Abstract
Understanding the way in which the immune system responds to infection is central to the development of vaccines and many diagnostics. To provide insight into this area, we fabricated a protein microarray containing 1,205 Burkholderia pseudomallei proteins, probed it with 88 melioidosis patient sera, and identified 170 reactive antigens. This subset of antigens was printed on a smaller array and probed with a collection of 747 individual sera derived from 10 patient groups including melioidosis patients from Northeast Thailand and Singapore, patients with different infections, healthy individuals from the USA, and from endemic and nonendemic regions of Thailand. We identified 49 antigens that are significantly more reactive in melioidosis patients than healthy people and patients with other types of bacterial infections. We also identified 59 cross-reactive antigens that are equally reactive among all groups, including healthy controls from the USA. Using these results we were able to devise a test that can classify melioidosis positive and negative individuals with sensitivity and specificity of 95% and 83%, respectively, a significant improvement over currently available diagnostic assays. Half of the reactive antigens contained a predicted signal peptide sequence and were classified as outer membrane, surface structures or secreted molecules, and an additional 20% were associated with pathogenicity, adaptation or chaperones. These results show that microarrays allow a more comprehensive analysis of the immune response on an antigen-specific, patient-specific, and population-specific basis, can identify serodiagnostic antigens, and contribute to a more detailed understanding of immunogenicity to this pathogen.
Keywords: antigen discovery, melioidosis, diagnostic, antigen prediction
Understanding the interaction of the immune system with bacteria is central to an understanding of the pathogenesis of infectious disease, and also to the development of diagnostics and vaccines. Yet there is an unmet need for a comprehensive and unambiguous approach for quantifying the immune response to infection in an antigen-specific manner and on a genome-wide scale. The development of antibodies to individual components of bacterial pathogens is 1 important element of the immune response, and is especially relevant to the development of diagnostics and for therapies where antibodies play direct or indirect roles in the killing of pathogens or pathogen-infected cells. Although protein antigens are generally assumed to make up the majority of the bacterial antigens that are recognized by antibodies, the reasons why only some proteins evoke antibody responses are poorly understood. There is a general and intuitive assumption that proteins that are displayed on the surface of the bacterium are more likely to evoke an antibody response. In the case of proteins able to induce protective immunity there is some evidence that this assumption is correct. An analysis of 72 bacterial proteins included in vaccines or shown to induce protective immunity in animal models of disease showed that 52 (72%) possessed signal sequences and were therefore likely to be located outside of the cell membrane (1). The assumption that proteins able to induce protective immunity possess signal sequences is exploited in reverse vaccinology, where candidate protective antigens are predicted from the genome sequence (2).
However, these studies fail to provide insight into the broader question of the differential abilities of bacterial proteins to induce antibody responses in a population. To a large extent, the difficulties in addressing this question are a reflection of the limitations of existing technologies for mapping the complete subset of proteins involved in the immune response against the infectious agent. These technologies rely on chromatographic or electrophoretic separation of proteins from bacteria or the screening of expression libraries. These approaches are limited by the different levels of expression of proteins in the native or recombinant hosts, specific bacteria growth conditions if cultivable, and are not conducive to high-throughput screening of a large collection of serum samples.
We have previously developed array technology that allows protein microarrays to be constructed from the predicted proteome of a microorganism (3–7). These arrays can be used to address basic questions about the interactions of a given pathogen with the host immune system (8–10), and allows the identification of proteins which can be used as diagnostic reagents or for inclusion in vaccines (3). The empirical data gathered from this type of array can also be used to evaluate and improve the accuracy of in silico antigen prediction of a proteome.
We report here the development of a Burkholderia pseudomallei protein array. B. pseudomallei is the causative agent of melioidosis, a serious and often fatal infectious disease of humans. It is an important medical problem across Southeast Asia and northern Australia, and is increasingly recognized in other tropical areas of the world including areas of South America (11, 12). The global incidence of disease is not known, but in northeast Thailand, the disease accounts for 40% of all deaths from community-acquired septicemia. The potential for the bacterium to cause disease after inhalation has also resulted in the inclusion of this pathogen on the CDC category B list of potential biological warfare and bioterrorism agents (12). Here we have used the protein array to map the antibody response in 747 serum samples from well-defined melioidosis positive and negative patients.
Results
Protein Microarray Design and Construction.
We devised a protein array of 1,205 proteins, including proteins predicted to be surface located using PSORTb (13), components of the 3 different type III systems, components of the flagella, proteins identified as immunoreactive from 2D gels, and 672 proteins selected at random. This array was probed with a collection of 88 sera from melioidosis patients in Singapore (SI Text). One hundred seventy antigens with average signal intensity greater than 2.5 times the standard deviation of the average negative control spots were considered seroreactive. Of these, 80 proteins were encoded on chromosome 1, and 90 on chromosome 2. Three of the seroreactive proteins were encoded in genomic islands (BPSL1705 in GI8, BPSS0663 in GI4, and BPSS1068 in GI15), and 2 proteins were encoded in possible genomic islands (BPSS0402 and BPSL0739). Some large CDSs (coding sequences) were expressed as overlapping fragments and for many of these, we found reactivity against several of the fragments, providing evidence that epitopes are distributed throughout these proteins. For example, 6 polypeptides from the BPSS2053 CDS, encoding a 3,103 amino acid protein similar to the Ralstonia solanacearum probable hemagglutinin-related protein, all reacted with melioidosis sera. Four polypeptides that form the BPSS1434 CDS, encoding a 2,178 amino acid protein similar to a Streptococcus pneumoniae cell wall surface anchor family protein, all reacted with patient sera. Nine polypeptides from the BPSL1661 CDS, encoding a 3,229 amino acid protein similar to a Ralstonia solanacearum putative hemagglutinin/hemolysin-related protein, all reacted with melioidosis sera. In a previous study, 12 B. pseudomallei proteins were identified as immunoreactive by 2D gels (14), and all of these proteins were identified as immunoreactive using the array.
Mapping the Antigenic Profile.
The seroreactive antigens were printed onto smaller arrays (Fig. 1), which we used to probe the entire collection of 747 serum samples from patients with melioidosis and control sera from individuals in Singapore, Thailand, and the USA (Table S1 and SI Text). The Singapore and Thailand melioidosis-positive and -negative cases were patients entering the hospital with symptoms of melioidosis that were later confirmed to be either positive or negative for melioidosis. The Thailand cases were from Ubon Ratchathani in Northern Thailand and healthy controls from the same region where the disease is endemic. There were also healthy controls from Southern Thailand (outside the endemic region) and patients diagnosed with leptospirosis, other bacteremia, or fungemia.
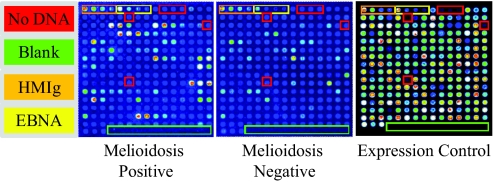
Fig. 1.
Construction of a B. pseudomallei Microarray. Arrays were printed containing 214 B. pseudomallei proteins, positive and negative control spots. The arrays were read in a laser confocal scanner, analyzed, and the data normalized as described in the Materials and Methods section. Protein expression efficiency was determined to be 99.2% by probing against a carboxy-terminal HA tag for quality control. Each array contains positive control spots printed from 4 serial dilutions of human IgG (HMIg), and the intensity of these spots was similar for both serum samples. Each array also contained 6 “No DNA” negative control spots, and the reactivity of these spots was low for both serum samples. There are also 4 serially diluted EBNA1 (EBNA) protein control spots that are reactive to varying degrees in different subjects, as expected, and provide a methodological control. The remaining spots on the array are in vitro transcription/translation reactions expressing 183 different B. pseudomallei proteins that were selected from our primary array analysis. The signal intensity of each antigen is represented by rainbow palette of blue, green, red, and white by increasing signal intensity. Representative microarray immunofluorescence images of individual patient sera are displayed.
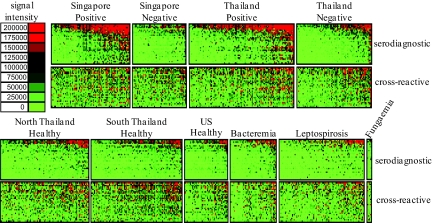
The reactivity of 747 sera from different individuals is shown as a heatmap (Fig. 2) and as a histogram (Fig. S1) with patient samples grouped according to their clinical description. Thirty-one of the most reactive serodiagnostic antigens and 31 cross reactive antigens are shown. ‘Serodiagnostic’ antigens are defined as significantly differentially reactive between the Singapore melioidosis-positive and -negative groups with Benjamini and Hochberg adjusted Cyber-T P values <0.05, and ‘cross-reactive’ antigens had a P value >0.05 (Fig. 3). Analysis of variance was performed to detect differences in signal intensity between groups. For the serodiagnostic antigens there is a significant difference when comparing melioidosis-positive patients to all other groups (P = 3.6 × 10−7), but the Singapore and Thailand melioidosis patients are not different from each other (P = 0.55), and all melioidosis negative patients are not different from each other (P = 0.91) (Fig. S1). All of the sera react similarly to the cross-reactive antigens whether from melioidosis-positive or -negative individuals, healthy subjects from endemic or nonendemic areas, or patients with other infections (P value = 0.08).
Fig. 2.
Probing a collection of B. pseudomallei infected, uninfected, and healthy control sera from Singapore, Thailand, and the USA. Arrays containing 214 B. pseudomallei proteins were probed with 747 melioidosis and nonmelioidosis sera organized into 11 groups as described in the text. The normalized intensity is shown according to the colorized scale with red strongest, bright green weakest, and black in between. The antigens are in rows and are grouped according to serodiagnostic and cross-reactive. The patient samples are in columns and sorted left to right by increasing average intensity to serodiagnostic antigens.
Fig. 3.
Serodiagnostic antigen discovery of melioidosis-positive patients from Singapore. The mean sera reactivity of the 214 antigens was compared between the Singapore melioidosis-positive and Singapore melioidosis-negative groups. Antigens with Benjamini Hochberg corrected P value less than 0.05 are organized to the left and cross-reactive antigens to the right. The 31 most reactive serodiagnostic and 31 of the most reactive cross-reactive antigens are shown.
Determining a Set of Serodiagnostic Antigens.
There were 2 distinct collections of melioidosis-positive and -negative specimens used for this work, from different clinical investigators and clinical sites in Thailand and Singapore, and different definitions of the cases and controls were used in each location. The incidence of melioidosis is relatively low in Singapore, and exposure to B. pseudomallei is considered to be an infrequent event. The Singaporean cases were clinically confirmed melioidosis and positive by the indirect hemagglutination assay (IHA) and about 40% of these samples were proven culture positive. All of the controls for this collection lacked clinical signs of melioidosis disease and were also IHA negative. The Northeast Thailand collection is from a region where the disease is considered endemic. All of the cases are proven culture positive, and the controls are culture negative but IHA positive subjects are not excluded, because it is known that healthy individuals in the general population in Northeast Thailand can be IHA positive. For all of the work reported here we used the Singaporean cases and controls to develop a classifier that can distinguish melioidosis-positive and -negative subjects, and tested sensitivity and specificity of the classifier independently on the Thailand collection.
Our goal was to establish a collection of antigens that could be used as a multiplex set to accurately distinguish the melioidosis cases from controls. As such, we studied the discriminatory power of different sets of ORFs using receiver operating characteristic (ROC) curves. First, ROC curves were generated for individual serodiagnostic antigens to assess their ability to separate the control and disease cases. The serodiagnostic antigens were ranked by decreasing single antigen area under the ROC curve (AUC) (Table S2). The top 5 ORFs all have an AUC greater than 0.81, with GroEL (BPSL2697; AUC 0.89; Benjamini and Hochberg adjusted Cyber-T P value <10e-14) giving the best single antigen discrimination. Heat shock proteins like GroEL are often dismissed a priori for serodiagnostic utility because of perceived cross-reactivity with heat shock proteins from other organisms that would theoretically result in low specificity of the test; here, we find the heat shock protein GroEL is the most significantly differentially reactive antigen and have similarly found serodiagnostic power from heat shock proteins from other organisms (7). The 31st antigen has an AUC of 0.622 which still exceeds the upper 95% confidence interval for random expectations for the AUC. To extend the analysis to combinations of antigens, we used kernel methods and support vector machines (15, 16) to build linear and nonlinear classifiers. As input to the classifier, we used the highest-ranking 1, 2, 5, 10, 20, 25, 30, and 31 ORFs on the basis of either P value or single antigen AUC and the results were validated with 10 runs of 3-fold cross-validation. The boxplots for these predictions were plotted and the results show that increasing the antigen number from 1 to 2, 2 to 5, and 5 to 10 produced an improvement in the classifier and a reduction in accuracy as the antigens increase above 20 due to over-fitting (Fig. 4A and Table S3).
Fig. 4.
Development of a melioidosis classifier for improved diagnosis. (A) The graph shows 9 Boxplots for nonlinear classifiers with increasing number of antigens. As the number of antigens increases up to 5 antigens, the classifier becomes more accurate. (B) Ten serodiagnostic antigens were printed onto nitrocellulose paper in adjacent stripes using a BioDot jet dispenser (SI Text). Strips were probed with patient sera diluted 1/200 followed by alkaline phosphatase conjugated secondary antibody and enzyme substrate. Weak reactivity in the naïve healthy controls can be readily distinguished from the strong reactivity in infected subjects. Ten representative strips for each group are shown. (C) The graph shows the immunostrip ROC curves compared to the microarray ROC curve generated for with the same 10 antigens. (D) Percent diagnostic accuracy for each assay is listed. Microarray accuracy was calculated for 1, 2, 5, and 10 antigens. Thailand sera are from a previously well-characterized cohort of patients and represent a selected population of samples with definite melioidosis compared to patients with an alternative diagnosis. Thailand sera samples were used for direct comparison of the microarray, ELISA, IHA, and immunostrip assays.
The classifier threshold that yielded the highest accuracy in the Singapore data set was then used to predict the samples in the Thailand collection. Using 20 antigens the classifier predicts 86% of the true positives and 98% of the true negatives from Singapore, and 76% and 72% of the Thailand positives and negatives, respectively. The healthy controls from North and South Thailand are predicted with 78% and 86% accuracy, respectively. Patients with other infections are predicted as melioidosis negative with 69% to 100% accuracy, depending on the group.
Improving Serodiagnostic Accuracy.
To test the feasibility of using the serodiagnostic antigens on an alternative analytical platform, 10 serodiagnostic proteins were printed onto an Optitran nitrocellulose membrane (typically used for western blots) using a BioDot jet dispenser (SI Text). These 10 serodiagnostic proteins were chosen for being highly significant and highly seroreactive in the protein microarray assay. The paper was then cut into 3 mm strips to produce ‘immunostrips’ (also called ‘line blots’) (Fig. 4B). The individual strips were probed with 60 different melioidosis positive sera and 67 melioidosis negative sera from the Singapore collection. Reactive bands were visualized after incubation with alkaline phosphatase conjugated anti-human secondary antibody followed by substrate, and the band intensities quantified with ImageJ (17). Melioidosis patients reacted strongly against the serodiagnostic antigens although the intensity pattern varied depending on the patient. Naïve and healthy control subjects had low reactivity against these serodiagnostic antigens.
A classifier was trained to accurately diagnose the melioidosis-positive and -negative samples from Singapore, and the classifier was tested against strips probed with the independent collection of positive and negative samples from Thailand. ROC curves were generated and compared to the microarray results (Fig. 4C). The area under the immunostrip ROC curve was greater than the microarray, indicating a more accurate test can be obtained by transferring the serodiagnostic antigens discovered by microarray to the immunostrip format. The results are summarized in the contingency table (Fig. 4D). Increasing the number of antigens from 1 to 5 on the microarray results in a more accurate discrimination between positive and negative samples, with maximum sensitivity and specificity of 90% and 55%, respectively, for the Thailand samples. However, immunostrips containing the top 10 serodiagnostic antigens discovered by microarray discriminate the Thailand positive and negative patient samples with 95% sensitivity and 83% specificity. This accuracy is greater than previously published assays using crude B. pseudmallei antigen ELISA and affinity-purified antigen ELISA, and a substantial improvement over the clinical standard IHA assay. (Table S3). This proof-of-concept diagnostic assay validates the set of serodiagnostic antigens and demonstrates the feasibility of transferring the antigens discovered by microarray to the immunostrip format to correctly classify B. pseudomallei positive sera using 10 differentially reactive serodiagnostic antigens.
Functional Classification of the Reactive Antigens.
We next classified the serodiagnostic and cross-reactive antigens according to annotated and computationally predicted features. The complete analysis is in Table S4 and a summary in Table 1. Annotated features were taken from the Artemis database on the Sanger website which has 2 categories of functional definitions called ‘Colour’ and ‘Class.’ Each of the 13 colors has a functional definition, and each of the 5,853 proteins in the database is assigned exactly 1 color. The results in Table 1 show that only 2 of the 13 color classifications are significantly enriched in both the serodiagnostic and cross-reactive antigen sets. ‘Pathogenicity/adaptation/chaperone’ and ‘surface’ molecules account for 75% of all of the reactive antigens whereas these functional categories account for only 38% of the proteins printed on the array (and 32% of the whole proteome). There are 180 class definitions in Artemis, and proteins can belong to more than 1 class; proteins classified as ‘chaperones’ and ‘outer membrane localized’ are significantly enriched in the serodiagnostic antigen set by 12.3- and 4.6-fold, respectively. Protein classified as membrane/exported/lipoproteins are enriched among the cross reactive antigens, but not the serodiagnostic antigens. The PSORTb computational predictor shows that cytoplasmic proteins are significantly underrepresented in the cross-reactive antigen set and cytoplasmic membrane proteins are significantly enriched. Proteins predicted to be extracellular and outer membrane localized are significantly enriched, and the presence of a signal peptide is a highly significant enrichment feature. We have expressed and probed 20% of the B. pseudomallei proteome and identified 49 serodiagnostic antigens and 59 cross-reactive antigens. By probing the remaining 4,648 proteins in the complete proteome, we may expect to identify a total of 295 cross-reactive and 240 serodiagnostic antigens. Pathogenicity associated molecules and membrane structures account for 32% of the entire proteome, but these 2 categories account for 74% of the reactive antigens. Thus by fabricating protein arrays containing 1901 molecules in only these 2 categories, we expect to identify more than 70% of the reactive antigens.
Table 1.
Enrichment table
| Counts |
Cross-reactive |
Serodiagnostic |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Whole proteome | Large chip | Hits | Fold enrich | P value | Hits | Fold enrich | P value | |||
| Artemis 'colour' definitions pathogenicity/adaption/chaperones | 368 | 106 | 13 | * | 2.50 | 1.2E-03 | 11 | * | 2.55 | 2.4E-03 |
| Surface (outer membrane, inner membrane, secreted, surface structures) | 1533 | 352 | 31 | * | 1.80 | 1.8E-04 | 25 | * | 1.75 | 1.1E-03 |
| All other categories | 3952 | 747 | 15 | — | — | 13 | — | — | ||
| Total | 5853 | 1205 | 59 | 49 | ||||||
| Artemis 'class' definitions | ||||||||||
| Chaperones | 30 | 6 | 0 | 0.00 | 1.0E + 00 | 3 | * | 12.30 | 1.2E-03 | |
| Membrane/exported/lipoproteins | 397 | 174 | 15 | * | 1.76 | 2.1E-02 | 12 | 1.70 | 5.8E-02 | |
| Outer membrane | 81 | 48 | 2 | 0.85 | 1.0E + 00 | 9 | * | 4.61 | 7.2E-05 | |
| Computational predictions | ||||||||||
| PSORTb cytoplasmic | 1975 | 293 | 2 | * | 0.14 | 1.6E-05 | 7 | 0.59 | 1.2E-01 | |
| PSORTb cytoplasmicmembrane | 1054 | 94 | 11 | * | 2.39 | 4.3E-03 | 5 | 1.31 | 5.8E-01 | |
| PSORTb extracellular | 37 | 11 | 5 | * | 9.28 | 8.8E-05 | 3 | * | 6.71 | 8.3E-03 |
| PSORTb outermembrane | 150 | 71 | 10 | * | 2.88 | 1.6E-03 | 14 | * | 4.85 | 2.2E-07 |
| PSORTb periplasmic | 130 | 30 | 5 | * | 3.40 | 1.3E-02 | 2 | 1.64 | 3.5E-01 | |
| PSORTb unknown | 2507 | 706 | 26 | * | 0.75 | 2.2E-02 | 18 | * | 0.63 | 1.8E-03 |
| SignalP >= 0.7 | 1041 | 340 | 40 | * | 2.40 | 1.1E-10 | 30 | * | 2.16 | 9.7E-07 |
The majority of these serodiagnostic antigens (36/49) were encoded on chromosome 2 of B. pseudomallei K96243. (Table S2) Chromosome 2 has previously been suggested to be associated with genes required for adaptation and survival in different niches (18), and our finding that serodiagnostic antigens were preferentially encoded on chromosome 2 is consistent with this hypothesis. The serodiagnostic proteins included a range of known or putative virulence associated proteins including components of TTSS3 (BPSS1532, BipB and BPSS1525, BopE) (19–21) and a type IV pilus protein PilO (BPSS1599) (22).
We have also interfaced our dataset with data on genes which are up-regulated in the hamster model of melioidosis (23). Genes encoding 6 of the 31 diagnostic signature proteins (BPSS0476; GroES: BPSL1445; putative lipoprotein BPSL2017; Di-heme cytochrome c peroxidase; BPSL2697; GroEL: BPSL3319; flagellin; and BPSL2520: putative exported protein) were up-regulated in vivo. Chaperones and stress response proteins from a range of pathogens have frequently been found to react with the appropriate convalescent sera (14, 24–28). Both GroEL and DNA K have previously been identified as reactive with sera from melioidosis patients (14, 25).
Discussion
This study is the most extensive use of a diverse collection of patient sera on a protein microarray. Of 1,205 proteins probed, 49 antigens were identified that are significantly more reactive in patients with melioidosis and 59 antigens were cross-reactive in healthy people and patients with other infections. Because melioidosis is confined to relatively small localized regions of the world, it might have been anticipated that immunoreactivity against this species would be localized as well. Our finding that these cross-reactive antigens also reacted equally with sera from individuals in the USA suggests that cross-reactive antibodies may be responsible for these signals. This finding contrasts markedly to our previous studies with Francisella tularensis (7, 29), Borrelia burgdorferi (5), Plasmodium falciparum (6), and vaccinia (8), where reactivity of arrayed proteins with naïve patient sera was minimal or not seen.
The antibodies in melioidosis negative sera may be indicative of past exposure to bacteria related to B. pseudomallei which possess similar proteins. For example, BPSS0542 (glycosyl hydrolase), BPSL1207 (polyribonucleotide nucleotidyltransferase), and BPSS2136 (family S43 nonpeptidase homologue) are identical in a range of Burkholderia species such as Burkholderia thailandensis, B. vietnamiensis, and B. cenocepacia. B. thailandensis is found in many regions of the world including SE Asia and the USA (30) and the B. pseudomallei and B. thailandensis genomes are highly syntenic (31). Bacteria of the B. cepacia complex are also widely distributed worldwide and are frequently the cause of complications in cystic fibrosis. All of these bacteria rarely cause disease in healthy individuals but it is possible that subclinical disease results in the induction of an immune response to these bacteria and the development of antibodies that react with the protein array. Even though cross-reactivity is widespread, some individuals within any region were essentially immunologically naïve. The results reported here suggest widespread exposure and cross-reactivity to nonpathogenic Burkholderia sp., but these conclusions warrant corroborating data from additional endemic and nonendemic regions and comparative data on arrays from nonpathogenic strains.
The information from the array was used to devise an accurate prototype diagnostic test, using differentially reactive antigens while avoiding the cross-reactive ones. The microarray platform may not necessary be the most sensitive to produce the most accurate test results. Serodiagnostic antigens discovered by microarray were transferred to the immunostrip platform, the Singaporean specimen collection was used to train the classifier, and the classifier tested on the Thailand collection. The immunostrip test allowed for the correct identification of 95% of melioidosis-positive samples and 83% of melioidosis-negative samples, a major improvement over the most commonly used diagnostic test (the IHA test) which identifies 79% and 72% of positive and negative samples, respectively. We regard the microarray as a serodiagnostic antigen discovery platform. Antigens discovered by microarray can be transferred to other more sensitive platforms to give more accurate test results.
The data we report here also provide a broader insight into the antigens that are recognized by the immune system after natural infection. Reactivity was not evenly distributed across the proteome and the cross-reactive and serodiagnostic antigens are selectively enriched with proteins from specific functional categories. Membrane and secreted molecules accounted for 52% of the reactive antigens, but this class of molecule constitutes only 29% of the proteins on the array. Similarly, pathogenicity-island related proteins and chaperones (as annotated in the Artemis database) accounted for 22% of the reactive antigens but only 9% of the proteins printed on the large array were from this category. Three of the 6 annotated chaperones printed on the array were serologically reactive, and all 3 of these were serodiagnostic for melioidosis. PSORTb predicts 82 proteins (7% of the large array) to be extracellular or outmembrane localized, and 32 (39%) are antigenic. Proteins with a signal peptide are enriched on the reactive antigen lists and proteins lacking a signal peptide sequence are underrepresented. Proteins involved in central and intermediary metabolism are also significantly underrepresented on the reactive antigen list. These examples illustrate that molecular recognition by the immune system is not stochastic. There is a preference for recognition of surface molecules, chaperones, and pathogenicity-related factors, and the PSORTb outer membrane and extracellular computational tool predicts antigenicity. These results classifying antigenicity by protein function are not out of line with expectations but they are far more quantitative and informative than previously reported.
Seventy-four percent of the antigens fall into functional annotation categories that are enriching features, but the other 26% of the reactive antigens are from categories with proteomic features that are underrepresented on the immunoreactive antigen list. We have not yet identified predictive features of these molecules that selectively target them for immune recognition. Some of the molecules with proteomic features that are underrepresented on the hit list may be expressed at high levels in vivo making them targets for immune recognition independent of their functional category. Although certain protein categories are enriched in the immunoreactive antigen list, there is no category that is entirely reactive. For example, there are 458 proteins on the array in the pathogenicity and outer membrane categories, but only 17% of these molecules are reactive. Part of the explanation for nonreactivity may be that the proteins expressed in the cell free in vitro expression system are not in the proper conformation to be recognized by the antibodies. For example, we have shown that proteins requiring disulfide bonds for their native conformation may not be seen unless in vitro expression is carried out under oxidizing conditions (4). Lack of posttranslational modification, incomplete folding of proteins expressed in vitro, or unfolding of proteins bound to nitrocellulose are other factors that could result in an underestimate of the total number of reactive antigens determined by this method, but our experience with other agents indicates that this effect is actually marginal (4). At this point, any in silico algorithm for predicting the antigenic profile based on sequence data alone will be imperfect because the majority of proteins predicted are nonreactive. Furthermore, there are categories of reactive molecules with proteomic features that are not enriched in the antigenic profile, so they would not be predicted. The data presented here highlight the need to improve in silico prediction algorithms for antigen discovery and vaccine development.
These results provide significant insight into the antigenicity of a bacterial pathogen, while both confirming and overturning assumptions that have previously been made about the nature of immunoreactive proteins. In addition to contributing to a more profound understanding of the humoral immune response to infection, this work could lead to the development more accurate tests for clinical diagnosis and for worldwide disease surveillance.
Materials and Methods
Protein Microarray Chip Fabrication and Probing Methods.
Protein microarray chips consisting of 1,205 B. pseudomallei antigens were fabricated as described previously (SI Text) (3, 4, 7).
SI.
The description of the serum samples, immunostrip fabrication and probing, and data analysis methods are in the SI Text.
Supplementary Material
Acknowledgments.
We thank Eng-Eong Ooi and Jimmy Loh (Defence Medical and Environmental Research Institute, Singapore) for providing serum samples; Allen C. Cheng (University of Melbourne) for sera collection; Mrs. Vanaporn Wuthiekanun and Dr. Narisara Chatratita (Mahidol-Oxford Tropical Medicine Research Unit, Bangkok) for providing samples from Thailand and for ELISA and IHA assays, respectively; Drs. Gavin Koh and Rapeephan Rattanawongnara for collection of clinical data; and Xiaolin Tan for assistance with ANOVA analysis. The primer design and statistical analyses of this work were supported primarily by National Institutes of Health Biomedical Informatics Training Program Grant 5T15LM007743 (to M.K.) and National Science Foundation Grants MRI EIA-0321390 and NSF 0513376 (to P.B.) and in part by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants U01AI061363 and U54065359 (to P.F.).
Footnotes
Conflict of interest statement: P.L.F. and D.H.D. have patent applications related to protein microarray fabrication and have stock positions with Antigen Discovery, Inc. D.M.M. is an employee with Antigen Discovery, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812080106/DCSupplemental.
References
- 1.Mayers C, et al. Analysis of known bacterial protein vaccine antigens reveals biased physical properties and amino acid composition. Comp Funct Genomics. 2003;4:468–478. doi: 10.1002/cfg.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 3.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies DH, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 5.Barbour AG, et al. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolan DL, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2009 doi: 10.1002/pmic.200800194. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyles JE, et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 8.Davies DH, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sette A, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: Deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benhnia MR, et al. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol. 2008;82:3751–3768. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dance DA. Melioidosis: The tip of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dance DA. Melioidosis. Curr Opin Infect Dis. 2002;15:127–132. doi: 10.1097/00001432-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Gardy JL, et al. PSORTb v. 2.0: Expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 14.Harding SV, et al. The identification of surface proteins of Burkholderia pseudomallei. Vaccine. 2007;25:2664–2672. doi: 10.1016/j.vaccine.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Baldi P, Brunak SR. Bioinformatics: The Machine Learning Approach. 2nd Ed. xxi. Cambridge, MA: MIT Press; 2001. p. 452. [Google Scholar]
- 16.Vapnik V. The Nature of Statistical Learning Theory. New York, New York: Springer; 1995. [Google Scholar]
- 17.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 18.Holden MTG, et al. Genornic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warawa J, Woods DE. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol Lett. 2005;242:101–108. doi: 10.1016/j.femsle.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 20.Stevens MP, et al. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology. 2004;150:2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 21.Burtnick MN, et al. Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect Immun. 2008;76:2991–3000. doi: 10.1128/IAI.00263-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essex-Lopresti AE, et al. A type IV pilin, PilA, contributes to adherence of Burkholderia pseudomallei and virulence in vivo. Infect Immun. 2005;73:1260–1264. doi: 10.1128/IAI.73.2.1260-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuanyok A, Tom M, Dunbar J, Woods DE. Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect Immun. 2006;74:5465–5476. doi: 10.1128/IAI.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amemiya K, et al. Detection of the host immune response to Burkholderia mallei heat-shock proteins GroEL and DnaK in a glanders patient and infected mice. Diagn Microbiol Infect Dis. 2007;59:137–147. doi: 10.1016/j.diagmicrobio.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Woo PC, Leung PK, Wong SS, Ho PL, Yuen KY. groEL encodes a highly antigenic protein in Burkholderia pseudomallei. Clin Diagn Lab Immunol. 2001;8:832–836. doi: 10.1128/CDLI.8.4.832-836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Campillo M, et al. Identification of immunoreactive proteins of Chlamydia trachomatis by western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis. 1999;20:2269–2279. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2269::AID-ELPS2269>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.Lemos JA, Giambiagi-Demarval M, Castro AC. Expression of heat-shock proteins in Streptococcus pyogenes and their immunoreactivity with sera from patients with streptococcal diseases. J Med Microbiol. 1998;47:711–715. doi: 10.1099/00222615-47-8-711. [DOI] [PubMed] [Google Scholar]
- 28.Hinode D, Grenier D, Mayrand D. Purification and characterization of a DnaK-like and a GroEL-like protein from Porphyromonas gingivalis. Anaerobe. 1995;1:283–290. doi: 10.1006/anae.1995.1028. [DOI] [PubMed] [Google Scholar]
- 29.Sundaresh S, et al. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 30.Glass MB, et al. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J Clin Microbiol. 2006;44:4601–4604. doi: 10.1128/JCM.01585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, et al. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.