Abstract
Background/aims
Clinical studies indicate that primary proinflammatory cytokines, such as interleukin-1β (IL-1β) are elevated in the gingival crevice around teeth with periodontitis but the secondary cytokines and chemokines, IL-6 and IL-8, are not. The human gingival epithelial cells (HGECs) lining the gingival sulcus respond to perturbation by microbes of dental plaque by releasing a wide range of cytokines. Porphyromonas gingivalis, a putative periodontal pathogen, possesses numerous virulence factors some of which directly impact on the host response. In the present study, we sought to determine how P. gingivalis influences the inflammatory cytokine responses.
Methods
HGECs were challenged with P. gingivalis and other putative periodontal pathogens, and the resultant production of IL-1β, IL-6, and IL-8 was assayed by enzyme-linked immunosorbent assay (ELISA). Culture supernatants and recombinant human cytokines were challenged with live P. gingivalis wild-type and gingipain-deficient strains and the resultant cytokine profile was assessed by ELISA and Western blot.
Results
We show here that primary HGECs challenged with live P. gingivalis result in high levels of IL-1β but not the related secondary cytokines IL-6 and IL-8. We further demonstrate that cytokine response differences are the result of the action of P. gingivalis proteases, with lysine gingipain being the most effective.
Conclusion
We conclude that P. gingivalis, through lysine gingipain, can subvert the protective host proinflammatory response by direct cytokine degradation. Changes in the crevicular cytokine profile have consequences in periodontal disease pathogenesis that should be considered in the development of diagnostic and therapeutic modalities.
Keywords: cytokine degradation, epithelial cells, gingipains, Porphyromonas gingivalis
The host–bacterial interaction that initiates periodontal disease takes place within the gingival crevice. The gingival epithelial cells that line the periodontal tissues are the first line of defense eliciting a wide array of responses to bacteria that includes cytokines and chemokines. Clinical studies have shown that proinflammatory cytokines and chemokines are present in the gingival crevicular fluid (GCF), both in health and disease, and are elevated in sites exhibiting clinical signs of inflammation (23). Interleukin-1β (IL-1β), a primary proinflammatory cytokine, was found in higher concentrations in the GCF of patients with periodontitis compared to that of healthy controls (7, 30). In contrast, IL-8, a secondary chemokine, was found in lower concentrations in the GCF of patients with either chronic (3, 13, 16) or aggressive (19) periodontitis. Furthermore, IL-6, another secondary cytokine, was detected at a low frequency in patients with periodontitis (22), and was found in lower concentrations in the GCF of sites with unresolved defects compared to resolved defects following treatment (9).
The fact that primary and secondary cytokines are at different levels in the periodontal pocket is surprising because at the cellular level they are linked (5). In vitro, IL-1β has been shown to be secreted early after challenge, and so is characterized as ‘primary’; it then modulates secretion of IL-6, IL-8, and other so-called ‘secondary’ cytokines in an autocrine manner (5). Inhibition of IL-1β secretion or blockage of the IL-1 receptor significantly reduces IL-6 and IL-8 production in different cell types, indicating that IL-1β is a key regulator of the inflammatory response (5). It would be expected that upregulation of IL-1β in the GCF would be followed by increased levels of IL-6 and IL-8. Since there is a disconnect between primary and secondary cytokines in vivo, we hypothesized that Porphyromonas gingivalis, a putative periodontal pathogen with powerful proteases, might modulate the inflammatory cytokine responses (10, 14).
Consistent with the published clinical data, our preliminary work reveals that gingival epithelial cells, challenged with live P. gingivalis, mount a primary cytokine response that is not followed by a secondary cytokine response. P. gingivalis possess several virulence mechanisms including the gingipains, a family of cysteine proteases that are either secreted or membrane-bound and arginine-specific or lysine-specific. The gingipains have numerous pathological actions such as dysregulation of clotting and fibrinolytic pathways, activation of the kallikrein/kinin pathway, modulation of cytokine networks, and activation of matrix metallo-proteinases (12, 21, 28, 29). Our hypothesis is that the decreased level of secondary cytokines, despite an elevated primary response in vivo and in vitro, is the result of cytokine degradation by P. gingivalis gingipains. The aim of the current study was therefore to determine the rates of IL-1β, IL-6, and IL-8 degradation by P. gingivalis and identify the role of gingipains in this process.
Materials and methods
Cell isolation and culture
Gingival tissue biopsies were obtained with informed consent from periodontally healthy patients undergoing crown-lengthening procedures at the University of Louisville School of Dentistry Graduate Periodontics Clinic, according to an Institutional Review Board approval. The gingiva was treated with 0.025% trypsin and 0.01% ethylenediaminetetraacetic acid overnight at 4°C and human gingival epithelial cells (HGECs) were isolated as previously described (25). The authenticity of the HGECs was confirmed by immunohistochemistry with monoclonal antibody against human pankeratin (Dako, Carpinteria, CA) and histologically by cell morphology. The HGECs were seeded in 60-mm plastic tissue culture plates coated with type-I collagen (BD Biocoat, Franklin Lakes, NJ) and incubated in 5% CO2 at 37°C using K-SFM medium (Invitrogen, Carlsbad, CA) containing 10 μg/ml of insulin, 5 μg/ml of transferrin, 10 μM 2-mercaptoethanol, 10 μM of 2-aminoethanol, 10 mM of sodium selenite, 50 μg/ml of bovine pituitary extract, 100 units/ml of penicillin/streptomycin, and 50 ng/ml of fungizone (complete medium). When the cells reached sub-confluence, they were harvested and sub-cultured as previously described (6).
Bacterial strains and conditions
P. gingivalis ATCC 33277 was purchased from the American Type Culture Collection (ATCC; Manassas, VA) and the derivative KDP128, an RgpA/RgpB/Kgp triple mutant (24), was kindly provided by Dr K. Nakayama (Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Tokyo). P. gingivalis W50 (ATCC 53978), and the derivative mutants E8, an RgpA/RgpB double mutant, and K1A, a Kgp mutant (1), were kindly provided by Dr M. Curtis (Barts and The London, Queen Mary’s School of Medicine and Dentistry, London, UK). All P. gingivalis strains at low passage were grown in GAM media (Nissui Pharmaceutical, Tokyo, Japan) under anaerobic conditions (85% N2, 10% CO2 and 10% H2; Coy Laboratory, Grass Lake, MI) for 2 days. Aggregatibacter actinomycetemcomitans Y4 and Fusobacterium nucleatum 364 were kindly provided by Dr D. Demuth (University of Louisville School of Dentistry). A. actinomycetem-comitans was grown in brain–heart infusion (Difco Laboratories, Detroit, MI) supplemented with 40 mg NaHCO3 per liter at 37°C in an atmosphere of 5% CO2. F. nucleatum were grown in brain–heart infusion broth (Difco Laboratories, Detroit, MI) supplemented with 0.2% yeast extract (Difco Laboratories), 0.5 mg of L-cysteine hydrochloride, and freshly prepared 0.5% sodium bicarbonate under anaerobic conditions for 3 to 4 days. After cultivation, the bacteria were harvested by centrifugation, washed in phosphate-buffered saline (PBS; pH 7.4) and used immediately for the live cell challenge or heat-inactivated for 1 h at 60°C.
Gingipain inhibitors
For the gingipain inhibition assays, live P. gingivalis 33277 was incubated with gingipain inhibitors for 15 min at 37°C, just before the cytokine degradation assay. zFKck, a specific lysine gingipain (Kgp) inhibitor (20) was kindly provided by Dr J. Potempa (University of Georgia) and used at a final concentration of 10 μM. Leupeptin (Sigma, St Louis, MO), a specific arginine-gingipain (Rgp) inhibitor, was used at a final concentration of 100 μM. The final concentrations used were the maximum inhibitory doses that retained specificity, as determined by a dose–response using the enzymatic substrate hydrolysis of N-α-benzoyl-DL-arginine-p-nitroanilide (BAPNA; Sigma), for Rgp activity, or acetyl-lysine-p-nitroanilide (ALNA; Bachem, King of Prussia, PA), for Kgp activity.
Bacterial challenge
HGEC cultures at the fourth passage were harvested and seeded at a density of 0.5 × 105 cells/well in a six-well culture plate coated with type-I collagen, and maintained in 2 ml complete medium. When they reached confluence (approximately 106 cells/well), the cells were washed twice with fresh medium and were challenged with live or heat-inactivated bacteria in antibiotic-free medium at a multiplicity of infection (MOI):100 (108 bacteria/well) at 37°C in 5% CO2 for 4 or 24 h. For each experiment the final concentration of the suspension was determined by measurement of absorbance at 600 nm (A600) and appropriate dilutions were made to achieve the desired MOI. The bacterial number was confirmed by viable counting of colony-forming units (CFU) on blood agar plates incubated anaerobically at 37°C. The ratio of live to dead bacteria was determined using a Petroff–Hausser counting chamber. Each assessment confirmed that live bacteria comprised 95% of total bacterial cells counted.
Cytokine degradation assay
Culture supernatants produced after a 24-h incubation of HGECs with heat-killed P. gingivalis 33277 were incubated with 5 × 107 live bacteria/ml of the various strains at 37°C. The reaction was stopped at 1 min, 30 min, 1 h, 2 h, and 4 h. Alternatively, solutions of recombinant human IL-1β, IL-6, and IL-8 (BD Biosciences, San Diego, CA) were also incubated with 5 × 107 live bacteria/ml of the various strains at 37°C, and the reactions were carried out for the same length of time. After incubation, the enzymatic activity of lysine gingipain was terminated by addition of zFKck (10 μM), and those of arginine-gingipains were terminated by addition of leupeptin (100 μM). The initial concentration of the recombinant cytokine was determined by the concentration of the respective cytokine in the above culture supernatant. IL-1β, IL-6, and IL-8 were measured by enzyme-linked immunosorbent assay (ELISA) using a commercially available kit (BD OptEIA; BD Biosciences) according to the manufacturer’s instructions. The absorbance was read at 450 nm.
Western blot analysis
Recombinant human (rh) IL-1β and rhIL-6 were incubated with live bacteria as described above and the samples were separated in a 15% polyacrylamide gel by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene difluoride membrane (BioRad, Hercules, CA). Briefly, the membranes were incubated for 1 h in the blocking buffer (5% non-fat milk in Tris-buffered saline–0.1% Tween-20) followed by the primary antibodies human IL-1β (Cell Signaling Technologies, Danvers, MA) and human IL-6 (R&D Systems, Minneapolis, MN) (1: 1000) at 4°C overnight. Then the membranes were incubated in the secondary antibody anti-mouse immunoglobulin G (IgG) and anti-goat IgG, respectively (horseradish peroxidase-conjugated) (1: 2000) for 1 h at room temperature. Both the primary and secondary antibodies were diluted in the blocking buffer, and Tris-buffered saline with Tween-20 was used for washing the membranes before, between, and after their incubation with the antibody solutions. Antibody-reactive proteins were detected using enhanced chemiluminescence reagents (ECL Plus kit; GE healthcare, Pittsburg, PA).
Quantitative real-time polymerase chain reaction
RNA was extracted from cultured cells using TRIzol (Invitrogen), and quantified by spectrometry at 260 and 280 nm. Ten micrograms from each RNA extract was used to perform first-strand complementary DNA (cDNA) synthesis using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) in a total volume of 100 μl. Real-time polymerase chain reaction (PCR) was performed using 100 ng cDNA with an ABI 7500 system (Applied Biosystems). Taq-Man probes, sense and antisense primers for gene expression of human IL-1β, IL-6, IL-8, and GAPDH as an endogenous control were purchased from Applied Biosystems. Using a Universal PCR Master Mix (Applied Biosystems), the reactions were carried out according to the manufacturer’s protocol.
Statistical analysis
All data are expressed as the mean ± SD. Statistical analyses were performed by one-way analysis of variance (anova) using the instat program (GraphPad, San Diego, CA) with Bonferroni correction. Statistical differences were considered significant at the P < 0.05 level.
Results
HGECs challenged with live P. gingivalis appear unable to elicit a secondary cytokine response, while the primary cytokine response is elevated
HGECs were challenged with live or heat-killed P. gingivalis 33277 at an MOI:100 for 4 or 24 h. Unchallenged cells were used as a negative control. Secreted IL-1β, IL-6, and IL-8 were measured in the supernatant by ELISA. At 24 h, HGECs challenged with heat-killed bacteria elicited a secondary cytokine (IL-6) and chemokine (IL-8) response, while the primary cytokine (IL-1β) response remained at a low level, comparable to that of the unchallenged control (Fig. 1), controlled possibly by a negative feedback mechanism. Surprisingly, almost no IL-6 and IL-8 were detected in the supernatant of the HGECs challenged with live bacteria, while IL-1β was increased more than four-fold (Fig. 1). This phenomenon was only observed with live P. gingivalis, because HGECs challenged with other putative periodontal pathogens under the same conditions elicited a normal primary and secondary response (Fig. 1).
Fig. 1.
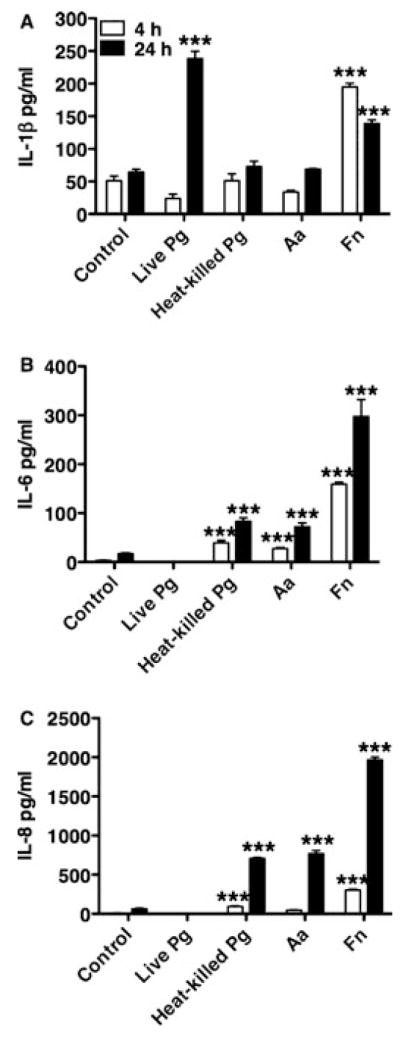
Enzyme-linked immunosorbent assay of supernatant from HGECs challenged with live or heat-killed Porphyromonas gingivalis 33277 (Pg), Aggregatibacter actinomycetemcomitans (Aa), or Fusobacterium nucleatum (Fn) MOI:100 for 4 and 24 h for interleukin-1β (IL-1β) (A), IL-6 (B), and IL-8 (C). The negative control was unchallenged human gingival epithelial cells in media. Values represent the mean ± SD of at least two experiments. Statistical comparisons are to the unchallenged negative control cells (***P < 0.001).
HGECs challenged with live P. gingivalis upregulate secondary cytokines at the transcriptional level
To identify if the downregulation of secondary cytokines after challenge with live P. gingivalis happens at the transcriptional or translational level, IL-1β, IL-6, and IL-8 messenger RNA (mRNA) levels were measured by quantitative real-time PCR (Fig. 2). HGECs were challenged with live P. gingivalis 33277 at an MOI:100 for 4 or 24 h and unchallenged cells were used as a negative control. The mRNA for all three cytokines was elevated up to two-fold after the 24-h challenge compared to the unchallenged negative control, indicating that the lack of IL-6 and IL-8 in the final supernatant (Fig. 1) was not the result of downregulation of these molecules at the transcriptional level.
Fig. 2.
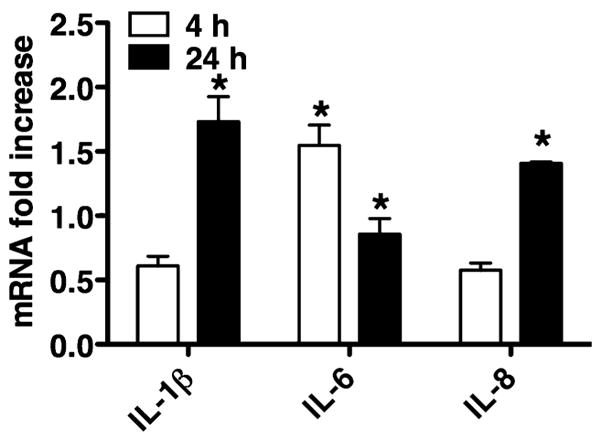
Interleukin-1β (IL-1β), IL-6, and IL-8 messenger RNA (mRNA) was measured by quantitative real-time polymerase chain reaction. Human gingival epithelial cells were challenged with live Porphyromonas gingivalis 33277 MOI:100 for 4 and 24 h. The negative control was unchallenged human gingival epithelial cells in media. The data are expressed as mRNA fold increases compared to the unchallenged negative control. Values represent the mean ± SD of at least two experiments. Statistical comparisons are to the unchallenged negative control cells (*P < 0.05).
Live P. gingivalis induce cytokine degradation and the rate of IL-6 and IL-8 degradation is higher than that of IL-1β
HGECs were challenged with heat-killed P. gingivalis 33277 for 24 h, the supernatant was collected and subsequently incubated with live P. gingivalis 33277 for 1 min, 30 min, 1 h, 2 h, and 4 h. Supernatant alone was used as a negative control. IL-1β, IL-6, and IL-8 were measured in the final supernatant by ELISA. IL-6 and IL-8 degradation began immediately and reached 100% after 30 min of exposure to P. gingivalis (Fig. 3). In contrast, there was no IL-1β degradation up to 1 h after exposure to P. gingivalis, while degradation reached 40 and 90% after 2 and 4 h, respectively. The slower rate of IL-1β degradation, compared to those for IL-6 and IL-8, could explain the persistence of IL-1β and the absence of IL-6 and IL-8 previously observed in the culture supernatant 24 h after challenge with live P. gingivalis (Fig. 1).
Fig. 3.
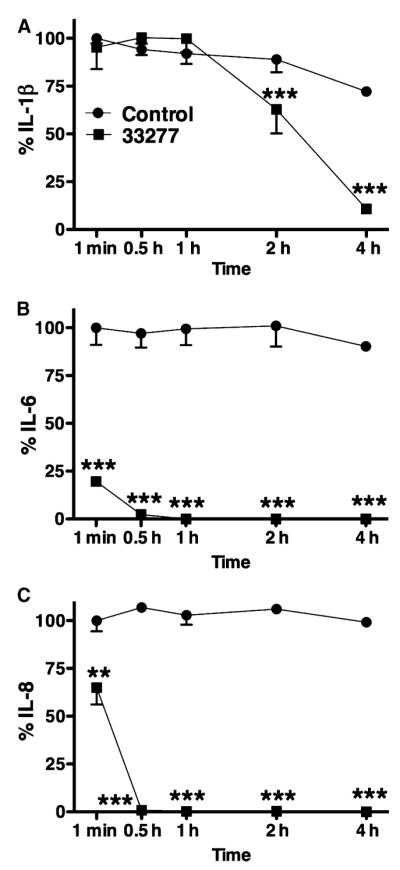
The induction of interleukin-1β (IL-1β) (A), IL-6 (B), and IL-8 (C) degradation by live Porphyromonas gingivalis was detected by enzyme-linked immunosorbent assay. Human gingival epithelial cells were challenged with heat-killed P. gingivalis 33277 for 24 h and the supernatant was subsequently incubated with live P. gingivalis 33277 for up to 4 h. The negative control was 24-h supernatant alone. The data are expressed as percentages of the initial cytokine concentration of the negative control. Values represent the mean ± SD of at least two experiments. Statistical comparisons are to the negative control (**P < 0.01, ***P < 0.001).
Porphyromonas gingivalis-induced cytokine degradation in HGECs is dependent mainly on Lys-gingipain
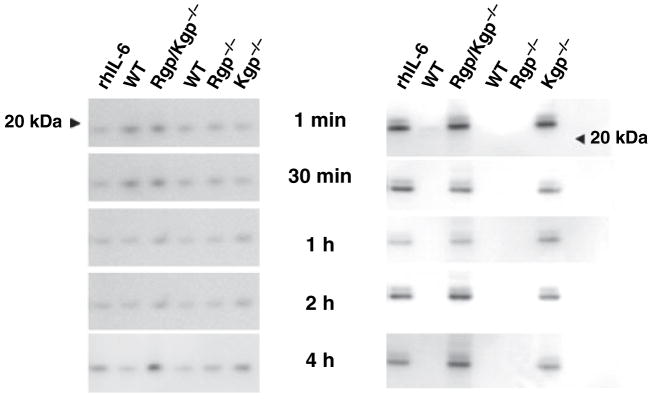
HGECs were challenged with heat-killed P. gingivalis 33277 for 24 h, the supernatant was collected and subsequently incubated with the triple gingipain mutant KDP128 or the double Arg-gingipain (RgpA/RgpB) mutant E8 or the Lys-gingipain (Kgp) mutant K1A for 1 min, 30 min, 1 h, 2 h, and 4 h. Supernatant alone was used as a negative control and live wild-type P. gingivalis 33277 and W50 were used as a positive control. IL-1β, IL-6, and IL-8 were measured in the final supernatant by ELISA. Lack of both Arg- and Lys-gingipains resulted in inability of the bacteria to induce IL-1β, IL-6, and IL-8 degradation (Fig. 4). More specifically, Lys-gingipain was responsible for degradation of all cytokines, while Arg-gingipains were only partially responsible for IL-1β and IL-8 degradation (Fig. 5). When Lys-gingipain was absent, cytokine degradation was prevented for up to 4 h, while lack of Arg-gingipains only temporarily prevented degradation of IL-1β and IL-6.
Fig. 4.
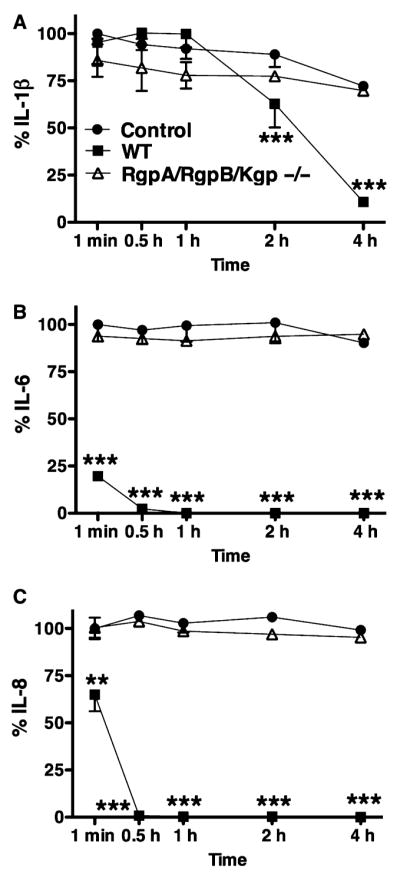
The inhibition of interleukin-1β(IL-1β) (A), IL-6 (B), and IL-8 (C) degradation by a Porphyromonas gingivalis triple gingipain-deficient mutant was detected by enzyme-linked immunosorbent assay. Human gingival epithelial cells were challenged with heat-killed P. gingivalis 33277 for 24 h and the supernatant was subsequently incubated with the live RgpA/RgpB/Kgp mutant KDP128. The negative control was 24-h supernatant alone. The positive control was supernatant incubated with live wild-type P. gingivalis 33277. The data are expressed as percentages of the initial cytokine concentration of the negative control. Values represent the mean ± SD of at least two experiments. Statistical comparisons are to the negative control (**P < 0.01, ***P < 0.001).
Fig. 5.
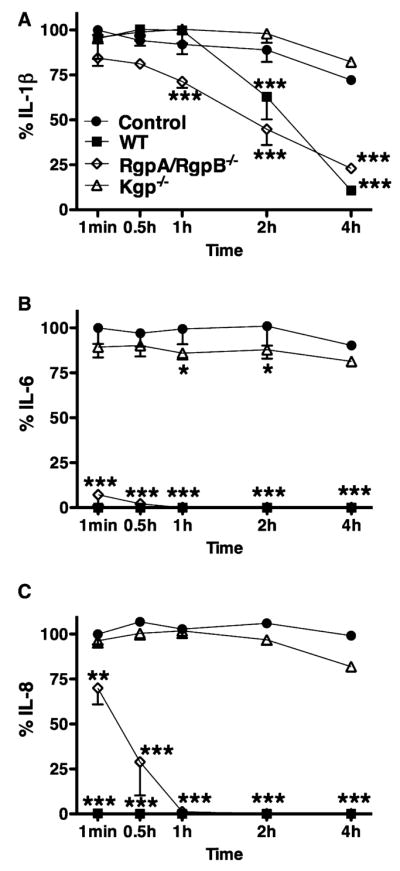
The inhibition of interleukin-1β (IL-1β) (A), IL-6 (B), and IL-8 (C) degradation by Porphyromonas gingivalis-specific gingipain-deficient mutants was detected by enzyme-linked immunosorbent assay. Human gingival epithelial cells were challenged with heat-killed P. gingivalis 33277 for 24 h and the supernatant was subsequently incubated with the live double RgpA/RgpB mutant E8 or the Kgp mutant K1A for up to 4 h. The negative control was 24-h supernatant alone. The positive control was supernatant incubated with live wild-type P. gingivalis W50. The data are expressed as percentages of the initial cytokine concentration of the negative control. Values represent the mean ± SD of at least two experiments. Statistical comparisons are to the negative control (**P < 0.01, ***P < 0.001).
In addition to the gingipain-deficient mutants, specific gingipain inhibitors were used to confirm the role of Arg-and Lys-gingipains. Preincubation of the wild-type P. gingivalis 33277 with the specific inhibitors leupeptin and zFKck or a combination of them prevented cytokine degradation in a manner similar to the respective mutants (data not shown).
Lower rate of IL-1β degradation is not linked to mode of secretion
The observed differences in cytokine degradation rates could be an artifact resulting from the different mode of secretion for IL-1β, which is in vesicles and not as a free molecule (15). To test this hypothesis, the previous experiment was repeated using purified recombinant IL-1β, IL-6, and IL-8 in single cytokine solutions. IL-1β, IL-6, and IL-8 were measured in the final solution by ELISA (data not shown) and Western blotting (Fig. 6). The results were similar to those of the previous experiment, confirming that cytokine degradation is mainly the result of Lys-gingipain and that the lower degradation rate of IL-1β is not an artifact linked to mode of secretion.
Fig. 6.
The induction of recombinant cytokine degradation by Porphyromonas gingivalis wild-type and gingipain-deficient strains was detected by Western blot. Purified recombinant human interleukin-1β (rhIL-1β) and IL-6 were incubated with live wild-type P. gingivalis 33277 or the RgpA/RgpB/Kgp mutant KDP128, and wild-type P. gingivalis W50, the RgpA/RgpB mutant E8 or the Kgp mutant K1A for up to 4 h. Purified rhIL-1β (rhIL-1β column) and rhIL-6 (rhIL-6 column), incubated alone under the same conditions, were the positive controls.
Discussion
We demonstrate that live P. gingivalis reduce the secondary cytokine and chemokine responses of primary HGECs while the primary cytokine response is elevated at 24 h. We further demonstrate that the lack of secondary response is caused by direct cytokine degradation by P. gingivalis’ proteases and that the rate of IL-6 and IL-8 degradation is significantly higher than that of IL-1β. The P. gingivalis protease, lysine gingipain, is particularly implicated in this secondary cytokine degradation.
In the present study, primary HGECs challenged with live wild-type P. gingivalis were able to elicit a high primary (IL-1β) response (Fig. 1). IL-1β is considered a ‘primary’ cytokine, because it has been previously shown to be secreted within the first minutes of challenge (27) and mediates the release of other ‘secondary’ proinflammatory cytokines, such as IL-6 and IL-8, in an autocrine manner (5). However, in our study, when live P. gingivalis were used to challenge HGECs, the secondary cytokine response was not elevated, despite the elevated primary response. The lack of secondary response was not observed at the transcriptional level because both IL-6 and IL-8 mRNA were elevated compared to the unchallenged control (Fig. 2). The fact that IL-6 and IL-8 were at higher levels indicates that the primary cytokine IL-1β is effective in enhancing the inflammatory response in an autocrine manner and that this may function to increase vascular permeability (IL-6) and to enhance leukocyte recruitment to the region (IL-8). Furthermore, HGECs challenged with heat-killed bacteria significantly upregulated IL-6 and IL-8 secretion (Fig. 1), indicating that the HGECs used are capable of mounting both a primary and a secondary response, in accordance with our previous publication (5). These data indicate that the lack of secondary cytokines after challenge with live P. gingivalis does not result from signal transduction, transcription, or translation defects in the specific HGECs but may be the result of protease activity by P. gingivalis postsecretion.
To test the hypothesis that secondary cytokine levels are reduced because of extracellular degradation, HGECs were challenged with heat-killed bacteria for 24 h and the supernatant was subsequently incubated with live wild-type P. gingivalis for up to 4 h (Fig. 3). In addition, purified recombinant human IL-1β and IL-6 were incubated with live P. gingivalis for up to 4 h (Fig. 6) to estimate degradation. IL-6 and IL-8 degradation began immediately and reached 100% after 30 min of exposure to live P. gingivalis, while there was no IL-1β degradation during the first hour of exposure (Fig. 3). These results are consistent with other reports (8, 26). The much lower rate of IL-1β degradation, compared to IL-6 and IL-8, explains the persistence of IL-1β and the absence of IL-6 and IL-8 in culture supernatants after challenge with live P. gingivalis (Fig. 1), as well as the discrepancies between different cytokines in the GCF of patients with periodontitis (3, 7, 9, 13, 16, 19, 22, 30).
The lack of a secondary response after bacterial challenge is a unique characteristic of P. gingivalis, because it was not observed after challenge with other putative periodontal pathogens, such as A. actinomycetemcomitans and F. nucleatum. (Fig. 1). It has been previously suggested that cytokine degradation by P. gingivalis involves bacterial proteases and more specifically the gingipains (2, 11, 17, 18, 26). Gingipains are cysteine proteases produced by P. gingivalis that are either secreted or membrane-bound and arginine-specific or lysine-specific. These proteases are unique to P. gingivalis and could be the causative agent of the fast cytokine degradation because other protease-producing bacteria, such as F. nucleatum, do not have a significant effect on cytokine degradation (Fig. 1). In the present study, the mechanism used by P. gingivalis to induce cytokine degradation was shown to be mainly Lys-gingipain-dependent. Gingipain-deficient P. gingivalis mutants lacking both Arg- and Lys-gingipains were unable to induce IL-1β, IL-6, and IL-8 degradation (Figs 4–6). More specifically, the Lys-gingipain-deficient mutant completely lacked the ability to degrade all cytokines, while the Arg-gingipain-deficient mutant induced cytokine degradation although at a slightly lower rate compared to the wild-type bacteria (Figs 5, 6). These results taken together indicate that cytokine degradation is mainly induced by Lys-gingipain and that the role of Arg-gingipains in the process is minimal.
The lower rate of IL-1β degradation compared to that of IL-6 or IL-8 could be due to a number of reasons. IL-1β may be degraded less because it can be secreted within vesicles, i.e. a protected mode of secretion (15); the variation in total number and spatial relationship of arginine and lysine residues might influence degradation; solvent accessibility to the molecules may differ; substrate specificity, optimal temperature, and pH may also affect degradation rates. The protected vesicle secretion of IL-1β does not appear to pertain here because purified recombinant human cytokines were assessed in the degradation assays (Fig. 6), giving results similar to the native cytokines found in the supernatants.
The possibility that the total number of arginine and lysine residues is lower for IL-1β compared to IL-6 and IL-8 is not correct because, according to the amino acid sequences, the number of predicted arginine cleavage sites is three for IL-1β, nine for IL-6, and five for IL-8. The number of lysine cleavage sites is 15 for IL-1β, 14 for IL-6, and 9 for IL-8 so although IL-1β has a lower number of arginine cleavage sites, the number of lysine cleavage sites is much higher and fails to explain the observed lack of IL-1β sensitivity to degradation by the lysine gingipain mutant (Fig. 5). Based on the crystal structure of IL-1β, IL-6, and IL-8, the arginine and lysine cleavage sites appear to be exposed and equally accessible to the solvent for all cytokines. However, the hydrolytic activity of lysine gingipain can be blocked when a lysine residue is followed by another lysine or arginine residue (4). Blockage of lysine gingipain activity in IL-1β as a result of its tertiary structure may be the reason for the observed differences in degradation rate between primary and secondary cytokines.
In conclusion, the present study provides evidence that P. gingivalis can induce a primary proinflammatory response without an elevated secondary cytokine response, as these cytokines are reduced by direct degradation, mainly by lysine gingipain. This appears to be a virulence attribute of P. gingivalis that subverts the protective host response; reduction in IL-6 will decrease the vascular and cellular inflammatory infiltrate and reduction in the chemokine IL-8 will reduce migration of phagocytes and subsequent bacterial phagocytosis and killing.
Acknowledgments
This work was supported by US Public Health Service, National Institutes of Health, NIDCR grant DE017384 to D.F.K.
References
- 1.Aduse-Opoku J, Davies NN, Gallagher A, et al. Generation of lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology. 2000;146:1933–1940. doi: 10.1099/00221287-146-8-1933. [DOI] [PubMed] [Google Scholar]
- 2.Banbula A, Bugno M, Kuster A, Heinrich PC, Travis J, Potempa J. Rapid and efficient inactivation of IL-6 gingipains, lysine-and arginine-specific proteinases from Porphyromonas gingivalis. Biochem Biophys Res Commun. 1999;261:598–602. doi: 10.1006/bbrc.1999.1075. [DOI] [PubMed] [Google Scholar]
- 3.Chung RM, Grbic JT, Lamster IB. Interleukin-8 and beta-glucuronidase in gingival crevicular fluid. J Clin Periodontol. 1997;24:146–152. doi: 10.1111/j.1600-051x.1997.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 4.Curtis MA, Aduse-Opoku J, Rangarajan M. Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med. 2001;12:192–216. doi: 10.1177/10454411010120030101. [DOI] [PubMed] [Google Scholar]
- 5.Eskan MA, Benakanakere MR, Rose BG, et al. IL-1β modulates proinflammatory cytokine production in human epithelial cells. Infect Immun. 2008;76:2080–2089. doi: 10.1128/IAI.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng L, Sun W, Xia Y, et al. Cloning two isoforms of rat cyclooxygenase: differential regulation of their expression. Arch Biochem Biophys. 1993;307:361–368. doi: 10.1006/abbi.1993.1601. [DOI] [PubMed] [Google Scholar]
- 7.Figueredo CM, Ribeiro MS, Fischer RG, Gustafsson A. Increased interleukin-1beta concentration in gingival crevicular fluid as a characteristic of periodontitis. J Periodontol. 1999;70:1457–1463. doi: 10.1902/jop.1999.70.12.1457. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher J, Reddi K, Poole S, et al. Interactions between periodontopathogenic bacteria and cytokines. J Periodontal Res. 1997;32:200–205. doi: 10.1111/j.1600-0765.1997.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 9.Guillot JL, Pollock SM, Johnson RB. Gingival interleukin-6 concentration following phase I therapy. J Periodontol. 1995;66:667–672. doi: 10.1902/jop.1995.66.8.667. [DOI] [PubMed] [Google Scholar]
- 10.Handfield M, Baker HV, Lamont RJ. Beyond good and evil in the oral cavity: insights into host–microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87:203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang GT, Kim D, Lee JK, Kuramitsu HK, Haake SK. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect Immun. 2001;69:1364–1372. doi: 10.1128/IAI.69.3.1364-1372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura T, Travis J, Potempa J. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr Protein Pept Sci. 2003;4:443–450. doi: 10.2174/1389203033487027. [DOI] [PubMed] [Google Scholar]
- 13.Jin L, Soder B, Corbet EF. Interleukin-8 and granulocyte elastase in gingival crevicular fluid in relation to periodontopathogens in untreated adult periodontitis. J Periodontol. 2000;71:929–939. doi: 10.1902/jop.2000.71.6.929. [DOI] [PubMed] [Google Scholar]
- 14.Kinane DF, Galicia JC, Gorr SU, Stathopoulou PG, Benakanakere M. P. gingivalis interactions with epithelial cells . Front Biosci. 2008;13:966–984. doi: 10.2741/2736. [DOI] [PubMed] [Google Scholar]
- 15.MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A. Rapid secretion of interleukin-1 beta by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 16.Mathur A, Michalowicz B, Castillo M, Aeppli D. Interleukin-1 alpha, interleukin-8 and interferon-alpha levels in gingival crevicular fluid. J Periodontal Res. 1996;31:489–495. doi: 10.1111/j.1600-0765.1996.tb01414.x. [DOI] [PubMed] [Google Scholar]
- 17.Mezyk-Kopec R, Bzowska M, Potempa J, et al. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect Immun. 2005;73:1506–1514. doi: 10.1128/IAI.73.3.1506-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 19.Ozmeric N, Bal B, Balos K, Berker E, Bulut S. The correlation of gingival crevicular fluid interleukin-8 levels and periodontal status in localized juvenile periodontitis. J Periodontol. 1998;69:1299–1304. doi: 10.1902/jop.1998.69.11.1299. [DOI] [PubMed] [Google Scholar]
- 20.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 21.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt RA, Masada MP, Payne JB, Allison AC, DuBois LM. Gingival fluid IL-1 beta and IL-6 levels in menopause. J Clin Periodontol. 1994;21:22–25. doi: 10.1111/j.1600-051x.1994.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 23.Sakai A, Ohshima M, Sugano N, Otsuka K, Ito K. Profiling the cytokines in gingival crevicular fluid using a cytokine antibody array. J Periodontol. 2006;77:856–864. doi: 10.1902/jop.2006.050340. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 25.Shiba H, Venkatesh SG, Gorr SU, Barbieri G, Kurihara H, Kinane DF. Parotid secretory protein is expressed and inducible in human gingival keratinocytes. J Periodontal Res. 2005;40:153–157. doi: 10.1111/j.1600-0765.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- 26.Steffen MJ, Holt SC, Ebersole JL. Porphyromonas gingivalis induction of mediator and cytokine secretion by human gingival fibroblasts. Oral Microbiol Immunol. 2000;15:172–180. doi: 10.1034/j.1399-302x.2000.150305.x. [DOI] [PubMed] [Google Scholar]
- 27.Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 28.Uehara A, Imamura T, Potempa J, Travis J, Takada H. Gingipains from Porphyromonas gingivalis synergistically induce the production of proinflammatory cytokines through protease-activated receptors with Toll-like receptor and NOD1/2 ligands in human monocytic cells. Cell Microbiol. 2008;10:1181–1189. doi: 10.1111/j.1462-5822.2008.01119.x. [DOI] [PubMed] [Google Scholar]
- 29.Uehara A, Naito M, Imamura T, et al. Dual regulation of interleukin-8 production in human oral epithelial cells upon stimulation with gingipains from Porphyromonas gingivalis. J Med Microbiol. 57:500–507. doi: 10.1099/jmm.0.47679-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Y, Slade GD, Beck JD, Offenbacher S. Gingival crevicular fluid interleukin-1beta, prostaglandin E2 and periodontal status in a community population. J Clin Periodontol. 2007;34:285–293. doi: 10.1111/j.1600-051X.2007.01057.x. [DOI] [PubMed] [Google Scholar]