Abstract
Both dramatic and subtle morphogenetic movements are of paramount importance in molding cells and tissues into functional form. Cells move either independently or as populations and the distance traversed by cells varies greatly, but in all cases, the output is common: to organize cells into or within organs and epithelia. In the developing Drosophila eye, a highly specialized, 90° rotational movement of subsets of cells imposes order by polarizing the retinal epithelium across its dorsoventral axis. This process was proposed to take place in two 45° steps, with the second under control of the gene nemo (nmo), a serine/threonine kinase. While our analysis confirms that these subsets of cells, the ommatidial precursors, do stall at 45°, we demonstrate that nmo is also required through most of the first 45° of rotation to regulate the speed at which the ommatidial precursors move. In addition, although the precursors reach only the halfway point by the end of larval life, this work demonstrates that patterning events that occur during pupal life move the ommatidial units an additional 15°. A re-analysis of nmo mosaic clones indicates that nmo is required in photoreceptors R1, R6 and R7 for normal orientation. This work also demonstrates that two major isoforms of nmo rescue the nmoP1 phenotype. Finally, a dominant modifier screen of a nmo misexpression background identified genomic regions that potentially regulate rotation. The results presented here suggest a model in which a motor for rotation is established in a nemo-dependent fashion in a subset of cells.
Background
Throughout the development of multicellular organisms, cells interpret molecular signals to evaluate their spatial coordinates within the organism. They use this information to sculpt cells into tissues and organs by initiating the appropriate programs for differentiation. In many cases, this level of organization is achieved by tissue polarity, or planar cell polarity, a process that organizes cells within the plane of an epithelial sheet. A set of core tissue polarity genes directs this process and employs a host of genes and programs to both execute and fine-tune the events that set up polarity in distinct tissues. In the Drosophila eye, signaling and cytoskeletal regulatory programs must be invoked to choreograph a tightly regulated series of differentiation events and morphogenetic movements necessary to polarize the tissue.
Between 750–800 ommatidia, or unit eyes, comprise the adult Drosophila compound eye (Fig. 1A). At the core of each 20 cell ommatidium lies a cohort of eight photoreceptor cells (R1–R8) that can be identified based on their unique and stereotyped position within the trapezoid (Fig. 1B, B′). The light-sensitive organelles of the photoreceptors, the rhabdomeres, are the most prominent features of the photoreceptor cells in tangential sections of adult eyes (Fig. 1B, B′). The rhabdomeres, which appear as circles in cross-section, form a re-iterative pattern of trapezoids across the epithelium. This pattern of trapezoids is reflected in mirror symmetric fashion across the dorsoventral midline, or equator (Fig. 1A, yellow line): in the dorsal half of the eye, the point of the trapezoidal array of rhabdomeres orients northward, and in the ventral half of the eye, the point orients southward.
Figure 1. Drosophila retinal tissue polarity arises in the eye imaginal disc.
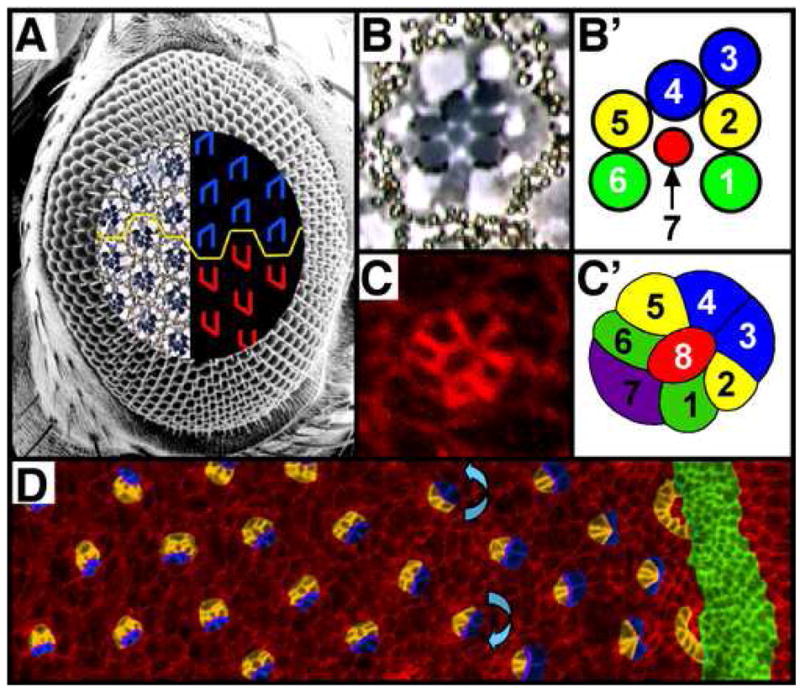
(A) Scanning electron micrograph of an adult Drosophila compound eye. The embedded tangential demi-section and corresponding schematic reveal the underlying cellular morphology of the eye. The rhabdomeres form chiral trapezoids (B, B′), shown schematically in blue (A: dorsally-oriented trapezoids) and red (A: ventrally-oriented trapezoids). The dorsal and ventral forms are separated by a horizontal line of mirror symmetry, the equator (yellow line). (C) Ommatidial precursor in eye imaginal disc with full complement of photoreceptor cells and (C′) accompanying schematic. (D) Eye imaginal disc from third instar larva immunostained with α-Arm to outline apical profiles of cells. Image is pseudocolored to highlight the morphogenetic furrow (green), arcs and photoreceptors R8, R2, R5, R1, R6 and R7 (orange) and R3 and R4 (blue). Blue arrows indicate the direction of rotation, which is opposite on opposite sides of the future equator. Anterior is to the right.
This symmetry is established during larval life, in the precursor tissue to the adult eye, the eye imaginal disc. Patterning in the eye disc propagates from posterior to anterior, following a front of differentiation that is demarcated by the morphogenetic furrow, an apical groove in the epithelium (Ready et al., 1976) (Fig. 1D, green). The key event in the establishment of polarity in the Drosophila eye is the process known as ommatidial rotation, in which the photoreceptor precursors (Fig. 1C, C′), and eventually the non-neuronal cone cells, rotate 90°, independently of their undifferentiated neighbors (Fig. 1D; reviewed in (Wolff and Ready, 1993); (Fiehler and Wolff, 2007)). The ommatidial precursors turn counterclockwise in the dorsal half of the eye and clockwise in the ventral half of the eye (anterior is defined as pointing to the right).
The mechanisms that drive the movement of rotating cells are unclear, however, several genes have been implicated in regulating rotation. These “rotation genes” differ from the tissue polarity genes in that they impact only the degree to which ommatidia rotate, not cell fates and chirality, as do the tissue polarity genes. Among these rotation-specific genes are scabrous (sca), members of the EGF signaling pathway, zipper and nemo (nmo) (Brown and Freeman, 2003; Choi and Benzer, 1994; Chou and Chien, 2002; Fiehler and Wolff, 2007; Freeman et al., 1992; Gaengel and Mlodzik, 2003; Lee et al., 1996; Mirkovic et al., 2002; Strutt and Strutt, 2003).
nmo mutant ommatidia fail to complete rotation, instead stopping prematurely at the halfway point, or 45°, whereas mutations in JNK pathway members have a tissue polarity phenotype, including rotation errors ((Choi and Benzer, 1994; Fanto et al., 2000; Mihaly et al., 2001; Strutt et al., 1997)). Although these two phenotypes are not identical, several lines of evidence link nmo to the Jun N-terminal kinase (JNK) signaling pathway as well as the Wnt/Wg signaling pathway. First, its molecular structure suggests a role for Nmo in JNK signaling: JNK is a MAP kinase and nmo encodes a serine/threonine kinase similar to members of the MAP kinase pathway (the MEKs) (Choi and Benzer, 1994). Second, members of the JNK signaling pathway modify nmo misexpression phenotypes in the wing (Mirkovic et al., 2002), whereas in the eye, misexpression of dTAK, a MAPKKK upstream of JNK, results in defects in rotation and chirality (Mihaly et al., 2001; Takatsu et al., 2000); this phenotype is dominantly suppressed by the loss of one copy of nmo (Mihaly et al., 2001). Third, in the Drosophila wing, nmo acts in a feedback loop to regulate Wg activity (Zeng and Verheyen, 2004). Finally, studies in worms and mice also link nmo to the Wnt/Wg and JNK signaling pathways: nmo plays a conserved role downstream of dTak to antagonize Wnt/Wg signaling (Ishitani et al., 2003; Ishitani et al., 1999; Kaletta et al., 1997; Meneghini et al., 1999; Rocheleau et al., 1999; Shin et al., 1999).
The observation that nmo mutant ommatidia rotate just 45° led to the hypotheses that rotation is a two-step process and that nmo is required for the second, but not the first, step (Choi and Benzer, 1994). Here, we present data that nmo in fact does play a role in the “first step:” our data show that nmo contributes to rotation, beginning at ~7° of rotation, by increasing the rate at which ommatidial precursors turn. Our data also support the finding of Choi and Benzer (1994) that nmo is essential for the second 45° of rotation. Furthermore, data presented here indicate that while there is a slowdown in the rate of rotation in wild type at roughly 45°, there is not a distinct pause, as previously reported (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003). We also report a requirement for Nmo in R1, R6 and R7 and propose a model that suggests nmo is required to either establish or move the motor for rotation to cells that comprise the “rotation interface,” including R1, R6, R7 and the cone cells. Finally, while the precise molecular pathway through which nmo acts to regulate rotation remains unclear, we have been unable to implicate the Wnt signaling pathway in nmo regulation of rotation but we do confirm previous reports that nmo interacts with the JNK signaling pathway (Mihaly et al., 2001).
Materials and Methods
Genetics and Deficiency screen
Fly strains used: Canton S, w1118, nmoP1/TM6, nmoadk1/TM6 (gift from E. Verheyen), UAS>eGFP.nmoI, UAS>eGFP.nmoII, UAS>nmoII (gift from E. Verheyen), nmoadk1, FRT80B/TM6B (gift from E. Verheyen), P(w+)70C, zip1/CyO, sev>GAL4, arm1/FM7a, fzR54/CyO, mδ0.5, argosrlt, bsk1/CyO, msn102/TM6, hepr75/FM7C, and pksple/CyO. nmoP1 is not a molecular null allele ((Zeng and Verheyen, 2004), and this work, Supp. Fig. 1), however, it is a reduced function allele that behaves as a genetic null with respect to rotation, as the phenotype of homozygous nmoP1 eyes is identical to that of nmoP1/Df(3L)ZIP1 eyes (data not shown).
sev>nmo flies were crossed to the 250 deficiency lines spanning chromosomes 2 and 3 (Bloomington Deficiency Kit). Three eyes from different individuals were fixed and sectioned (as described in Wolff, 2000) for each cross, and the phenotypes scored. Modifiers were confirmed by scoring three additional eyes, each from independent flies.
nmo transgenic lines
The following fragments of nmo were cloned into P{UAST} containing an N-terminal GFP tag (numbers indicate amino acid number): NmoI (2-414) and NmoII (2-430). Fragments were subcloned from the cDNA clones LD36031 (nmoI) and LD42550 (nmoII) (Drosophila Genomics Resource Center, Bloomington, IN). Injections were performed using standard protocols. Expression levels were assayed by Western blot analysis using rabbit α-GFP and mouse α-Actin antibodies.
RTPCR assay
RNA was isolated from 40 pairs of third instar eye imaginal discs for each genotype using the micro-RNeasy RNA isolation kit (Qiagen). The RTPCR reaction was done using the One-Step RTPCR kit (Qiagen) using a forward (ATGTGCAAATGCTGCTTCAC) and reverse primer (TCATTTTGCCGTCATTCCCAT) encompassing the nmoI-specific exon. Control reactions were done with no RNA input and primers to actin: (forward-GGGCATGTGCAAAGCCG), (reverse-GAAGGTCTCGAACATGATCTGGG).
Immunohistology
Third instar eye imaginal discs were dissected as described (Wolff, 2000). Early pupal eyes were dissected at 18 hours (25°) after pupal formation (apf) as described (Wolff, 2000). All primary antibody incubations were done at 4°C overnight at the following concentrations: 1:10 for α-Armadillo (mouse monoclonal, Developmental Studies Hybridoma Bank), 1:20 for α-Bar (rabbit polyclonal, generous gift of T. Kojima (Higashijima et al., 1992)), and 1:10 for α-ELAV (rat monoclonal, Developmental Studies Hybridoma Bank). Alexafluor-conjugated secondary antibodies (Molecular Probes) were diluted at 1:300 and incubated at room temperature for two hours. A Leica TCS SP2 confocal microscope was used to collect fluorescent images.
in situ hybridization to third instar eye imaginal discs was performed according to protocols adapted from (Tautz and Pfeifle, 1989). Nonradioactive single stranded RNA probes were made using SP6 and T7 RNA polymerase.
Phenotypic analyses
Adult eyes were fixed, embedded and sectioned according to standard protocols (Wolff, 2000). Angles were measured between two vectors, one running parallel to the equator and the second drawn through the rhabdomeres of photoreceptors (R) R1, R2, and R3, using the ImageJ software. The number of ommatidia and eyes scored is as follows: Canton S: 2103 ommatidia from 11 eyes; nmoP1: 1443 ommatidia from 11 eyes; nmoP1/Df: 1040 ommatidia from 6 eyes; UAS>eGFP.nmoII4.1;sev>Gal4;nmoP1/nmoP1: 1084 ommatidia from 9 eyes; sev>Gal4/UAS>eGFP.nmoII3.2;nmoP1/nmoP1: 691 ommatidia from 7 eyes; sev>Gal4/UAS>eGFP.nmoII4.2;nmoP1/nmoP1: 680 ommatidia from 6 eyes; sev>Gal4/UAS>nmoII;nmoP1/nmoP1: 931 ommatidia from 6 eyes; UAS>eGFP.nmoI22E;sev>Gal4;nmoP1/nmoP1: 2022 ommatidia from 12 eyes; sev>Gal4/UAS>eGFP.nmoI22B;nmoP1/nmoP1: 1059 ommatidia from 7 eyes; UAS>eGFP.nmoI10.2;sev>Gal4;nmoP1/nmoP1: 910 ommatidia from 8 eyes; UAS>nmoII4.1;sev>Gal4/UAS>nmoII;nmoP1/nmoP1: 1742 ommatidia from 11 eyes. UAS>zipDN;sev>Gal4/nmoP1: 568 ommatidia from 6 eyes.
Third instar larval eye discs were dissected and fixed as described (Wolff, 2000). Apical surfaces of cells were immunolabeled with mouse α-Armadillo antibody, as described above. Rotation angles were measured between vectors drawn parallel to the equator and running between R3/4 and through R8 using the ImageJ software. The number of ommatidia scored is as follows. w1118: row 2, 92 ommatidia; row 3, 101 ommatidia; row 4, 101 ommatidia; row 5, 101 ommatidia; row 6, 100 ommatidia; row 7, 100 ommatidia; row 8, 100 ommatidia; row 9, 99 ommatidia; row 10, 99 ommatidia; row 11, 99 ommatidia; row 12, 98 ommatidia; row 13, 98 ommatidia; row 14, 88 ommatidia; row 15, 80 ommatidia. nmoP1: row 2, 75 ommatidia; row 3, 76 ommatidia; row 4, 77 ommatidia; row 5, 77 ommatidia; row 6, 76 ommatidia; row 7, 74 ommatidia; row 8, 76 ommatidia; row 9, 75 ommatidia; row 10, 74 ommatidia; row 11, 72 ommatidia; row 12, 83 ommatidia; row 13, 76 ommatidia; row 14, 65 ommatidia; row 15, 42 ommatidia.
Pupal eyes were dissected and fixed using the same methods as described for larval eye discs. Nuclei were marked with α-ELAV and α-Bar as described above. Vectors were drawn parallel to the equator and through the R1/6 and R3/4 nuclei; angles were measured between these vectors using the ImageJ software. 282 ommatidia were scored for the 18 hr apf time point.
Mosaic Analysis
nmoadk1 clones were generated using X-ray irradiation (1000 rads) of nmoadk1/P(w+)70C second instar larvae. Mutant tissue was marked with w− and identified by the absence of pigment granules in the adult eye. Eyes were fixed and sectioned as described. The angles and genotypes of photoreceptors in 157 mosaic ommatidia from 10 clones were scored.
Results
Nemo regulates the speed of rotation
nmo mutant ommatidia were originally described as rotating just halfway through the wild-type 90° of movement before stalling at 45° (Fig. 2A,B) (Choi and Benzer, 1994). This observation led to the hypotheses that ommatidial rotation occurs in two 45° steps, that these steps can be genetically separated, and that nmo is essential for the second step (from 45° to 90°) (Brown and Freeman, 2003; Choi and Benzer, 1994; Gaengel and Mlodzik, 2003). Three alternative models for nmo function are equally plausible: 1) nmo is required for the first step (0° to 45°), and the 45° of rotation evident in nmo eyes is a consequence of the activity of the gene required for the second step; 2) Nmo regulates the speed at which ommatidia rotate, so the rate of rotation is slower in nmo mutant eyes; and 3) Nmo regulates the amount of time ommatidial precursors rotate.
Figure 2. Nmo regulates the speed of rotation.
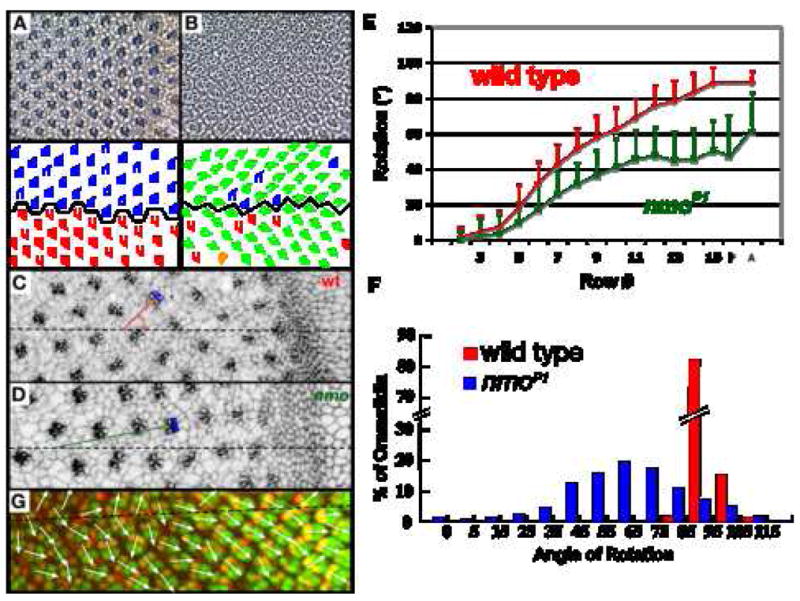
(A,B) Tangential sections of adult wildtype (A) and nmo (B) eyes and corresponding schematics. Most ommatidia in nmo mutant eyes under-rotate (green trapezoids) while occasional ommatidia over-rotate (orange trapezoid). Blue and red trapezoids denote ommatidia that rotate within one standard deviation of the average (92° +/−4). (C,D) Apical view of wild-type (C) and nmo (D) eye imaginal discs stained with α-Arm. The angle of rotation is measured by extending a line from the equator (dashed line) through a point that bisects the R3/R4 junction (R3 and R4 are highlighted in blue); the line is illustrated in red for wild type and green for nmo. (E) Rate of ommatidial rotation in wild-type (red) and nmo (green); rate is a function of distance traveled per row. P: 18 hour apf pupal eye (25°); A: adult eye. (F) Histogram illustrating the distribution of ommatidial angles in wild-type (red) and nmo (blue) adult eyes. nmo rotation angles are distributed in a bell-shaped curve with an apex at approximately 60°. (G) 18 hour pupal eye (25°) stained with α-Bar (red) and α-ELAV (green). Dashed line identifies the equator and white arrows indicate ommatidial angles. Anterior is to the right.
As a first step in distinguishing between these models, the rates of rotation in eye imaginal discs in wild type and nmoP1, a genetic null (see methods), were compared (Fig. 2). We analyzed the rotation phenotype in the nmoP1 allele rather than in one of the stronger alleles (nmoadk1, nmoadk2) because 1) the stronger alleles are lethal as homozygotes, and 2) the depth and precision of the analysis conducted here would not have been feasible in clones of the strong alleles. Importantly, the nmoP1 allele is a suitable allele for phenotypic analysis since the rotation phenotype in nmoP1 homozygous eyes is equivalent to the rotation phenotype in nmoadk1/Df (R. Fiehler, unpublished data).
In the eye disc, development proceeds as a wave that moves from posterior to anterior. As a consequence of this dynamic mode of development, successively posterior rows of ommatidial precursors represent increasingly older ommatidia. Angles of ommatidial orientation were therefore measured row-by-row in nmo eye imaginal discs and compared to equivalent rows in wild-type discs to determine the change in the average ommatidial angle from one row to the next, or the rate of ommatidial rotation. Angles between two lines, one drawn through the center of R8 and between R3 and R4 (blue cells, red/green lines, Fig. 2C,D), and the second defined by a line running parallel to the equator (dashed line, Fig. 2C,D), define the ommatidial angle. Angle measurements were grouped by row (where a row consists of contemporaneous ommatidia) and the average angle for each row determined. These data show that nmo precursors rotate at a slower rate than wild type (Fig. 2E). They also reveal that initiation of rotation is nmo-independent, whereas subsequent rotation is nmo-dependent: rotation initiates normally, but between rows 4 and 5, the nmo rate diverges from that of wild type (the average angle of rotation is significantly different between the two genotypes at row 5 (p=E-5)). For the first ~7° of the nmo-independent phase there is no overlap with the nmo-dependent phase (discussed below).
The standard deviations do not become statistically different until row 10, based on analysis using the F-test. The larger standard deviation in nmo suggests a looser regulation of the degree to which the precursors rotate. Finally, these data also confirm the report by Choi and Benzer (1994) that nmo mutant ommatidia stall at 45° (Fig. 2E).
The 45° of orientation in eye discs is discordant with our measurements of the average angle of rotation in adult nmo eyes: 61° +/−23 (Fig. 2B,E, F). The +15° change in orientation that occurs between larval and adult life (Fig. 2E) could be a consequence of any of several factors, including further, delayed rotation during pupal life or the morphological changes that occur throughout pupal life to transform the imaginal disc into a fully functional adult eye. The first major patterning events that occur in pupal life are a streamlining of the interommatidial cells from a two to three cell-thick layer to just a single layer between neighboring ommatidia, and cell death. These events occur at about 21 hours apf (Cordero et al., 2004). We cannot exclude the possibility that rotation, rather than an alternative patterning event, carries these cells from 45° to 61°. However, if the average angle increases prior to the onset of these other patterning events, delayed rotation is a likely mechanism for the additional 15° of movement. To determine if a mechanism that is unrelated to these pupal eye patterning events drives further ommatidial movement, ommatidial angles were measured in 18 hour pupal eyes (25°), a time point that precedes the onset of the major phase of cell death that participates in sorting out the pigment cell lattice (Cordero et al., 2004; Wolff and Ready, 1991a). Bar expression was used to label R1 and R6 to assign angle measurements (Fig 2G). These results reveal that the average angles in early pupal eyes are the same as in row 11 eye discs, the point at which rotation stalls in nmo discs (Fig. 2E). As stated above, while we cannot rule out the possibility that rotation underlies this post-larval movement, these data do indicate that the mechanism that moves nmo mutant ommatidia the last 15 degrees could be due to alternative patterning events, as the movement either coincides with, or begins after, cells sort into a single layer and cell death is initiated.
In summary, these data reveal both a previously undescribed role for nmo in the first 45° of rotation (regulation of the rate of rotation), as well as a second mechanism that operates during pupal life to continue to move nmo mutant ommatidia closer to the wild-type orientation of 90°. Finally, while these data support a role for nmo in the second 45 degrees of rotation, they do not distinguish between the possibilities that nmo regulates either the time or distance that ommatidial precursors rotate.
Nmo is required in R1, R6 and R7 for normal rotation
The deviation between the amount of rotation in wild type relative to nmo becomes significant at row 5. This time point correlates with the recruitment of photoreceptor cells, R1, R6, and R7 into the maturing ommatidial precursor (Fiehler and Wolff, 2007). This observation raises the intriguing possibility that photoreceptors R1, R6, R7 are important for maintaining the rate of rotation via Nmo activity. In this scenario, nmo should be required in photoreceptors R1, R6 and R7 for normal rotation. Since this hypothesis is at odds with the observation that Nmo is required in any single photoreceptor for normal rotation (Choi and Benzer, 1994), we repeated the mosaic analysis of nmo. Clones of mutant nmoadk1 tissue were generated using X-rays (Fig. 3A,B) and angles for mosaic ommatidia were measured between the equator and a line drawn through the R1, R2, R3 rhabdomeres using ImageJ software, as described earlier.
Figure 3. nmo is required in R1, R6 and R7.
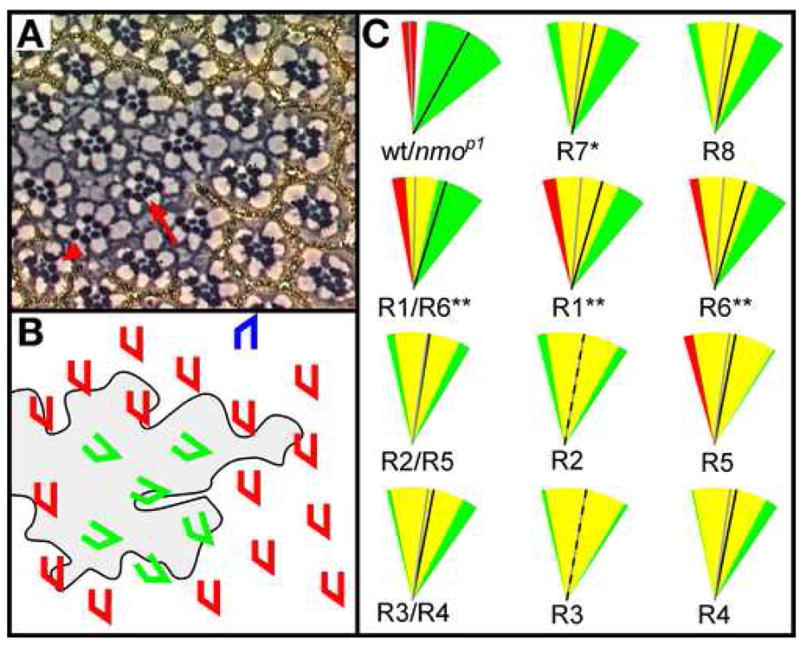
(A) Tangential section through nmoadk1 clone in adult eye. Clone is marked by w−, or the absence of pigment. nmo mutant ommatidia within the clone are mis-oriented, as are mosaic ommatidia on the clonal borders. Mosaic ommatidia with nmo− R1 and R6 photoreceptors often result in rotation defects (arrow), and the presence of nmo+ R1 and R6 photoreceptors in mosaic ommatidia (arrowhead) often improves rotation. (B) Schematic of clone shown in (A). Clonal border is outlined and clone is shaded. Phenotypically wild-type (red and blue) and mutant (green) ommatidia are identified. (C) Schematic representation of single or paired photoreceptor cell type/s and relative effect on ommatidial orientation. The standard deviations of individual and paired nmo-positive (red wedges) and nmo-mutant (green wedges) photoreceptor types are shown. Overlap of the standard deviations is shown in yellow. Black and gray lines represent the average angles of rotation for nmo and wild type, respectively. For example, the average angle of rotation for mosaic ommatidia with a genetically wild-type R1 cell, represented by the gray line, is 85°, whereas the average is 74° (black line) for mosaic ommatidia with a genetically mutant R1 cell. The distribution is skewed to the under-rotated end of the spectrum for mosaic ommatidia with a mutant R1 (green wedge) whereas the displacement of the red wedge to the left indicates a propensity to rotate further when R1 is wild type. While there is significant overlap in the two standard deviations (yellow wedge), there is a greater likelihood for ommatidia to rotate closer to wild type (gray line) when the R1 is wild type relative to when the R1 is mutant (black line). Wild type and nmo are shown in the first panel for reference. P-values (asterisks) pertain to a comparison of the nmo-positive distribution relative to the nmo-mutant distribution (*p < 0.01, **p < 0.001 in a Student’s t-test).
To determine the contribution of each photoreceptor type to the ommatidial phenotype, ommatidia were grouped into categories according to which photoreceptor type was genetically wild-type for nmo. In other words, all ommatidia with a wild-type R1 were placed into the R1+ category, regardless of the genotypes of the remaining photoreceptors. Average angles and standard deviations were calculated for each of these categories. These values were then compared to essentially equivalent groups in which ommatidia were scored for mutant, rather than wild-type, photoreceptor types. For example, the R1-positive category described above was compared to the R1-mutant category, which consists of ommatidia with a mutant R1 and photoreceptors R2 through R8 of either wild-type or mutant genotype. The positive category and mutant categories were compared using the Student’s t-test to determine if the average angle of orientation is linked to the genotype of the R1 photoreceptor (or the R2 photoreceptor, the R3 photoreceptor, etc.). Equivalent analyses were carried out for each photoreceptor type.
This comparison revealed that ommatidia with wild-type photoreceptors R1 and R6, and to a lesser extent, R7, generally rotate closer to the wild-type 90° than if these photoreceptor cells are mutant (p=6.3E-4, 5.6E-4 and 8.3E-3, respectively; Fig. 3C). P-values for the remaining photoreceptors were not significant, suggesting the genotypes of photoreceptors R8, R2, R5, R3, and R4 do not significantly influence the degree to which ommatidia rotate. Furthermore, ommatidia that have both a wild-type R1 and R6 rotate, on average, 89.5° +/− 12 whereas those with a mutant R1 and R6 rotate, on average, just 73° +/−21 (p=1.26E-5), so the combined positive effect of R1 and R6 is greater than either photoreceptor on its own (Fig. 3C). Notably, the SD also improves when both R1 and R6 are wild type in a single ommatidium. As expected, no differences are seen in parallel pairwise comparisons of R3/R4 and R2/R5. Finally, although increasing the number of nmo-positive R1, R6 and R7 cells increases the number of degrees an ommatidium rotates, a parallel effect is not observed with the remaining five photoreceptors, indicating the overall number of nmo-positive photoreceptors only improves rotation if the right subset of photoreceptors is wild type (data not shown).
These data suggest that nmo is not sufficient for normal rotation in R8, R2, R5, R3, and R4, but is required in R1 and R6, and, to a lesser extent, R7. Since R1, R6 and R7 are recruited after rotation is initiated, yet R8, R2, R5, R3 and R4 are present prior to rotation, the respective requirements for Nmo in these subsets of cells suggest a model in which R8, R2, R5, R3 and R4 initiate rotation in a nmo-independent fashion and, in combination with the rate of rotation data, that R1, R6 and R7 supplement the rate of rotation upon recruitment into the ommatidial precursor. Finally, nmo likely plays a role in other, non-photoreceptor cells, such as the cone cells or interommatidial cells, given that 1) R1, R6 and R7 do not completely rescue the phenotype, and 2) that the phenotype of ommatidia that are mutant for R1, R6 and R7 is not as strong as nmoP1.
nmoI and nmoII rescue the nmo mutant phenotype
Drosophila nmo is predicted to encode three protein isoforms (FlyBase), although it is not yet known which of these isoforms regulates ommatidial rotation. Two isoforms, NmoI and NmoII, differ only in their C-termini: the NmoI tail contains three amino acids whereas the NmoII tail contains 23 amino acids. The nmo orthologs Nlk (vertebrate) and lit-1 (C. elegans) do not have nmo I (Brott et al., 1998; Harada et al., 2002; Kaletta et al., 1997; Rocheleau et al., 1999); rather, the 3′ tails of Nlk and lit-1 are more similar to nmo II (Brott et al., 1998; Kaletta et al., 1997). The 23 amino acid tail is therefore expected to be functionally significant, perhaps in the regulation of rotation. The third predicted isoform likely represents a spurious transcript, because 1) it has an alternative start site that eliminates the ATP binding and catalysis domains and is therefore unlikely to be biologically active, and 2) it is supported by only one EST (FlyBase). The third isoform is therefore not included in the experiments that follow.
The nmo transcript is expressed at high levels in a narrow stripe posterior to the furrow (Choi and Benzer, 1994), a region that overlaps with the region where rotation occurs; the isoforms that constitute this pattern have not yet been identified. Both nmoI and nmoII are present as revealed by RTPCR of eye imaginal discs (Supplementary Fig. 1A). To determine which isoforms are expressed in this pattern, in situ hybridization was carried out. The nmoII expression pattern cannot be revealed directly because nmoII lacks a unique exon, precluding the opportunity to generate a nmoII-specific probe. Alternatively, the nmoII pattern of expression was inferred by subtracting the nmoI expression pattern from the nmoI/nmoII pattern that was revealed with a probe to an exon shared by nmoI and nmoII. This approach revealed that whereas the nmoI/nmoII probe reveals a stripe of high levels of nmoII expression posterior to the morphogenetic furrow (Supplementary Fig. 1B′), nmoI is expressed at low levels throughout the entire disc (Supplementary Fig. 1C′). Although these expression patterns suggest that nmoII regulates rotation, it is possible that the level of sensitivity of the probes is insufficient to detect the nmoI transcript, particularly in light of the RTPCR results. Furthermore, the small size of the nmoI exon (374bp) does not allow for multiple probes to confirm these results. Expression of the nmoI isoform in ommatidial precursors thus cannot be ruled out.
To identify potential differences in cellular localization between the two isoforms, transgenic lines carrying UAS>eGFP.nmoI and UAS>eGFP.nmoII were generated and expressed posterior to the furrow using sev>Gal4, which drives expression in a subset of cells behind the furrow. The two protein isoforms exhibit indistinguishable patterns of localization, indicating the 23 amino acid tail in NmoII does not regulate localization (Fig. S1D). Furthermore, GFP.Nmo is present in the nucleus (Fig. S1D, lower panels, arrowheads). One caveat to using GFP-tagged proteins to evaluate subcellular localization is that it is well-documented that a portion of an untagged GFP pool translocates to the nucleus on its own. The fusion of Nmo, a 46–48 kDa protein (FlyBase), to GFP, likely makes the macromolecule large enough to prevent diffusion through the nuclear pores (a proposed mechanism for entry of GFP into the nucleus; (Seibel et al., 2007)). If so, and GFP-tagged Nmo represents localization of endogenous protein, the nuclear localization pattern shown in Fig. S1 is consistent with the nuclear localization of Nlk and its proposed role in regulating transcription factors (Brott et al., 1998). In addition, the fact that both isoforms, neither of which has a nuclear localization signal, show nuclear localization, suggests that the mechanism for transporting Nmo into the nucleus in ommatidial cells does not discriminate between the isoforms.
As an in vivo test for the potential function of the nmoI and nmoII isoforms, UAS>GFP.nmoI and UAS>GFP.nmoII transgenes were used to rescue the nmoP1 mutant phenotype. sev>Gal4 was used to drive expression of the transgenes in an effort to approximate the endogenous pattern of nmo expression (Choi and Benzer, 1994). Whereas three nmoII transgenic lines partially rescue the mutant phenotype, improving the average angle of rotation anywhere from 21–24° (p-values range from E-90 to E-129; Table 1, Fig. 4A–C), the three nmoI lines show inconsistent rescue, as follows: two lines improve the average angle of rotation by 11° and 6° (p=E-28, p=E-12, respectively), whereas one line, 22E, almost completely rescues the average angle of rotation (p=E-200; Table 1, Fig. 4D–F). It is interesting to note that the standard deviation remains unchanged in all lines except sev>nmoI22E. Why this line fully rescues the nmoP1 phenotype is unclear, but it is unlikely that the enhanced rescue is due to higher levels of expression in this line, for two reasons. First, two copies of the nmoII transgene in a nmoP1 background provide only minimally better rescue than a single transgene (Table 1). Second, expression levels of sev>nmoI22E on Western blots are only slightly greater than those of the remaining sev>nmo lines (Supplementary Fig. 1E). The possibility that differences in activity are due to distinct subcellular localization can also be ruled out given the identical localization of all of the transgenes (Supplementary Fig. 1D). The expression and rescue results do not conclusively point towards a single transcript (nmoI or nmoI) as the essential isoform in rotation, although the ability of both isoforms to at least partially rescue the rotation phenotype rules out a role for the 23 amino acid tail in regulating rotation. Perhaps combinations of both isoforms are used in the regulation of this process.
Table 1.
nmoI and nmoII transgenes rescue the nmoP1 phenotype.
| Genotype | Angle (°) | Eyes | Ommatidia |
|---|---|---|---|
| wild type | 92 +/− 4 | 11 | 1909 |
| nmoP1 | 61 +/− 22 | 12 | 1443 |
| UAS>GFP.nmoI10.2;sev>Gal4; nmoP1 | 72 +/− 22* | 8 | 910 |
| UAS>GFP.nmoI22B/sev>Gal4; nmoP1 | 67 +/− 21* | 7 | 1059 |
| UAS>GFP.nmoI22E; sev>Gal4; nmoP1 | 92 +/− 9* | 12 | 2021 |
| UAS>GFP.nmoII4.1;sev>Gal4; nmoP1 | 82 +/− 21* | 9 | 1084 |
| UAS>GFP.nmoII3.2/sev>Gal4; nmoP1 | 85 +/− 18* | 6 | 691 |
| UAS>GFP.nmoII4.2/sev>Gal4; nmoP1 | 84 +/− 23* | 6 | 680 |
| UAS>nmoII/sev>Gal4; nmoP1 | 82 +/− 19* | 6 | 931 |
| UAS>GFP.nmoII4.1;UAS>nmoII/sev>Gal4; nmoP1 | 85 +/− 17* | 11 | 1742 |
The p-values for the rescue constructs are statistically significant (p≪0.001 t-test)
Figure 4. NmoI and NmoII function in eye development.
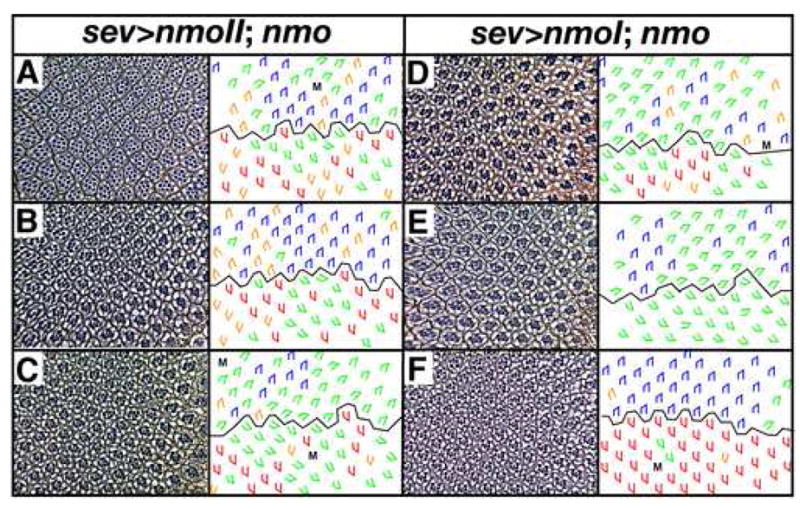
Tangential sections through adult eyes and accompanying schematics illustrate rescue/partial rescue of the nmo phenotype by two nmo transcripts. Three independent UAS>nmoII (A–C) and UAS>nmoI (D–F) transgenic lines rescue the nmoP1 phenotype to various degrees (compare to Fig. 2B). Note that rescue was also quantitated since qualitative assessment of green and orange trapezoids is not absolute (Table 1). Red, blue trapezoids: wild type; green: under-rotated ommatidia, orange: over-rotated ommatidia; M: missing photoreceptors. Anterior is to the right.
Several genes dominantly modify sev> nmoII
The molecular mechanisms and regulatory pathways by which nmo regulates development have been explored in various organisms, including vertebrates, worms and Drosophila. nmo has been demonstrated to interact with Wnt/Wg and JNK signaling in these systems. In the Drosophila wing, the juxtaposition of nmo and wg expression appears to be important for the regulation of these genes by each other in a feedback loop (Zeng and Verheyen, 2004). Genetic interactions in the fly also link nmo to the JNK signaling pathway. First, dTAK regulates JNK signaling by acting as a JNKKK, and second, it interacts with nmo in the eye (Mihaly et al., 2001; Takatsu et al., 2000). Furthermore, JNK signaling suppresses over-expression of Nmo in the wing (Mirkovic et al., 2002).
Several lines of evidence suggest that nmo interacts with JNK, but not Wnt/Wg, signaling to regulate ommatidial rotation. First, JNK acts downstream of the tissue polarity genes, which regulate ommatidial rotation, during eye development (Fanto et al., 2000; Strutt et al., 1997). Second, canonical Wg signaling has not been clearly implicated in regulating tissue polarity, suggesting it is unlikely to interact with nmo to regulate this process. Third, nmo expression does not flank wg expression as it does in the wing (Zeng and Verheyen, 2004), so the negative feedback loop postulated for Wnt/Wg and nmo expression in the wing is unlikely to drive ommatidial rotation. While these data suggest that nmo interacts with JNK, but not Wg, signaling to regulate ommatidial rotation, no direct genetic links have been made. In an effort to better understand this process, a dominant modifier screen was undertaken to identify genetic modifiers that cooperate with nmo to regulate ommatidial rotation.
Loss-of-function alleles of nmo exhibit decreased viability and fertility (Choi and Benzer, 1994) and are therefore not amenable to genetic screens, so a dominant modifier screen in a nmo misexpression background was carried out as an alternative. While misexpression of both the nmoI and nmoII transgenes under the sev>Gal4 driver produces a rotation phenotype suitable for conducting a screen, the nmoII transgene was selected for this screen since it exhibits a specific expression pattern posterior to the furrow and given its homology to nmo orthologs.
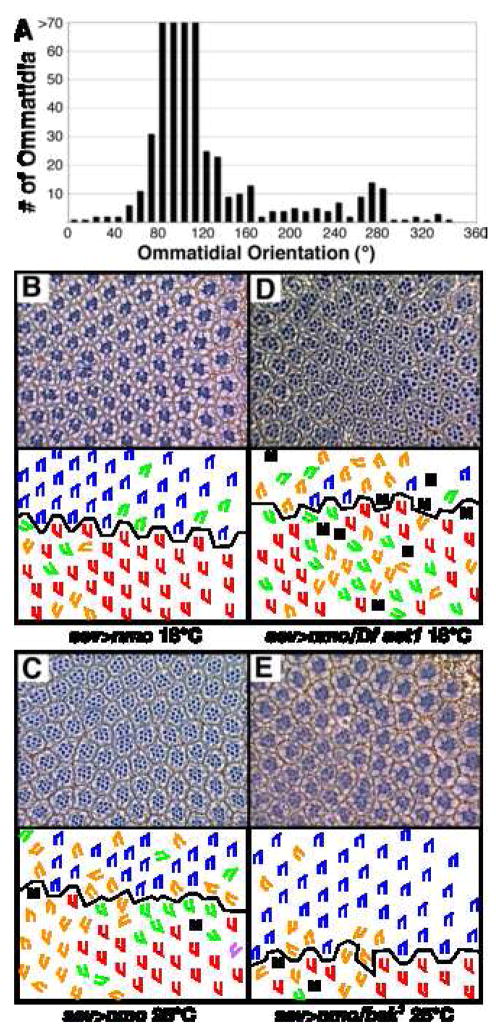
All ommatidia rotate in the correct direction in sev>nmoII transgenic eyes: sev>nmoII, mδ0.5 third instar eye discs co-stained with α-βgal and α-Elav revealed that all ommatidial precursors rotate counterclockwise in the dorsal half and clockwise in the ventral half of the eye (mδ0.5 is an R4 marker (Cooper and Bray, 1999); data not shown). The predominant defect in sev>nmoII eyes is the degree to which ommatidia rotate (Fig. 5A). It is interesting to note that a lack of nmo activity primarily results in rotations under 90° whereas an excess of nmo activity primarily moves ommatidia beyond the normal stopping point of 90°. It is also intriguing to note that sev>nmo ommatidia exhibit a propensity to align themselves along the north-south axis: the majority (~60%) of ommatidia rotate 90°; of the remaining 40% of ommatidia, more orient at 270° (10%) than at any other degree (Fig. 5A). This alignment of ommatidia along the north/south (or D/V) axis suggests the “stopping mechanism” is perpendicular to the equator. This preferential alignment with the poles further suggests that the stopping mechanism is extrinsic rather than an intrinsic clock that regulates the amount of time ommatidial precursors rotate in that if the clock was broken, ommatidia would be expected to stop over a range of angles rather than at 90° and 270°.
Figure 5. sev>nmoII exhibits a temperature-sensitive rotation phenotype.
(A) Histogram illustrating the distribution of ommatidial orientation in sev>nmo eyes. (B–E) Tangential sections through sev>nmo eyes reveal that these flies exhibit a weak phenotype at 18°C. (B) and a strong phenotype at 25°C (C). These phenotypes are enhanced by Df(2L)ast1 (D) and suppressed by basket1 (a JNK family member) (E), respectively. M, missing photoreceptors; E, extra photoreceptors; blue trapezoids: dorsal ommatidia; red trapezoids: ventral ommatidia; green trapezoids: under-rotated ommatidia; orange trapezoids: over-rotated ommatidia.
The sev>nmoII phenotype is temperature-sensitive and thereby enabled the screen to be sensitized to pick up enhancers and suppressors, as follows. The screen was conducted at two temperatures, 18°C and 25°C. At 18°C, the phenotype is weak (7% rotation errors), producing a background suitable for identification of enhancers of the sev>nmoII phenotype (Fig. 5B, Table 2). The phenotype of sev>nmoII flies raised at 25°C is significantly stronger (43% rotation errors), facilitating the identification of suppressors of the sev>nmoII phenotype (Table 2, Fig. 5C). A pilot screen was performed to demonstrate the validity of the screen, as follows. basket, misshapen, hemipterous, argos, and dTak either interact with nmo in other systems or are implicated in regulating rotation and were therefore tested in the pilot screen (Choi and Benzer, 1994; Chou and Chien, 2002; Mirkovic et al., 2002). These genes were tested for their ability to modify the sev>nmo phenotype; basket and a deficiency that uncovers dTak (Df(1)HM44) were found to suppress the 25° phenotype (Fig 5E, Table 2).
Table 2.
A dominant modifier screen of sev>nmoII identifies chromosomal regions that harbor potential enhancers and suppressors.
| sev>nmoII/* | Misrotated ommatidia (%) | n (eyes) | |
|---|---|---|---|
| 18°C | + | 7 +/− 5 | 17 |
| Df(2L) ast1 | 34 +/− 9‡ | 5 | |
| Df(2L) S2590 | 20 +/− 8† | 6 | |
| Df(2R) CX1 | 31 +/− 9‡ | 6 | |
| Df(2R) AA21 | 21 +/− 6‡ | 6 | |
| Df(3L) vin7 | 26 +/− 9‡ | 6 | |
| Df(3L) st-f 13 | 44 +/− 3‡ | 3 | |
| Df(3L) 81K19 | 25 +/− 9† | 6 | |
| Df(3R) 6–7 | 20 +/− 6‡ | 6 | |
| Df(3R) P14 | 21 +/− 8‡ | 6 | |
| 25°C | + | 43 +/− 16 | 21 |
| Df(2L) ed-dp | 21 +/− 7‡ | 6 | |
| Df(2L) TRF-C6R31 | 22 +/− 8† | 5 | |
| Df(2R) NP3 | 8 +/− 4‡ | 6 | |
| Df(2R) B5 | 15 +/− 3‡ | 6 | |
| Df(2R) BSC 19 | 15 +/− 7‡ | 6 | |
| Df(2R) M60E | 20 +/− 6‡ | 5 | |
| Df(2R) Kr10 | 15 +/− 4‡ | 6 | |
| Df(3L) HR119 | 12 +/− 4‡ | 5 | |
| Df(3R) Scr | 21 +/− 4‡ | 5 | |
| Df(3R) T-32 | 22 +/− 3‡ | 6 | |
| Df(3R) ry615 | 12 +/− 2‡ | 6 | |
| Df(3R) mbc-R1 | 13 +/− 3‡ | 5 | |
| Df(3R) ESPl3 | 23 +/− 7‡ | 6 | |
| Df(3R) Tl-P | 14 +/− 5‡ | 6 | |
| Df(3R) 3450 | 22 +/− 9‡ | 6 | |
| Df(3R) Dr-rv-1 | 23 +/− 7‡ | 5 | |
| Df(3R) L127 | 21 +/− 5‡ | 6 | |
| Candidate suppressors | argosrlt | 35 +/− 11 | 3 |
| bsk1 | 21 +/− 7‡ | 10 | |
| Df(1)HM44 (dTak) | 21 +/− 9‡ | 6 | |
| arm1 | 50 +/− 11 | 5 | |
| fzR54 | 38 +/− 17 | 3 | |
| msn102 | 35 +/− 16 | 6 | |
| hep175 | 31 +/− 15 | 6 | |
| pksple | 34 +/− 9 | 6 | |
| zip1 | 15 +/− 5‡ | 12 | |
| sca1 | 30 +/− 10† | 6 |
The p-values for the selected modifiers are statistically significant († p<0.05, ‡ p<0.01; t-test).
In the first round of screening, sev>nmoII virgin females were crossed to the deficiency kits (Bloomington stock center) for chromosomes 2 and 3, and three eyes, one from each of three flies, were scored to determine the number of ommatidia that mis-rotated. In the second phase of the screen, an additional three eyes from three independent flies were scored for those deficiencies that showed modification of the starting sev>nmoII phenotype in round one. Nine enhancers representing eight distinct chromosomal regions (Fig. 5D, Table 2) and 17 suppressors representing 13 distinct chromosomal regions were identified (Fig. 5E, Table 2). Candidate genes within these deficiency regions were then tested for their ability to recapitulate the original interaction.
This screen identified approximately 20 regions that either enhance or suppress the sev>nmo phenotype (Table 2). Several of these genes are known interactors of nmo and therefore validate the screen. Although the interacting genes within many of the deficiencies have not yet been identified, one deficiency, (Df(2R)Kr10), identified non-muscle myosin, which we have shown regulates the rate of rotation (Fiehler and Wolff, 2007). Future studies on these modifiers will likely identify new genes involved in various aspect of ommatidial rotation and will provide deeper insight into the mechanisms underlying ommatidial rotation.
Discussion
nmo/NLK/lit-1 regulates transcription in both vertebrates and invertebrates. In vertebrates, NLK modifies the ability of the transcription factor Tcf/Lef to bind DNA (Ishitani et al., 2003; Ishitani et al., 1999). In the Drosophila wing, nmo modifies the capacity of wg to regulate the transcription of its target genes (Zeng and Verheyen, 2004). As reported here, GFP-tagged Nmo becomes prominent in the nuclei, where it likely regulates transcription of key ommatidial rotation genes using a mechanism similar to the paradigms noted above.
The mechanism by which nmo expression is regulated in the eye likely differs from its mode of regulation in the wing, where nmo is a direct target of wg (Zeng and Verheyen, 2004). In the eye, nmo is unlikely to be a direct target of wg for two reasons. First, nmo expression in the eye does not flank wg expression as it does in the wing. Instead, nmo is expressed in a stripe posterior to the morphogenetic furrow, whereas wg is expressed at the dorsal and ventral poles ahead of the furrow (Ma and Moses, 1995). Second, overexpression of wg in the eye fails to produce ectopic nmo (R. Fiehler, unpublished results). Non-canonical Wnt signaling, however, has been implicated in driving nmo expression in the eye (Zheng et al., 1995).
The target of nmo activity also remains unclear. Possible targets, based on both published data as well as data provided here, include Tcf/Lef and JNK. Our data, in conjunction with published data (Fanto et al., 2000; Mihaly et al., 2001; Strutt et al., 1997) point towards JNK as the best candidate as a target for nmo in the eye. Alternatively, nmo may target a distinct transcription factor, or it may play a novel role in directing rotation, independent of transcriptional regulation.
The net movement of nmo mutant ommatidia in a positive direction during pupal eye development is curious, and, at this point, we do not know what mechanism underlies this directed movement. However, the fact that ommatidia do exhibit a net movement indicates that the overall direction of movement is not due to chance. Rather, perhaps there is an internal compass with which the ommatidia attempt to align. This phenotype is reminiscent of the sev>nmo phenotype, in which ommatidia also exhibit a tendency to align along the north-south axis.
At the commencement of rotation, the precluster consists of photoreceptors, R8, R2, R5, R3 and R4 (Fiehler and Wolff, 2007). Since nmo is not required in these five cells (Fig. 3), and rotation does initiate in nmo mutants (Fig. 2), this suggests the mechanism that starts rotation is nmo-independent. This nmo-independent mechanism appears to be capable of driving rotation for 45°, as ommatidial precursors rotate 45° in nmoP1 mutants. However, the rate of rotation is significantly slower in nmo mutants relative to wild type (Fig. 2). This indicates the nmo-independent mechanism is inefficient in its capacity to turn ommatidia at a normal pace in the absence of nmo. It further suggests that nmo supplements the nmo-independent mechanism, apparently by accelerating the rate. Rotation stops altogether at row 11 (45°) in the nmo mutant, suggesting that the nmo-independent mechanism does not operate beyond this point (perhaps because the mechanism gets blocked, see below), and that the nmo-dependent mechanism is a non-redundant driving force from rows 11–15. If a similar, supplementary mechanism operates from row 11 on, it apparently has no compensatory effect in the absence of nmo function, as movement stalls at this stage.
The specific role for nmo in ommatidial rotation has not yet been uncovered. We propose a model for nmo function in which nmo helps to establish the “rotation machinery,” or “motor,” at the interface between moving and stationary cells, as follows. This model is based on two hypotheses reported in Fiehler and Wolff (2007). First, we hypothesized that the motor resides at the interface between those cells that rotate (the photoreceptors and cone cells) and those that remain stationary (the undifferentiated cells between ommatidial precursors). Second, since this interface is dynamic, changing as cells are recruited into the assembling ommatidium, we hypothesized that the driving force for rotation should shift to the outermost rotating cells as new cells are incorporated into the cluster. In the model described below, it is important to note that details regarding both the rows at which photoreceptors are added and the number of degrees ommatidial precursors have rotated at the time subsets of cells are recruited are “best estimates.” The inherent nature of eye development – in particular the equator-lateral (Wolff and Ready, 1991b) and anterior-posterior gradients -- precludes the possibility of providing precise values, since these gradients introduce variability in terms of the timing of events. Given this variability, the relative timing of events is emphasized in discussing the model that follows rather than tying events to specific points in time.
Placing the results of the mosaic analysis and the rates of rotation in nmo mutants in the context of 1) what is currently known about eye development and 2) the hypotheses outlined above, leads to the following model. We propose that the nmo-independent component of the motor is housed in photoreceptors R8, R2, R5, R3 and R4, and that this motor resides at the interface between all or a subset of these five photoreceptors and the interommatidial cells (red line, Fig. 6). In this model, as additional cells get recruited, they block the nmo-independent mechanism and consequently affect the output of this nmo-independent motor. For example, the recruitment of R1, R6 and R7 at approximately row 5 (one row after initiation of rotation; (Fiehler and Wolff, 2007)) completely blocks R8, and partially to completely blocks R2 and R5, from contact with the stationary cells. This creates a new interface between rotating and stationary cells (Fig. 6). Given that the photoreceptor cluster continues to move an additional ~38° once R8, R2 and R5 are blocked, it seems likely that R3 and R4 play a more significant role in the first 45° of rotation than do R8, R2 and R5.
Figure 6. Model for nmo’s role in ommatidial rotation.
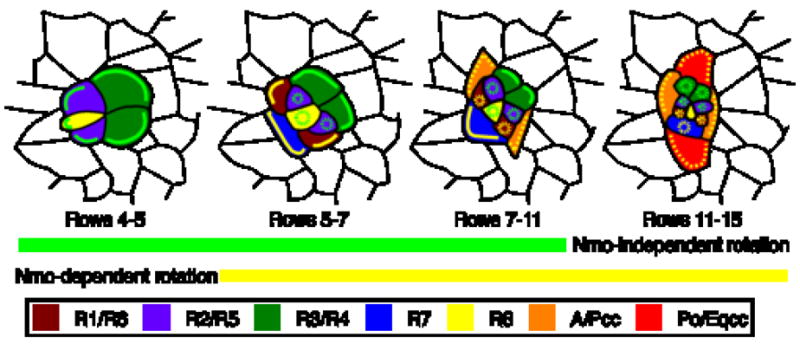
Two overlapping but independent mechanisms regulate ommatidial rotation. A nmo-independent mechanism (green lines) operates in R2, R3, R4, R5 and R8 whereas a nmo-dependent mechanism (yellow lines) functions in R1, R6, R7 and the cone cells. When contacts between rotating and stationary cells become occluded by newly recruited cells, the rotation machinery is no longer active (yellow and green dashed circles). Dashed yellow line in cone cells indicates hypothetical nmo activity. See text for details.
The mosaic analysis presented here indicates a requirement for nmo in photoreceptors R1, R6 and, to a less significant extent, R7. In addition, rotation proceeds more quickly for the first 45° in wild type than in nmo mutants, and in nmo mutants, rotation stalls at 45°. The data and arguments presented thus far suggest R1, R7 and R6 actively take place in rotation via a regulatory mechanism distinct from R3 and R4, and that this secondary, R1/R6/R7, nmo-dependent mechanism not only speeds up rotation during the first 45°, but is also essential to move ommatidial precursors the second 45°. We therefore propose that nmo is involved in either setting up or transferring the motor from R8/R2/R5 to R1, R7, and R6, and later from the photoreceptors to the cone cells, as described below.
Genetically wild-type R1, R6 and R7 cells do not rescue rotation, raising the interesting possibility that other cells also play a critical role in ommatidial rotation. The anterior and posterior cone cells are recruited following R1, R6, R7, at approximately row 6/7 in wild type (~55°). In nmo, the anterior and posterior cone cells are added well before 55° since nmo mutant ommatidia rotate more slowly. Addition of the anterior and posterior cone cells occludes contact between photoreceptors R1, R6 (and, in some cases, minor portions of R2/R5 and R3/R4) and the interommatidial cells (Fig. 6), so at this point in development, R3, R4, R7 and the anterior and posterior cone cells constitute the rotation interface. The polar cone cell is recruited at approximately 80% rotation in wild type, but, given the slower rate of rotation in nmo, this translates to only 45° of rotation in nmo mutant discs. Given that the polar cone cell blocks R3 and R4 from their contact with the interommatidial cells, what we propose to be the only functional motor in nmo, the nmo-independent motor in R3 and R4, can no longer provide the driving force for rotation in nmo mutants. We therefore argue that the mechanism underlying the derailment of rotation is that nmo either fails to relocate or otherwise establish the motor in the new interface cells, initially R1, R6 and R7, and ultimately the anterior, posterior and polar cone cells.
It is also interesting to note that rotation in wild type consists of an early, fast phase and a later, slow phase, and that the slow phase correlates with addition of the anterior and posterior cone cells (Fiehler and Wolff, 2007). This observation is consistent with our model in that addition of the cone cells in wild type blocks cells that we propose to house the more efficient or faster rotation machinery.
Supplementary Material
(A) RTPCR using primers embedded in exons that flank the nmoI exon. RNA was purified from wild type (wt) eye discs and wild-type and nmoP1 adults. Both the nmoI and nmoII transcripts are evident in nmoP1 mutants. Actin was used as a loading control. in situ hybridization of a nmoI-specific probe (B, B′) and a nmoI/nmoII probe (C,C′). Expression immediately behind the morphogenetic furrow is not seen with the nmoI-specific probe but is apparent with the nmoI/nmoII probe (white arrow). (D) Localization of each transgene expressed under the sev>Gal4 driver (GFP.nmo: green; α-Elav: red). High magnification images of single ommatidial precursors illustrates nuclear localization of the GFP.Nmo transgene. (E) Western blot analysis of NmoI and NmoII transgenic protein. Protein levels are normalized to an actin control. Relative expression levels of the transgenes compared to actin expression levels are shown at the bottom as percent actin expression. Anterior is to the right.
Acknowledgments
We gratefully acknowledge K. Choi, E. Verheyen and the Bloomington Stock Center for fly stocks, and S. Takeda and M. Das Thakur for research assistance. This investigation was supported by National Institutes of Health, Institutional National Research Service Award 5-T32-EY13360-05 to R.W.F. and NIH grant R01 EY13136 to T.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brott BK, et al. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci U S A. 1998;95:963–8. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–12. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–36. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–50. doi: 10.1016/s1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–30. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- Cordero J, et al. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech Dev. 2004;121:1523–30. doi: 10.1016/j.mod.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Fanto M, et al. Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Curr Biol. 2000;10:979–88. doi: 10.1016/s0960-9822(00)00645-x. [DOI] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–62. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M, et al. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–75. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–23. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- Harada H, et al. Genomic structure of the human NLK (nemo-like kinase) gene and analysis of its promoter region. Gene. 2002;285:175–82. doi: 10.1016/s0378-1119(02)00412-2. [DOI] [PubMed] [Google Scholar]
- Ishitani T, et al. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–89. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Kaletta T, et al. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390:294–8. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- Lee EC, et al. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–88. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Moses K. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development. 1995;121:2279–89. doi: 10.1242/dev.121.8.2279. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–7. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Mihaly J, et al. The role of the Drosophila TAK homologue dTAK during development. Mech Dev. 2001;102:67–79. doi: 10.1016/s0925-4773(01)00285-4. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, et al. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Ready DF, et al. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–40. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, et al. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–26. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Seibel NM, et al. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem. 2007;368:95–9. doi: 10.1016/j.ab.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Shin TH, et al. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol Cell. 1999;4:275–80. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- Strutt DI, et al. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–5. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–7. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- Takatsu Y, et al. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol Cell Biol. 2000;20:3015–26. doi: 10.1128/mcb.20.9.3015-3026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–5. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological Techniques for the Drosophila Eye. In: Sullivan William, Ashburner Michael, Hawley R Scott., editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. pp. 201–244. [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991a;113:825–39. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991b;113:841–50. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate Michael, Martinez-Arias Alfonso., editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 1277–1325. [Google Scholar]
- Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–20. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
- Zheng L, et al. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–55. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) RTPCR using primers embedded in exons that flank the nmoI exon. RNA was purified from wild type (wt) eye discs and wild-type and nmoP1 adults. Both the nmoI and nmoII transcripts are evident in nmoP1 mutants. Actin was used as a loading control. in situ hybridization of a nmoI-specific probe (B, B′) and a nmoI/nmoII probe (C,C′). Expression immediately behind the morphogenetic furrow is not seen with the nmoI-specific probe but is apparent with the nmoI/nmoII probe (white arrow). (D) Localization of each transgene expressed under the sev>Gal4 driver (GFP.nmo: green; α-Elav: red). High magnification images of single ommatidial precursors illustrates nuclear localization of the GFP.Nmo transgene. (E) Western blot analysis of NmoI and NmoII transgenic protein. Protein levels are normalized to an actin control. Relative expression levels of the transgenes compared to actin expression levels are shown at the bottom as percent actin expression. Anterior is to the right.