of the 1S and 2S ensembles reshuffle fast enough to treat these ensembles as a single kinetic entity.
Under all conditions, with an example shown in Fig. 5, the concentration (calculated with the observed rate constants of Table 1 together with the concentrations of DTTox) of I1 is always greater than that of 2S. This is consistent with the mechanism of Figure 4 in which the pathway from I1 to N (through I2 and I3) is favored over the pathways from 2S to ONC (through I3).
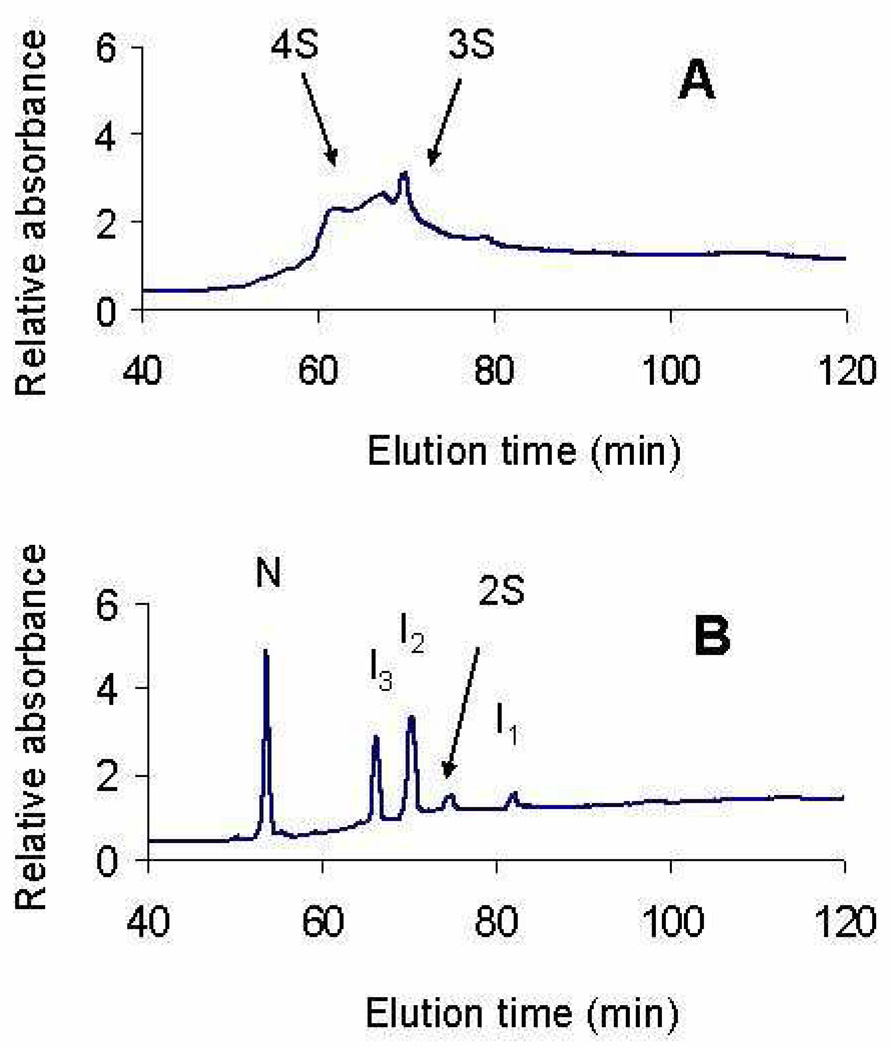
One interesting omission from the mechanism for oxidation of ONC [different from that of RNase A (2)] is the absence of unstructured ensembles containing three and four disulfide bonds (3S and 4S). To verify the omission of these species in the mechanism of Fig. 4, it was necessary to identify the chromatographic elution times of 3S and 4S ensembles of ONC, in which the concentration of N is very small. For this purpose, GdnHCl was added to R-ONC in an oxidative-folding experiment. The resulting chromatogram in Fig. 6A shows the elution pattern of 3S and 4S. On the other hand, in the absence of GdnHCl, no 3S or 4S peaks appear during oxidative folding. By avoiding 3S and 4S ensembles, which would have non-native interactions, ONC can explore conformational space more efficiently than RNase A does.
Figure 6.
A. Elution of the 3S and 4S ensembles that were populated by performing oxidative folding with 115 mM DTTox at 25 °C, pH 8.0, in the presence of GdnHCl. B. Oxidative folding without GdnHCl under the same redox condition; no 3S or 4S ensemble is observed under this condition.
Kinetic Mechanism – Structured Intermediates
A critical step in the oxidative folding mechanism of ONC is the formation of the structured intermediate I1, des-[19–68, 30–75], which (along with I2, I3 and N) are identified as those that survive a reduction pulse (14), applied to the oxidation mixture. Once I1 is formed, two of four native disulfide bonds are in place and, from this intermediate, the remaining native disulfide bonds are formed in I2, des-[30–75], and I3, des-[19–68], and finally in N. In Fig. 5, I2 and I3 can be observed forming after I1 is populated. Under both conditions in Fig. 5, I2 is preferentially formed over I3 at the same concentration of DTTox as indicated by its more rapid rate of formation (k8 > k9). The decay of I2 and I3 are also different. The more rapid decay of I2 relative to I3 (k10> k11) under both conditions in Fig. 5 indicates that I2 is more readily oxidized to N than I3 at the same concentration of DTTox. For the formation and oxidation of I2, the observed rate constants corresponding to these two steps are larger for I2. In the next section, the importance of I2 at different temperatures is discussed. Similar relative reactivity between these structured intermediates is observed when oxidative folding to form native ONC is initiated from unblocked I1; I2 and I3 and N are formed by oxidation of I1 with DTTox, as shown in Fig. 7. The amount of I2 present at the beginning is due to the overlapping of the I1 and I2 peaks during the preparative-separation of I1 by RP-HPLC.
Figure 7.
Oxidation of unblocked I1 at 70 mM DTTox, 25 °C and pH 8.0 I2 is also present at the beginning of the oxidation because it eluted close to I1 during its preparation by RP-HPLC. As I1 is being oxidized, N, I3, and more I2 are also formed as oxidation proceeds.
An interesting observation about the formation and further oxidation of I1, seen under both conditions in Fig. 5, is that the rate of formation of I1 in a bimolecular reaction from a relatively high concentration of an unstructured ensemble (1S) is faster than the rate of further oxidation of a relatively lower concentration of I1 compared to 1S to form I2 and I3. RNase A exhibits the opposite behavior in that its first structured species, des-[40–95] and des-[65–72] (1 and 2) appears more slowly in a unimolecular reshuffling rate-determining step than its more rapid oxidation in a bimolecular step to N. As a result, the observed populations of these structured species in RNase A are very small because, once they are formed in the rate-determining steps, they are subsequently oxidized very rapidly. Because of the rapid formation of I1 in ONC and its slower subsequent oxidation, the structured intermediates dominate the population of intermediate species during the folding pathway and play more of a role in the oxidative folding mechanism.
Effects of Temperature on the Oxidative Folding Pathway
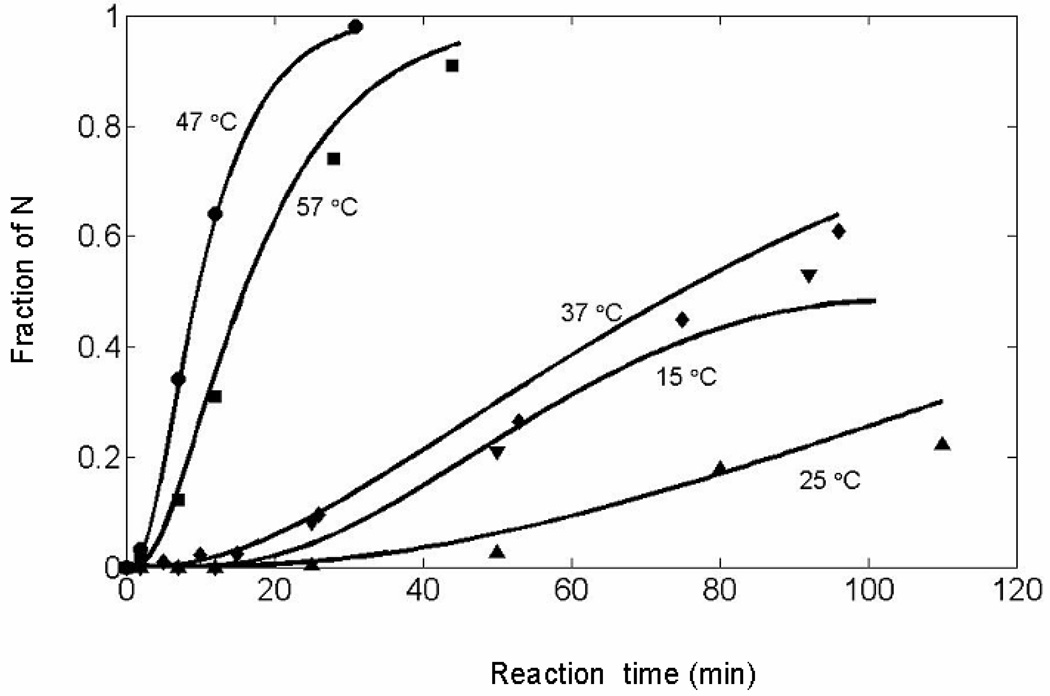
The oxidative folding of ONC was studied at various temperatures, 15 ° to 57 °C (Table 3) in order to identify the key steps that drive the formation of structure. By performing oxidative folding at higher temperatures, the less stable species would be destabilized and only the more stable ones would drive the folding. The effects of different temperatures on the recovery of ONC are shown in Fig. 8. Aside from the decrease in rate between 15 °C and 25 °C, the rate of formation of N increases with temperature until 47 °C and, at 57 °C, the rate decreases because of thermal destabilization of intermediate species such as the 1S ensemble and I1.
Table 3.
Observed rate constants for each step in the oxidative folding mechanism at different temperatures at pH 8.0
| k (M−1 min−1) | |||||
|---|---|---|---|---|---|
| 15 °C | 25 °C | 37 °C | 47 °C | 57 °C | |
| k1 | 26 ± 1.3 | 14.0 ± 0.14 | 26 ± 2.2 | 40.0 ± 0.8 | 53.0 ± 0.46 |
| k2 | 160 ± 43 | 2770 ± 51 | 6800 ± 782 | - | - |
| k3 | 0.67 ± 0.01 | 0.19 ± 0.01 | - | - | - |
| k4 | 70 ± 15 | 14 ± 4.3 | - | - | - |
| k5 | 1.94 ± 0.04 | 0.62 ± 0.01 | 0.83 ± 0.01 | 14.0 ± 0.20 | 3.10 ± 0.01 |
| k6 | 130 ± 34 | 45 ± 7.5 | 2 ± 1.8 | - | - |
| k7 | 4.70 ± 0.9 | 1.00 ± 0.07 | - | - | - |
| k8 | 1.08 ± 0.08 | 1.28 ± 0.02 | 12 ± 1.6 | 85.00 ± 0.01 | 78.00 ± 0.01 |
| k9 | 1.15 ± 0.03 | 0.39 ± 0.03 | 0.3 ± 0.12 | - | - |
| k10 | 0.64 ± 0.03 | 0.59 ± 0.01 | 2.2 ± 0.3 | 5.80 ± 0.09 | 5.90 ± 0.01 |
| k11 | 0.24 ± 0.03 | 0.28 ± 0.01 | 18.0 ± 0.8 | - | - |
Figure 8.
Recovery of N-ONC at different temperatures, 15 °, 25 °, 37 °, 47 ° and 57 °C at pH 8.0 at each temperature (25 mM DTTox, 16 µM DTTred).
The rate of formation of N decreases as the temperature increases from 15 °C to 25 °C because undetermined interactions in R and/or 1S are stabilized at the lower temperature, as indicated by considering the following equilibrium:
| (23) |
with equilibrium constant,
| (24) |
The value of K decreases from 0.0024 at 15 °C to 7.0 × 10−5 at 25 °C, and increases to 0.0013 at 37 °C. Even though K is less at 37 °C than at 15 °C, the greater rate of formation of N at 37 °C is due to the larger rate constant, k8, at 37 °C compared to 15 °C, by a factor of 10 for the formation of I2 from I1 (Also, see Fig. 9).
Figure 9.
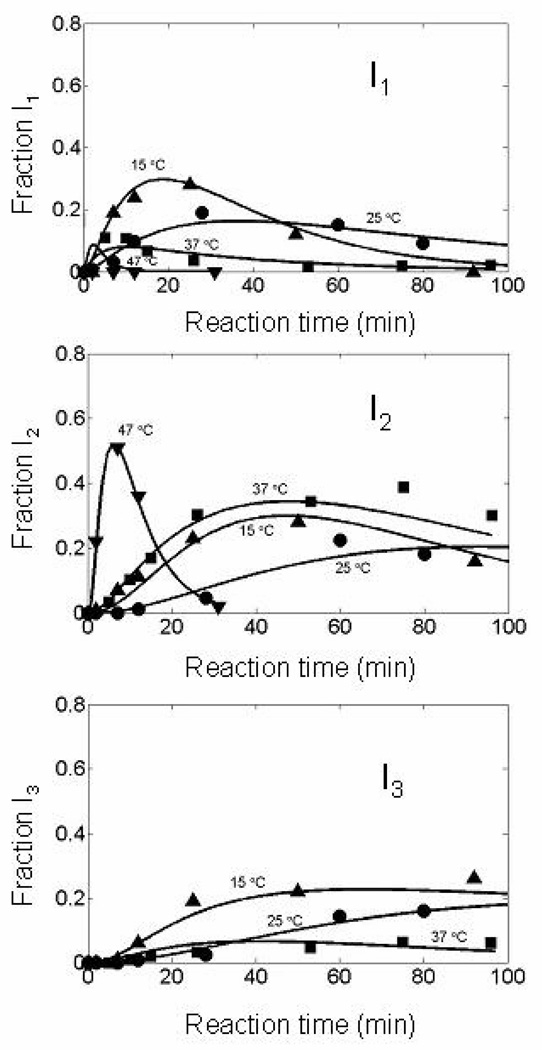
Kinetic fate of the structured intermediates at different temperatures [15 ° (triangles), 25 ° (circles), 37 ° (squares), and 47 °C (upside-down triangles)] under the same conditions as in Fig. 8. Corresponding observed rate constants are shown in Table 3. As the temperature is increased, I1 becomes more reactive to DTTox and is oxidized to I2 faster than to I3 to form N-ONC. I1 and I3 are populated to a very low extent at higher temperatures.
At temperatures higher than 37 °C, the distribution of species changes so that only species that are stable at higher temperatures are populated to recover N-ONC. At these temperatures, these fewer species fold faster.
The oxidation of I1 to form I2 (with rate constant k8) preferentially over I3 (with rate constant k9) is observed in the formation of N-ONC at higher temperatures (Table 3). The rate constants k8 and k9 refer to the oxidation of I1 to I2 and I3, respectively. From 15 °C to 47 °C, the ratio k8/k9 increases considerably, favoring the formation of I2 relative to I3.
Thermodynamic Stability of Intermediates I1, I2, I3 and N
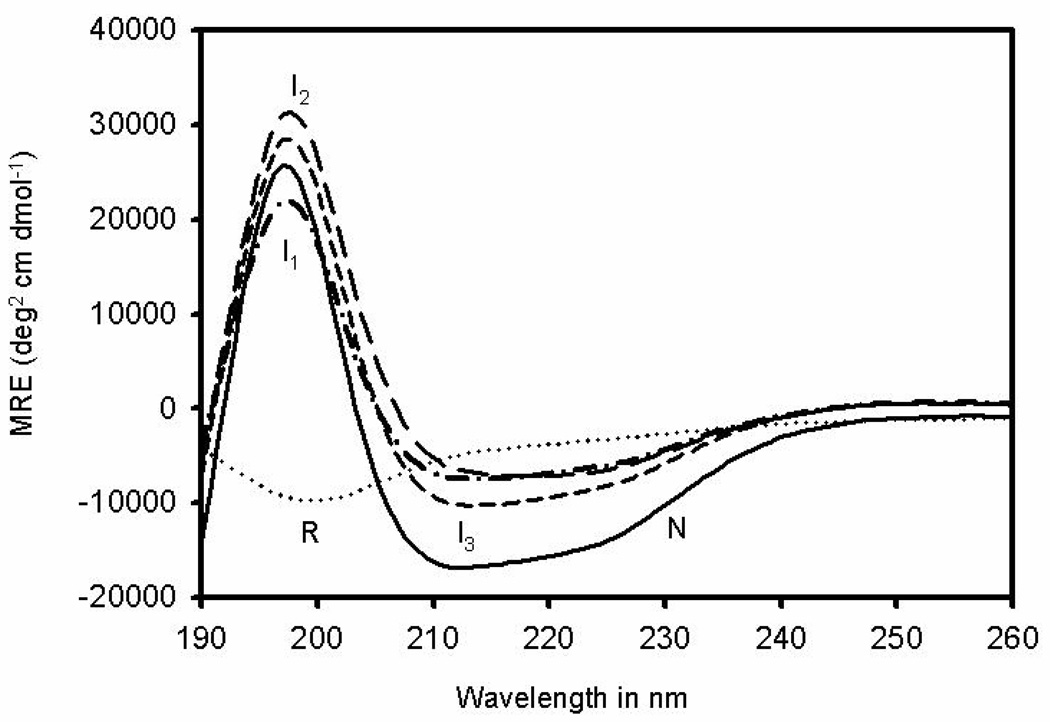
The extent of global folding of I1, I2, I3 and N was determined by far-UV circular dichroism spectroscopy, a probe for secondary structure. The spectra, shown in Fig. 10, are compared to that of the unfolded species R-ONC. Each of the intermediates is observed to have a degree of native-like structure as indicated by the maxima at 198 nm and the broad minima from 208 to 222 nm. Even the two-disulfide-bond intermediate, I1, contains a substantial amount of secondary structure. The broad minima from 208 to 222 nm for the intermediates are not as substantial, compared to N-ONC, which indicates that there is only partial native structure.
Figure 10.
Far-UV CD spectra of different species populated along the oxidative folding pathway at 25 °C and pH 3.0. Unblocked R-ONC, I1 – AEMTS blocked, I2 – AEMTS blocked, I3 – AEMTS blocked, N-ONC
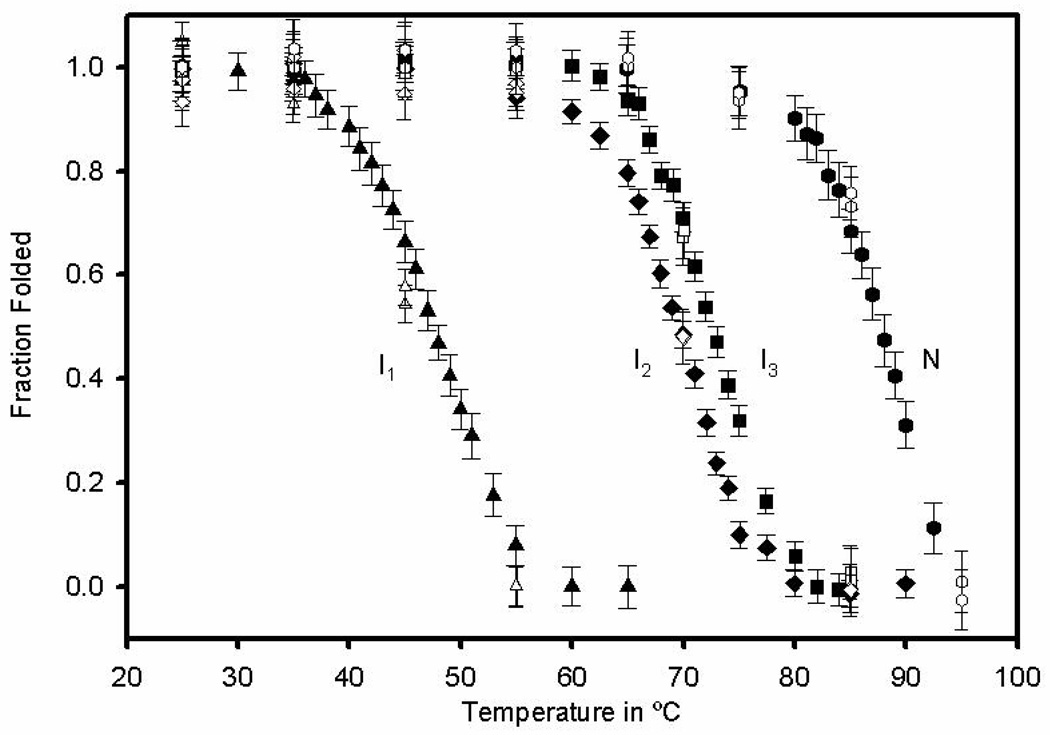
To measure the thermodynamic stability of these partially folded intermediates, temperature denaturation experiments were carried out by monitoring the loss of structure at 198 nm. The results are shown in Fig. 11. The solid characters were used to extract thermodynamic data and the unfilled characters show heating and cooling curves to demonstrate reversibility. The remarkable stability of ONC can be observed when comparing thermodynamic data for ONC with existing thermodynamic data for RNase A (12, 13, 27, 28). The results are summarized in Table 4. Essentially, the three-disulfide-bond intermediates in ONC (I2 and I3) at pH 3.0 are as stable (ΔGunf ~ 10 kcal mol−1) as native RNase A at pH 4.6 with four disulfide bonds. The two-disulfide-bond intermediate, I1, at pH 3.0 in ONC is as stable as the three-disulfide bond intermediates in RNase A at pH 4.6 as indicated by similar magnitudes of their melting temperatures, Tm, and their free energies of unfolding. The stability of RNase A decreases when the pH is reduced from 4.6 to 3.0 (28).
Figure 11.
Temperature denaturation at pH 3.0 of different species populated along the oxidative folding pathway. I1 – AEMTS blocked (triangles). I2 – AEMTS blocked (diamonds). I3 – AEMTS blocked (squares). N-ONC (hexagons). Unfilled characters correspond to heating and cooling curves of each of the species.
Table 4.
Thermodynamic data obtained from temperature denaturation of each species, and compared to similar data for RNase A
| Tm (°C) | ΔGunf (kcal mol−1 at 25 °C) | |
|---|---|---|
| WT RNase A | 47a | 3a |
| WT RNase A | 56 ± 0.2b | 10.7 ± 0.2b |
| des-[65–72] – RNase A | 39 ± 0.2c | 3.5 ± 0.4c |
| des-[40–95] – RNase A | 34 ± 0.2d | 2.0 ± 0.1d |
| WT ONC | 94 ± 4.4e | 14.0 ± 0.5e |
| I1- des-[19–68, 30–75] – ONC | 45 ± 2.6e | 4.1 ± 0.2e |
| I2 - des-[30–75] – ONC | 71 ± 3.5e | 9.8 ± 0.9e |
| I3 - des-[19–68] – ONC | 76 ± 7.7e | 9.5 ± 0.8e |
The free energy of unfolding, ΔGunf, was calculated from equation (18), with the terms, fU and ff, determined from the CD signal, compared to the CD signals for the folded and unfolded protein, at each temperature. By plotting ΔGunf against T, -ΔSunf and ΔHunf were evaluated as the slope and intercept, respectively, of this plot. The transition temperature, Tm, was evaluated as the temperature at which ΔGunf = 0 (eq. 20).
Discussion
Comparison of Oxidative Folding Rates of ONC and RNase A
The oxidative folding of ONC to recover its biologically active form is faster than that of its structural homolog, RNase A. By avoiding formation of 3S and 4S ensembles, present in RNase A, ONC can explore conformational space more efficiently. This could be facilitated by interactions which lead to very stable intermediates compared to those of RNase A. These interactions could also account for the preferential populations of key species at higher temperatures, such as I1 and I2, Fig. 9.
Another aspect of the more efficient folding mechanism of ONC is the presence of a two-disulfide-bond intermediate, I1. While it is not as stable as a three-disulfide–bond species, it aids in attaining three-disulfide bond intermediates (I2 and I3) from an unstructured 1S ensemble. In RNase A, the structured species, des-[40–95] and des-[65–72], are populated from a 3S ensemble by a reshuffling reaction. In ONC, by utilizing a 1S → I1 → I2 + I3 pathway, the search for a structured species (I1) from an unstructured ensemble involves roughly 15 times fewer species; i.e., rather than searching for 2 species out of 420 theoretical ones in the 3S ensemble of RNase A, the structured species, I1 in ONC, is formed by oxidation of either of two 1S species out of 28 theoretical ones. In addition, the native disulfide bonds in I1 are [87–104] and [48–90]. The [87–104] disulfide bond connects the shortest portion of the peptide chain and, thus, incurs the lowest entropic penalty in loop formation of ONC compared to the other disulfide bonds. Also, there are hydrophobic contacts among residues 86–104 (with hydrophobic residues in boldface type: fcvtcenqapvhfvgvgsc) at the C-terminus that could possibly form stable contacts without the need for a disulfide bond (29). In RNase A, the [65–72] disulfide bond is the first one to form, despite the fact that within this loop there are no hydrophobic residues. The non-native disulfide bond, [58–65], with similar loop size, is also populated in the early stages of oxidative folding (4), but not to the same extent as the [65–72] disulfide bond. Both of these disulfide bonds form the same size loop; so presumably, the favorable enthalpic interactions in the native [65–72] disulfide loop account for why this disulfide bond is preferred in a ratio of 4:1 (4) in the oxidative folding pathway over the [58–65] disulfide bond.
In addition, these nonpolar residues could be responsible for the hydrophobic stabilization at higher temperatures, and the increase in folding rate above 25 °C, in contrast to the decrease observed in RNase A, Table 5. In RNase A, the oxidative folding rate slows down as the temperature is increased. It is known that hydrophobic interactions are stabilized as the temperature increases (30 – 32).
Table 5.
Rate of oxidative folding of RNase A at different temperatures, pH 8.0, obtained from Ref. 11
| k(× 104 min−1)a | T in °C |
|---|---|
| 7.2 ± 0.5 | 15 |
| 12.5 ± 0.6 | 25 |
| 1.3 ± 0.2 | 37 |
The rate of oxidative folding of RNase A can be approximated by a first-order rate equation, ln[1-N] = - kt, to which these rate constants correspond.
Other hydrophobic interactions could also influence the preferential oxidation of I1 to I2 over I3 at elevated temperatures. To form I2 from I1, residues Cys19 and Cys68 must be oxidized to form the [19–68] disulfide bond, and to from I3 from I1, residues Cys30 and Cys75 must be oxidized to form the [30–75] disulfide bond. Therefore, the preference to form I2 or I3 is based on the penalty of forming or not forming the [19–68] or [30–75] disulfide bond, respectively. The [19–68] disulfide bond is buried with other hydrophobic residues, according to the structure in Fig. 1. However, the [30–75] disulfide bond is almost 4 times more exposed than the other disulfide bonds in RNase A (33). Therefore, at higher temperatures, greater hydrophobic tendencies could be responsible for oxidizing the Cys19 and Cys68 instead of residues Cys30 and Cys75.
Role of disulfide bonds in stabilizing intermediates
After considering the thermodynamic stability of the structured intermediates and N-ONC, the origin of the increased stability can be accounted for in terms of the entropy of formation of overlapping loops (34) by eq. 23,
| (25) |
where m is the number of disulfide-bonded loops in a particular species of ONC, a is the length of a chain element, and |C| is the determinant of a matrix whose elements depend on the size of the overlapping loops. If the addition of the disulfide bond does not affect the three-dimensional structure of the folded state of a species, then the addition of the disulfide bond stabilizes the protein by decreasing the entropy of the unfolded state. The extent that the unfolded state is destabilized, ΔGU = ΔΔGunf, can be taken as the difference between the free energy of unfolding of a protein with a disulfide bond, minus the free energy of unfolding of a protein without the disulfide bond. The temperature at which to calculate this difference must be chosen so that one protein is completely unfolded and the other is completely folded at this temperature.
This observed destabilization free energy, ΔGU, can be compared to the calculated destabilization free energy by multiplying the differences in the entropy loss due to disulfide bond formation of two species, ΔΔSx, by the same temperature to obtain a theoretical free energy of unfolding. The calculated destabilization entropy, ΔSx of each of the structured intermediates, I1, I2, I3 and N was calculated and is shown in Table 6. These values of ΔSx were used to compute ΔΔSx which, multiplied by the appropriate T from Fig. 11, led to the calculated values of ΔGU shown in Table 6. These calculated destabilization free energies were compared to the observed destabilization free energies for these transitions.
Table 6.
Free energy penalty, ΔGU, from formation of a disulfide bond calculated from theory and compared to experiment.
| ΔSxa (kcal mol−1 K−1) | ||
|---|---|---|
| I1 | −0.033 | |
| I2 | −0.050 | |
| I3 | −0.051 | |
| N | −0.065 | |
| Loop Formation | ΔGU (kcal mol−1) | |
| Theoretical Valueb | Experimental Valuec | |
| I1 to I2 60 °C | 5.69 | 5.11 ± 0.8 |
| I1 to I3 60 °C | 5.99 | 6.01 ± 0.8 |
| I2 to N 80 °C | 5.29 | 4.87 ± 0.9 |
| I3 to N 80 °C | 4.94 | 3.97 ± 0.9 |
| I1 to N 60 °C | 10.66 | 10.18 ± 0.5 |
Theoretical calculation of the entropy loss due to loop closure for each intermediate according to ref. (34)
Theoretical ΔGU = -TΔΔSx at 60 ° or 80 °C and ΔΔSx according to the loop formation indicated
Comparison of the calculated and observed destabilization free energies, ΔGU, of Table 6 indicates that the stability of each of the intermediates relative to I1 is due mostly to the entropic destabilization of the unfolded state of each of I1, I2, and I3 by the addition of a disulfide bond. In other words, I1 has a substantial amount of native-like structure which is further stabilized by the addition of native disulfide bonds of I2, I3, and N. Therefore, the formation of I1 plays an important part in stabilization of the native structure in ONC because most of the necessary interactions to form the biologically active structure have already formed in I1. The folding mechanism of ONC is contingent upon forming structured species that oxidize to N, and by forming an intermediate early in the oxidative folding mechanism that contains the necessary structure to obtain the biologically active form. Therefore, the oxidative folding pathway of ONC becomes more efficient than that of RNase A.
Conclusions
This study has demonstrated how the oxidative folding mechanism of ONC is different from that of RNase A, a structural homolog. ONC is more efficient at recovering biologically-active structure than RNase A. ONC has more stabilizing interactions that restrict the conformational search to 1S and 2S ensembles without including the unstructured 3S and 4S ensembles, which would increase the number of non-native disulfide bonds. The cysteines that form the [87–104] and [48–90] disulfide bonds in I1 are most likely aligned by hydrophobic interactions, as seen in the crystal structure. Further evidence for the importance of these hydrophobic interactions is the observation that the rate of folding increases with temperature as a combination of increasing reaction rate with temperature plus increased stabilization of hydrophobic interactions. At higher temperatures, the intermediate I2 is preferentially formed over I3 presumably because of the hydrophobic residues surrounding Cys19 and Cys68 in I1, which drive their oxidation. The addition of disulfide bonds also provides additional stability by decreasing the entropy in the unfolded state. Most of the native structure in N is formed in I1, and biologically-active stability is obtained for N by destabilizing the unfolded state of I1 with native disulfide bonds. From this kinetic and thermodynamic study of the oxidative folding of ONC, we have learned that the sequence of ONC, when compared to that of RNase A, exhibits increased folding efficiency and thermodynamic stability. This comparison of two structurally homologous proteins shows how sensitive protein-folding mechanisms are to details of the amino acid sequence.
Acknowledgements
We thank R. J. Youle for providing the cDNA of ONC, and B. R. Crane for access to his CD spectrometer. This work was supported by NIH grant GM-24893.
Footnotes
This work was supported by NIH grant GM-24893
Abbreviations: ONC, frog onconase (Rana pipiens); R-ONC, onconase with all its disulfide bonds reduced; RNase A, bovine pancreatic ribonuclease A; AEMTS, 2-aminoethylmethylthiosulfonate; DTTox and DTTred, oxidized and reduced dithiothreitol, respectively; GdnHCl, guanidine hydrochloride; Tris, tris(hydroxymethyl)aminomethane; HEPES, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid); DTNB - 5, 5'-dithiobis-2-nitrobenzoic acid; TNB- 2-nitro-5-mercaptobenzoic acid; NTSB- disodium 2-nitro-5-thiosulfobenzoate; MRE, mean residue ellipticity; TFA, trifluoroacetic acid; SCX, strong cation-exchange; RP, reversed phase; HPLC, high performance liquid chromatography; des-[a–b, c–d], a species that contains all native disulfide bonds except for the a–b and c–d disulfide bonds; nS, an unstructured ensemble of disulfide-containing intermediates each having n disulfide bonds; nS* a single structurally folded intermediate species containing n native disulfide bonds; N, native onconase.
References
- 1.Rothwarf DM, Li Y-J, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A: identification of two nativelike three-disulfide intermediates involved in separate pathways. Biochemistry. 1998;37:3760–3766. doi: 10.1021/bi972822n. [DOI] [PubMed] [Google Scholar]
- 2.Rothwarf DM, Li Y-J, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A: detailed kinetic analysis of two independent folding pathways. Biochemistry. 1998;37:3767–3776. doi: 10.1021/bi972823f. [DOI] [PubMed] [Google Scholar]
- 3.Leland PA, Raines R. Cancer chemotherapy – ribonucleases to the rescue. Chem. Bio. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X, Rothwarf DM, Scheraga HA. Nonrandom distribution of the one-disulfide intermediates in the regeneration of ribonuclease A. Biochemistry. 1996;35:6406–6417. doi: 10.1021/bi960090d. [DOI] [PubMed] [Google Scholar]
- 5. [5/28/2008]; Press Release, http://www.ir-site.com/alfacell/press.html.
- 6.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol. Lett. 2006;28:1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 7.Ardelt W, Shogen K, Darzynkiewicz Z. Onconase and amphinase, the antitumor ribonucleases from Rana pipiens oocytes. Curr. Pharm. Biotechnol. 2008;9:215–225. doi: 10.2174/138920108784567245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notomista A, Catanzano F, Graziano G, Dal Piaz F, Barone G, D’Alessio G, Di Donato A. Onconase: an unusually stable protein. Biochemistry. 2000;39:8711–8718. doi: 10.1021/bi000415x. [DOI] [PubMed] [Google Scholar]
- 9.Lee JE, Raines R. Contribution of active-site residues to the function of onconase, a ribonuclease with antitumoral activity. Biochemistry. 2003;42:11443–11450. doi: 10.1021/bi035147s. [DOI] [PubMed] [Google Scholar]
- 10.Rothwarf DM, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A. 1. Steady-state distribution. Biochemistry. 1993;32:2671–2679. doi: 10.1021/bi00061a027. [DOI] [PubMed] [Google Scholar]
- 11.Rothwarf DM, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A. 4. Temperature dependence of the regeneration rate. Biochemistry. 1993;32:2698–2703. doi: 10.1021/bi00061a030. [DOI] [PubMed] [Google Scholar]
- 12.Shimotakahara S, Rios CB, Laity JH, Zimmerman DE, Scheraga HA, Montelione GT. NMR structural analysis of an analyog of an intermediate formed in the rate-determining step on one pathway in the oxidative folding of bovine pancreatic ribonuclease A: automated analysis of 1H, 13C, and 15N resonance assignments for wild-type and [C65S, C72S] mutant forms. Biochemistry. 1997;36:6915–6929. doi: 10.1021/bi963024k. [DOI] [PubMed] [Google Scholar]
- 13.Laity JH, Lester CC, Shimotakahara S, Zimmerman DE, Montelione GT, Scheraga HA. Structural characterization of an analog of the major rate-determining disulfide-folding intermediate of bovine pancreatic ribonuclease A. Biochemistry. 1997;36:12683–12699. doi: 10.1021/bi970878b. [DOI] [PubMed] [Google Scholar]
- 14.Gahl RF, Narayan M, Xu G, Scheraga HA. Dissimilarity in the oxidative folding of onconase and ribonuclease A, two structural homologs. Protein Eng., Des. Sel. 2008;21(4):223–231. doi: 10.1093/protein/gzm093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. Role of the N terminus in RNase A homologues: differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J. Mol. Bio. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 16.Laity JK, Shimotakahara S, Scheraga HA. Expression of wild-type and mutant bovine pancreatic ribonuclease A in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1993;90:615–619. doi: 10.1073/pnas.90.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 18.Bates RG. Amine buffers for pH control. Ann. N.Y. Acad. Sci. 1961;92:341. doi: 10.1111/j.1749-6632.1961.tb44985.x. [DOI] [PubMed] [Google Scholar]
- 19.Lide David R., editor. CRC Handbook of Chemistry and Physics, 88th Edition (Internet Version 2008) Boca Raton, FL: CRC Press/Taylor and Francis; “Values of pH(SS) of Some Secondary Standards from Harned Cell I Measurements”. [Google Scholar]
- 20.Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes: The Art of Scientific Computing. 3rd Ed. Cambridge University Press; 2007. [Google Scholar]
- 21.Straume M, Johnson ML. Monte Carlo method for determining complete confidence probability distributions of estimated model parameters. Methods in Enzymol. 1992;210:117. doi: 10.1016/0076-6879(92)10009-3. [DOI] [PubMed] [Google Scholar]
- 22.Box GEP, Muller ME. A note on the generation of random normal deviates. Annals of Math. Stat. 1958;29:610–611. [Google Scholar]
- 23.Thannhauser TW, Rothwarf DM, Scheraga HA. Kinetic studies of the regeneration of recombinant hirudin variant 1 with oxidized and reduced dithiothreitol. Biochemistry. 1997;36:2154. doi: 10.1021/bi962340w. [DOI] [PubMed] [Google Scholar]
- 24.Thannhauser T, Konishi Y, Scheraga HA. Analysis for disulfide bonds in peptides and proteins. Methods in Enzymol. 1987;143:115–119. doi: 10.1016/0076-6879(87)43020-6. [DOI] [PubMed] [Google Scholar]
- 25.Jocelyn PC. Chemical reduction of disulfides. Methods in Enzymol. 1987;143:246–256. doi: 10.1016/0076-6879(87)43048-6. [DOI] [PubMed] [Google Scholar]
- 26.Rothwarf DM, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A. 3. Dependence on the nature of the redox agent. Biochemistry. 1993;32:2690–2697. doi: 10.1021/bi00061a029. [DOI] [PubMed] [Google Scholar]
- 27.Harrington WF, Schellman JA. Evidence for the instability of hydrogen-bonded peptide structures in water, based on studies of ribonuclease and oxidized ribonuclease. C. R. Trav. Lab. Carlsberg [Chim] 1956;30:21–43. [PubMed] [Google Scholar]
- 28.Hermans J, Jr, Scheraga HA. Structural studies of ribonuclease. V. Reversible change of configuration. J. Am. Chem. Soc. 1961;83:3283–3292. [Google Scholar]
- 29.Matheson RR, Jr, Scheraga HA. A method for predicting nucleation sites for protein folding based on hydrophobic contacts. Macromolecules. 1978;11:819–829. [Google Scholar]
- 30.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 31.Nemethy G, Scheraga HA. The structure of water and hydrophobic bonding in proteins. II. A model for the thermodynamic properties of aqueous solutions of hydrocarbons. J. Chem. Phys. 1962;36:3401–3417. [Google Scholar]
- 32.Nemethy G, Scheraga HA. The structure of water and hydrophobic bonding in proteins. III. The thermodynamic properties of hydrophobic bonds in proteins. J. Phys. Chem. 1962;66:1773–1789. [Google Scholar]
- 33.Narayan M, Xu G, Ripoll DR, Zhai H, Breuker K, Wanjalla C, Leung HJ, Navon A, Welker E, McLafferty FW, Scheraga HA. Dissimilarity in the reductive unfolding pathways of two ribonuclease homologues. J. Mol. Bio. 2004;338:794–809. doi: 10.1016/j.jmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Lin SH, Konishi Y, Denton ME, Scheraga HA. Influence of an extrinsic cross-link on the folding pathway of ribonuclease A. Conformational and thermodynamic analysis of cross-linked (lysine7-lysine41)-ribonuclease A. Biochemistry. 1984;23:5504–5512. doi: 10.1021/bi00318a019. [DOI] [PubMed] [Google Scholar]