Abstract
The Saccharomyces cerevisiae Rlp7 protein has extensive identity and similarity to the large ribosomal subunit L7 proteins and shares an RNA-binding domain with them. Rlp7p is not a ribosomal protein; however, it is encoded by an essential gene and therefore must perform a function essential for cell growth. In this report, we show that Rlp7p is a nucleolar protein that plays a critical role in processing of precursors to the large ribosomal subunit RNAs. Pulse–chase labeling experiments with Rlp7p-depleted cells reveal that neither 5.8SS, 5.8SL, nor 25S is produced, indicating that both the major and minor processing pathways are affected. Analysis of processing intermediates by primer extension indicates that Rlp7p-depleted cells accumulate the 27SA3 precursor RNA, which is normally the major substrate (85%) used to produce the 5.8S and 25S rRNAs, and the ratio of 27SBL to 27SBS precursors changes from approximately 1:8 to 8:1 (depleted cells). Because 27SA3 is the direct precursor to 27SBS, we conclude that Rlp7p is specifically required for the 5′ to 3′ exonucleolytic trimming of the 27SA3 into the 27SBS precursor. As it is essential for processing in both the major and minor pathways, we propose that Rlp7p may act as a specificity factor that binds precursor rRNAs and tethers the enzymes that carry out the early 5′ to 3′ exonucleolytic reactions that generate the mature rRNAs. Rlp7p may also be required for the endonucleolytic cleavage in internal transcribed spacer 2 that separates the 5.8S rRNA from the 25S rRNA.
Ribosomes are large ribonucleoproteins that translate mRNA into protein. They are ribozymes, possessing solely RNA in their active site (1). The large and small ribosomal subunits harbor distinct RNA and protein components. In the yeast Saccharomyces cerevisiae, the small subunit bears only 1 rRNA, the 18S, whereas the large subunit bears 3 rRNAs, the 5S, 5.8S, and 25S. The 18S, 5.8S, and 25S rRNAs are processed from a polycistronic RNA polymerase I transcript and assembled with their respective small and large subunit proteins in the cell nucleolus. Ribosome biogenesis is highly regulated during the cell cycle and under different growth conditions (2). An insufficient number of ribosomes may be a checkpoint for cell growth in size and therefore for cell cycle progression (3).
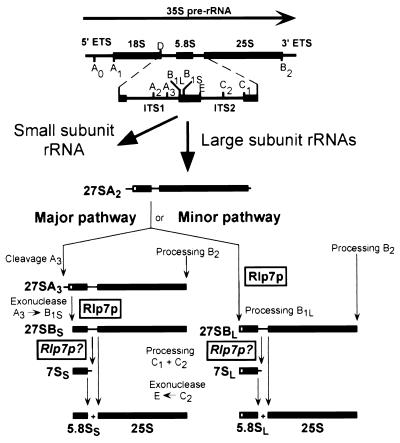
Ribosome biogenesis in S. cerevisiae begins with transcription of a 35S precursor rRNA (pre-rRNA) transcript that is modified at the nucleotide level and processed at a number of specific sites to release the mature 18S, 5.8S, and 25S rRNAs (Fig. 1; refs. 4, 5). Processing of RNA precursors to the 18S rRNA requires a number of endonucleolytic cleavage reactions. The four small nucleolar ribonucleoproteins (snoRNPs), U3, U14, snR10, and snR30 play an essential role in these reactions, but none of them have yet been shown to possess the cleaving enzyme. Processing of the large subunit RNAs, 5.8S and 25S, requires both endonucleolytic cleavage and exonucleolytic processing of the cleavage products and begins on the 27SA2 precursor, which results from endonucleolytic cleavage at the A2 site in internal transcribed spacer (ITS) 1 (Fig. 1). Two forms of 5.8S rRNAs (5.8SS and 5.8SL) differing slightly in length are made via two different processing pathways known as the major and minor pathways (6). In both pathways, the initial 5′ end processing reactions occur concurrently with 3′ end processing at the B2 site. In the major pathway, which generates 85% of the 5.8S and 25S rRNA, the 27SA2 transcript is first cleaved by RNase MRP at the A3 site. The 5′ end of the resulting 27SA3 transcript is then processed exonucleolytically by the 5′ to 3′ exonucleases, Rat1p and Xrn1p, to generate the 27SBS transcript (6). In the minor pathway, which generates 15% of the 5.8S and 25S rRNA, little is known about the factors involved in the initial processing step that generates the 27SBL pre-rRNA from the 27SA2 precursor. However, it is clear that the 5′ ends of the 27SBS and 27SBL precursors are the mature 5′ ends of what will become the 5.8SS and 5.8SL rRNAs, respectively. From this point on, the processing steps in both the major and minor pathways are identical. Endonucleolytic cleavage of the 27SBS/27SBL precursors at the C2 site in ITS2 generates the 7S precursors to the 5.8S rRNAs. The 7S precursor is processed at its 3′ end by a series of 3′ to 5′ exonucleases, which includes the Rex proteins and the exosome to generate 5.8SS and 5.8SL (7, 8). The 5′ end of the 25S precursor generated by cleavage at C2 is also matured by Rat1p and Xrn1p to produce the mature 25S rRNA (9).
Figure 1.
The pre-rRNA processing pathway in S. cerevisiae. Yeast pre-rRNA is synthesized as a 35S primary transcript that gets cleaved and trimmed at several sites to produce the mature 18S, 5.8S, and 25S rRNAs of the ribosome. Pre-rRNA processing results in a number of recognizable pre-rRNA processing intermediates. The first processing steps occur in the 5′ portion of the 35S RNA and lead to the production of precursors to the small ribosomal subunit RNA; these processing reactions in turn release the 27SA2 precursor, which will produce the large ribosomal subunit RNAs. The 27SA2 precursor can enter two different pathways to generate the mature 5.8S and 25S rRNAs. Processing through the major pathway is initiated by RNase MRP cleavage at the A3 site, and the 27SA3 RNA gets further trimmed by 5′ to 3′ exonucleases; this exonucleolytic reaction extends to the 5′ end of the 27SBS precursor (site B1S), which is also the 5′ end of mature 5.8SS rRNA. Processing of the 27SA2 precursor through the minor pathway may also be initiated by 5′ to 3′ exonucleolytic enzyme(s); this reaction extends to site B1L and generates the 5′ end of the 27SBL, which is the mature 5′ end of 5.8SL rRNA. In both the major and minor pathways, endonucleolytic cleavages in ITS2 generate the mature 5′ end of 25S rRNA and the 7S precursors that get trimmed by 3′ to 5′ exonucleases to produce the mature 3′ end of 5.8S rRNAs. This figure has been adapted from Kressler et al. (5).
To date, the only proteins so far implicated in 5′ to 3′ exonucleolytic processing of rRNA precursors are the Rat1 and Xrn1 proteins. Their subcellular localization is difficult to resolve with their proposed collaborative role in pre-rRNA processing in the nucleolus. Rat1p, encoded by an essential gene, is nuclear, whereas Xrn1p, encoded by a nonessential gene, is cytoplasmic (6, 10, 11). Both participate in additional exoribonucleolytic reactions in their respective cellular compartments (6, 10–13). These exoribonucleases act on multiple substrates and therefore may require other proteins as specificity factors to direct them to particular substrates.
One of the U3 snoRNP components required for ribosome biogenesis is a protein called Imp3p (14). Imp3p is a member of the S4 family of ribosomal proteins (S9 in yeast and humans), which are RNA-binding proteins of remarkable versatility (15). We reasoned that there might be other uncharacterized proteins in S. cerevisiae that would have extensive identity and similarity to a ribosomal protein but that were not components of cytoplasmic ribosomes. By using the key features we gleaned from our study of Imp3p as a guide, we identified a protein named Rlp7p in the Yeast Proteome Database (16). Rlp7p had been previously discovered because of its extensive identity and similarity with the large ribosomal subunit protein L7, but its function had not been elucidated (17, 18). Because Imp3p, a member of a family of small ribosomal subunit proteins, was found to be required for processing of the small subunit pre-rRNA, we hypothesized that Rlp7p might, by analogy, be required for processing of the RNA precursors to the large subunit RNAs.
Here we report that Rlp7p is indeed a nucleolar protein required for an early step in the processing of the precursors to the 5.8S and 25S rRNAs in both the major and minor processing pathways. Because it possesses a conserved RNA-binding domain, we propose that it may serve as a specificity factor for the 5′ to 3′ exonucleases that are involved in 5′ end maturation of the pre-rRNAs. Rlp7p may also be required for the essential endonucleolytic cleavage that occurs in ITS2 to separate the 5.8S from the 25S rRNA.
Materials and Methods
Disruption of RLP7 and Expression of Rlp7 Protein from a Conditional Promoter.
Studies were performed with the diploid strain YPH260 or its haploid derivatives, YPH258 and YPH259, and grown as previously described (14).
One allele of the RLP7 gene was disrupted in YPH260 by using a PCR-based strategy (14, 19, 20). The two oligonucleotides used to create the disruption cassette were synthesized to be complementary to the RLP7 5′ and 3′ flanking regions and to a HIS3 gene from plasmid pAD11. The sequences of the oligonucleotides are: 5′-TAGAAAGAAGAGGCTGCCACAGGGAAAAAGAATAAGATATACAAGTCACACTCTTGGCCTCCTCTA (rlp7.1) and 5′-TTAGATAACTAACAACCTATGTACTATACAATTTTAAAATACTCTCTTAATCGTTCAGAATGACACG (rlp7.2). Confirmation that the RLP7 gene had been replaced with HIS3 was carried out by using the PCR on chromosomal DNA with oligonucleotides that flank the RLP7 locus. The oligonucleotides used for confirmation are 5′-GGTACTTTCGTTATT (rlp7.3) and 5′-TCTGCTCCTTCAATG (rlp7.4).
To create a conditionally expressed RLP7 gene, it was cloned under the control of a GAL1–10 promoter in the centromeric-based plasmid pGAD3 (14, 19, 21). The GAL1–10 promoter is activated in the presence of galactose but is repressed in the presence of glucose. The RLP7 gene was amplified by PCR from yeast chromosomal DNA with a 5′ oligonucleotide 5′-CCGCGGATCCATGTCTTCAACTCAAGAC (rlp7.5) that contains a BamHI restriction site and a 3′ oligonucleotide 5′-GCATAGTTTAGCGGCCGCTTAATTCAACTTGGCTAATAAAG (rlp7.6) that contains a NotI restriction site. The PCR product was digested with BamHI and NotI and ligated into the same sites of plasmid pGAD3 to make pGAD3-RLP7. This plasmid was transformed into a heterozygous disrupted strain by lithium acetate transformation. Diploids were sporulated, and tetrads were dissected on plates containing galactose. The strains from the resulting spores were streaked onto 5-fluoroorotic acid to identify a haploid strain that harbors the plasmid-borne RLP7 gene and the chromosomally disrupted RLP7 gene (22). We call this strain pGAL1∷RLP7.
Indirect Immunofluorescence Microscopy.
To obtain an HA-tagged Rlp7 protein, the ORF of RLP7 was amplified by using the polymerase chase reaction with oligonucleotides 5′-TCCCCGGGCATGTCTTCAACTCAAGAC (19.72) and 5′-CCGCCTCGAGTAATTAATTCAACTTGGC (20.20) bearing XmaI and XhoI sites, respectively, and cloned into the XmaI and XhoI sites of pACT2 (CLONTECH). The BglII DNA fragment coding for HA-Rlp7p was subcloned into the BamHI site of plasmid p415 ADH (23) to generate pFD207. Proper orientation of the insert was checked by restriction digestion. Strain pGAL1∷RLP7 was transformed with pFD207 and grown in the presence of glucose and 5-fluoroorotic acid to shuffle the plasmid pFD207 for pGAD3-RLP7 (22). Expression of HA-Rlp7p in different transformants was checked by Western blotting, and one strain (YFD44) was chosen for further analysis. Strain YFD44 was grown in yeast extract/peptone/dextrose (YPD) to an OD600 of 0.5 and 10 ml of cells was processed for indirect immunofluorescence microscopy, as described by Michael Snyder's laboratory (http://ygac.med.yale.edu/triples/Immu_protocols.html), except that glusulase/zymolase treatment was reduced to 30 min. Fixed cells were incubated overnight at 4°C with rabbit anti-HA.11 antibodies (Covance, Richmond, CA) diluted 1:250 and mouse anti-Nop1p mAb A66 (a kind gift of John Aris, University of Florida; ref. 24) diluted 1:2,000. Secondary antibodies (Jackson ImmunoResearch) were used at dilution of 1:400 for FITC-conjugated anti-rabbit and 1:200 for tetramethylrhodamine B isothiocyanate-conjugated anti-mouse antibodies and incubated for 2 h at room temperature. Nuclear DNA was stained by incubation with 1 μg/ml 4′,6-diamidin-2-phenylindole (Sigma) for 5 min followed by two washes. Cells were mounted with Fluoromount-G (Southern Biotechnology Associates) containing 2% 1,4-diazobicyclo-(2,2,2)-octane (Sigma). Specimens were observed with a Zeiss Axiophot 2 microscope using a ×100 oil-immersion objective. Images were acquired with a Photometrics Quantix charge-coupled device camera, and digitized images were treated with Scanalytics ip lab (Billerica, MA) software.
Pulse–Chase Labeling of rRNA.
Pulse–chase labeling with l-[methyl-3H]methionine was carried out on the pGAL1∷RLP7 and YPH259 strains as previously described (14, 19). Twenty thousand cpm per sample was resolved on a 1.2% formaldehyde–agarose gel. Labeled RNAs were transferred to a zeta-probe membrane (Bio-Rad), sprayed with EN3HANCE (Dupont), and exposed to x-ray film.
For pulse–chase labeling with [5,6-3H] uracil, a control strain was made, YPH259-pGAD3, where YPH259 was transformed with an empty pGAD3 plasmid that contains URA3. Both pGAL1∷RLP7 and YPH259-pGAD3 cells were grown to an OD600 of approximately 1.0 in SGal–uracil (synthetic medium with galactose and supplements but without uracil) media and diluted 1:500 into fresh SD–uracil media (synthetic medium with dextrose and supplements but without uracil). Approximately 50 OD600 units of pGAL1∷RLP7 cells and 10 OD600 units of YPH259-pGAD3 cells was resuspended in 0.4 ml SD–uracil media and labeled with 25 μCi of [5,6-3H] uracil per milliliter (47 Ci/mmol) at room temperature for 2 min. The chase was initiated by adding the pulsed-labeled cells to 4 ml of SD medium containing 1 mg/ml of cold uracil. At 0, 5, 30, and 60 min after the chase, 1-ml aliquots of cells were pelleted, resuspended in cold dH2O, repelleted, and quick frozen in a dry-ice ethanol bath. Total labeled RNAs were isolated by the hot-phenol technique (25). Thirty thousand cpm per sample was resolved on either a 1.2% agarose gel or an 8% polyacrylamide gel. Labeled RNAs were transferred to a zeta-probe membrane, sprayed with EN3HANCE (Dupont), and exposed to x-ray film.
Mapping of the 5′ Ends of Mature and pre-rRNAs by Primer Extension.
The pGAL1∷RLP7 strain was grown in galactose medium (yeast extract/peptone/galactose) YPGal to an OD600 of 1.0 and diluted 1:500 into fresh glucose medium (YPD). Cells were then grown in glucose medium for different periods of time. Cells were pelleted, and total RNA was isolated by the hot-phenol technique (25). The oligonucleotide A2/A3 (5′-GGCCAGCAATTTCAAGTTA) was used to map cleavages at the A2, A3, B1S, B1L processing sites. The C1 oligonucleotide (5′-TACTAAGGCAATCCGGTTGG) was used to map the 5′ ends of the 25S rRNA. These oligonucleotides have been used previously to map the 5′ ends of yeast precursor and mature rRNAs (26). Primer extensions were carried out on total RNA from pGAL1∷RLP7 cells grown in galactose medium (YPGal) and glucose medium (for depletion; YPD) and compared with total RNA from YPH259 cells grown in the same media. For each sample, 5 μg of total RNA was annealed to 15 ng of 32P-labeled oligonucleotides for 10 min at 70°C. Primer extensions were carried out in 20 μl of a buffer containing 50 mM Tris⋅HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, and 10 mM dTT in the presence of 10 units/microliter Superscript reverse transcriptase (GIBCO/BRL) and 1 mM dNTPs at 42°C for 1 h. For primer extensions using the 5.8S and C1 oligonucleotides, the extension products were resolved on a 10% sequencing gel. For primer extensions using the A2/A3 oligonucleotide, the extension products were resolved on a 5% polyacrylamide gel.
Results
Mining the S. cerevisiae Genome for Proteins Required for Ribosome Biogenesis.
We have previously characterized Imp3p, a U3-specific protein that is a member of the S4 family of ribosomal proteins. The S4 ribosomal protein (S9 in yeast and humans) is a primary RNA-binding protein. Like the depletion of the U3 small nucleolar RNA (snoRNA), depletion of Imp3p prevents the synthesis of mature 18S rRNA (14). Analysis of the Yeast Proteome Database (16) indicated there were other uncharacterized essential ORFs with extensive identity (greater than 20%) to ribosomal proteins. These proteins had a low codon bias, indicating that they are expressed at much lower levels than ribosomal proteins. RLP7 is one such ORF with extensive homology to the rRNA-binding protein, ribosomal protein L7 (17, 18). Rlp7p is 32% identical to the yeast ribosomal protein L7 and 29% identical to the rat ribosomal protein L7. It does not cosediment with ribosomes and cannot substitute for ribosomal protein L7. Both the yeast and rat L7 proteins have a conserved RNA-binding domain located at the carboxy terminus (27) that is also conserved in Rlp7p. We hypothesized that Rlp7p may represent another example of a transacting nucleolar factor that has “borrowed” an ancient RNA-binding domain from a ribosomal protein for use in ribosome biogenesis in the nucleolus.
Rlp7p Is Localized to the Nucleolus.
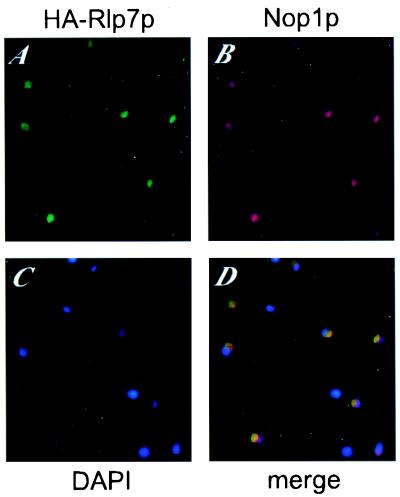
To determine the subcellular localization of Rlp7p, a yeast strain expressing an HA epitope-tagged Rlp7p was used for indirect immunofluorescence microscopy (Fig. 2). The N-terminally tagged Rlp7p (HA-Rlp7p) localizes to a crescent-shaped region of the nucleus (Fig. 2A) that is characteristic of the yeast nucleolus. Cells were stained for Nop1p (yeast fibrillarin; refs. 28, 29) as a nucleolar marker (Fig. 2B). The merge of the images from the green channel (HA-Rlp7p) and red channel (Nop1p) clearly shows that HA-Rlp7p and Nop1p colocalize in the nucleolus, which appears in yellow in Fig. 2D. Therefore, we conclude that Rlp7p is a nucleolar protein and must have a function in that compartment.
Figure 2.
Rlp7p localizes to the nucleolus. Immunofluorescence microscopy was performed with a strain expressing an HA-tagged Rlp7p. Rabbit anti-HA antibodies were used to detect HA-Rlp7p (A) and the mouse mAb A66 was used to detect the nucleolar protein Nop1p (fibrillarin) (B). Nuclear DNA was stained with 4′,6-diamidin-2-phenylindole (C). The images were merged (D); the yellow staining results from overlapping of the green and red signals.
Rlp7p Is Required for Processing of the Precursors to the 5.8S and 25S rRNAs.
To test whether Rlp7p plays a role in pre-rRNA processing, we constructed the strain pGAL1∷RLP7. This haploid strain bears a disrupted chromosomal copy of RLP7 but maintains a plasmid with the RLP7 gene under the control of a galactose-inducible/glucose-repressible promoter. Rlp7p is expressed when this strain is grown in galactose but not when it is grown in glucose. We assessed the growth characteristics of pGAL1∷RLP7 and found that growth in glucose begins to slow 10–15 h after the switch from galactose (data not shown), as has been observed previously on depletion of other essential proteins required for ribosome biogenesis (14, 19).
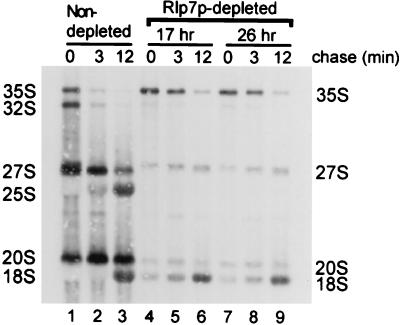
We investigated whether Rlp7p is required for processing of the large subunit rRNAs by analysis of newly made rRNAs via l-[methyl-3H] methionine pulse–chase labeling in vivo. l-[methyl-3H] methionine labels the 2′-O-methyl groups of the ribose moieties on newly synthesized rRNA. In the isogenic YPH259 strain, the 35S precursor was processed rapidly into the 27S and 20S pre-rRNAs and then into the mature 25S and 18S rRNAs (Fig. 3, lanes 1–3). In the Rlp7p-depleted strain, the overall incorporation of label into the methyl groups of ribose moieties is reduced, as has been observed previously (14, 19), perhaps because growth of this strain is greatly slowed. In contrast to the nondepleted strain, pre-rRNA processing defects were observed on Rlp7p depletion. The 27S precursor was made but was not processed into the mature 25S rRNA (Fig. 3, lanes 4–9). The depletion of Rlp7p did not significantly affect processing of the 18S rRNA, because 20S and 18S rRNAs were present at all time points (Fig. 3A, lanes 4–9). Small effects on pre-18S processing, probably secondary to the defect in large subunit biogenesis, can be seen in the lag of 35S to 32S conversion. Similar results were obtained by using [5,6-3H] uracil pulse–chase labeling (data not shown). Therefore, Rlp7p depletion inhibits processing of the 27S rRNA precursor to the mature 25S rRNA and suggests that Rlp7p is required for processing of the 27S rRNA precursors.
Figure 3.
Rlp7p depletion prevents the synthesis of the 25S rRNA. The pGAL1∷RLP7 strain was depleted of Rlp7p for 17 and 20 h (depleted) and compared with an otherwise isogenic strain, YPH259 (nondepleted). Cells were pulse labeled for 2.5 min with l-[methyl-3H] methionine and then chased for 3 and 12 min with an excess of cold methionine. Total RNA was extracted, and 20,000 cpm per lane was loaded and resolved on a 1.2% formaldehyde–agarose gel. Labeled RNAs were transferred to ζ-probe membrane and visualized by fluorography.
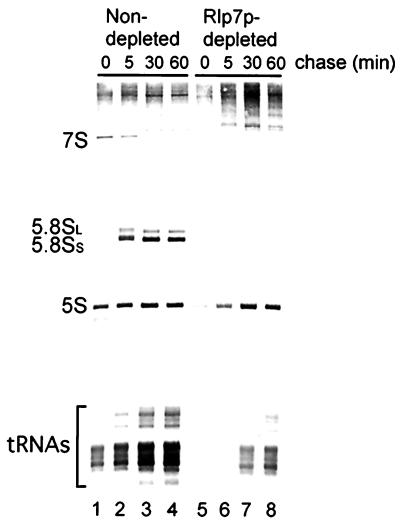
The processing of precursors to the small rRNAs (5.8S and 5S) was assessed by analysis of newly synthesized rRNA by using [5,6-3H] uracil pulse–chase labeling in vivo. In the non-Rlp7p-depleted strain (YPH259-pGAD3), the 7S pre-rRNA is processed into the mature 5.8SL and 5.8SS rRNAs (Fig. 4, lanes 1–4). In the Rlp7p-depleted cells, which are growing much more slowly than the nondepleted, incorporation of [5,6-3H] uracil into newly synthesized RNAs is also slowed. However, when Rlp7p is depleted, the 7S precursor and mature 5.8S rRNAs are absent even at the latest time points (Fig. 4, lanes 5–8), suggesting that Rlp7p is required for the synthesis of both mature 5.8SL and 5.8SS rRNAs. Because the levels of newly synthesized 5S and tRNA are similar in Rlp7p-depleted and nondepleted cells after a long chase, Rlp7p is not required for their synthesis (Fig. 4, lanes 4 and 8). Taken together, this analysis of pre-rRNA processing indicates that Rlp7p is required for processing of precursors to the large subunit RNAs, 5.8S, and 25S. Furthermore, because neither form of the 5.8S rRNAs is made, Rlp7p is required for pre-rRNA processing in both the major and minor pathways.
Figure 4.
Rlp7p depletion prevents the synthesis of both 5.8SS and 5.8SL rRNA. The pGAL1∷RLP7 cells were depleted of Rlp7p for 17 h (depleted) and compared with an otherwise isogenic strain YPH259-pGAD3 (nondepleted). Cells were pulse labeled for 2 min with [5,6-3H] uracil and then chased for 5, 30, and 60 min with an excess of cold uracil. Total RNA was extracted, and 30,000 cpm per lane was resolved on an 8% polyacrylamide gel. Labeled RNAs were transferred to zeta-probe membrane and visualized by fluorography.
Rlp7p Is Required for the Reaction That Processes the 27SA3 Precursor.
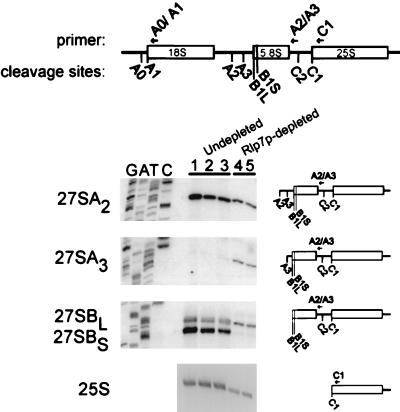
To pinpoint the precise step(s) at which pre-rRNA processing is blocked, the 27S precursors were mapped at their 5′ ends by primer extension analysis. Using an oligonucleotide that hybridizes to a region in ITS2, primer extensions were performed to map the levels of the 27SA2, 27SA3, 27SBL, and 27SBS transcripts after Rlp7p depletion. Rlp7p depletion results in slightly decreased levels of the 27SA2 precursor, which has been observed previously when large ribosomal subunit biogenesis is interrupted and has been attributed to negative feedback control of pre-18S processing (4). An increased signal at the A3 cleavage site was apparent, indicating that Rlp7p depletion results in increased levels of the 27SA3 precursor (Fig. 5, lanes 4 and 5). A large reduction in the signal at the B1S cleavage site was observed, indicating a large reduction in the levels of the 27SBS precursor when Rlp7p is depleted (Fig. 5, lanes 4 and 5). The ratio of 27SBL to 27SBS precursors changes from approximately 1:8 (wild-type cells) to 8:1 (depleted cells). The oligonucleotide used for this primer extension also hybridizes to the 7S precursor. On Northern blots, the 7S steady-state levels were greatly reduced on Rlp7p depletion, and the ratios of 7S long to short were unchanged (data not shown). Because of this, it is likely that primer extension with this oligonucleotide reflects the levels of 27SB precursors. The increased levels of the 27SA3 precursor and the decreased levels of the 27SBS precursor, which is the exonucleolytic digestion product of the 27SA3 precursor, suggest that Rlp7p depletion prevents the exonucleolytic processing of the 27A3 to the 27SBS pre-rRNA (Fig. 1).
Figure 5.
The 27SA3 pre-rRNA accumulates during Rlp7p depletion. RNA was isolated from the pGAL1∷RLP7 strain after growth in galactose (lane 3) or 17 h (lane 4) and 20 h (lane 5) after switching to growth in glucose. RNA was isolated from an otherwise isogenic strain, YPH259, after growth in galactose (lane 2) or 24 h after the switch to growth in glucose (lane 1). Primer extension with the oligonucleotide A2/A3 that hybridizes to ITS2 was used to reveal processing sites A2, A3, B1L, and B1S. Primer extension with the oligonucleotide C1 that hybridizes to the 25S rRNA was used to map the 5′ end of the 25S rRNA. Products of the primer extension reactions were resolved on polyacrylamide gels next to a DNA sequencing ladder (not shown for 25S) and exposed to x-ray film.
Primer extensions were also performed to map the mature 5′ end of the 25S rRNA. Reduced signal at the 5′ end of 25S (C1 processing site) was observed by using an oligonucleotide that hybridizes to 25S rRNA, indicating that Rlp7p depletion leads to diminished amounts of the mature 25S rRNA (Fig. 5, lanes 4 and 5). This result is similar to the results obtained by pulse–chase labeling (Fig. 3) and by Northern blot analysis of steady-state RNA (data not shown).
Discussion
The nucleolar protein, Rlp7p, plays a unique role in processing of the precursors to the large subunit rRNAs, 5.8S and 25S. Analysis of newly synthesized rRNA after Rlp7p depletion indicates that Rlp7p is required for the production of 5.8S and 25S rRNAs but not 18S rRNA. Primer extension analysis indicates that Rlp7p depletion leads to the accumulation of the 27SA3 pre-rRNA and the subsequent decrease in its 27SBS processing product, with a concomitant reversal in the 27BSS to 27BL ratio (Fig. 1). The results from the pulse–chase analysis and appearance of these specific processing intermediates after Rlp7p depletion indicate that Rlp7p is required for a very early step in processing of the pre-rRNA precursors.
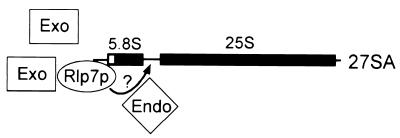
Processing of the 27SA3 to the 27SBS rRNA precursor in the major pathway involves a 5′ to 3′ exonucleolytic processing reaction. This reaction matures the 5′ end of the 5.8SS rRNA and involves the Rat1p and Xrn1p exonucleases. Our results suggest that Rlp7p is also required for this 5′ to 3′ exonucleolytic processing reaction. As these two exonucleases are promiscuous and not solely nucleolar, Rlp7p may act as a specificity factor that brings the exonucleases to the 27SA3 precursor via its proposed RNA-binding domain (Fig. 6). Rlp7p may also interact with other unidentified 5′ to 3′ exonucleases that participate in this reaction. In addition, our results indicate that Rlp7p is required for the production of the long form of 5.8S and is therefore an essential component of the minor processing pathway (Fig. 1). Although it has not been experimentally demonstrated, processing in the minor pathway may also be carried out by 5′ to 3′ exonucleases (4). If so, Rlp7p may serve as a specificity factor in this pathway for maturation of the 5′ end of 5.8SL as well. On the basis of the results presented in this report, Rlp7p is the only known factor shared by both the major and minor pathways at this early processing step in large ribosomal subunit biogenesis.
Figure 6.
Model for the role of Rlp7p in processing of 27SA precursor RNAs. Rlp7p interacts with the pre-rRNA through its RNA-binding domain and tethers exonucleases (Exo) that carry out the 5′ to 3′ exonucleolytic reactions at the 5′ end of 27SA precursors. Rlp7p could also interact with ITS2 and attract the endonuclease (Endo) that cleaves it, as symbolized by the arrow and question mark.
Rlp7p may also be required for the proposed endonucleolytic cleavage in ITS2 at the C2 site and is the first such candidate protein required for this processing event (Fig. 6). In a cell where normal pre-rRNA processing is occurring, cleavage in ITS2 at the C2 site results in the appearance of the 7S precursors, which is the direct precursor to the 5.8S rRNAs. Because no 7S RNA is detectable either by analysis of newly synthesized rRNA (Fig. 4) or by analysis of steady-state rRNA (data not shown) in Rlp7p-depleted cells, we find that Rlp7p is also essential for this processing event (Fig. 1).
We cannot distinguish between a primary role for Rlp7p in 5′ to 3′ exonucleolytic processing or in endonucleolytic cleavage in ITS2. One possibility is that disruption of the 5′ to 3′ exonucleolytic processing at 27SA3 leads to a defect in ITS2 cleavage. However, previous results suggest that 5′ end maturation of the 5.8S rRNAs is not required for cleavage at C2. Studies on both the RNA and protein components of the MRP ribonucleoprotein, which is required for cleavage at the A3 site, suggest that cleavage can occur normally in ITS2 if 5′ end maturation of 5.8SS is blocked (reviewed in ref. 4). In these cases, extended forms of 5.8S or 7S rRNA are detectable by Northern blot hybridization. After Rlp7p depletion, we were unable to detect any 5′ extended forms of 5.8S or 7S rRNA by Northern blot hybridization (data not shown). Rlp7p may, in fact, be required for both the maturation of the 5′ ends of the 5.8S rRNAs and the crucial C2 cleavage event that leads to the release of the 25S rRNA, ensuring that production of these two RNA components of the large ribosomal subunit are coordinately regulated.
In addition to their role as trans-acting factors in ribosome biogenesis, the 5′ to 3′ exonucleases Rat1p and Xrn1p have also been implicated in the maturation of other small, stable RNAs such as U18 and U24 (12, 13). We examined whether Rlp7p was required for these reactions by probing for 5′ extended U18 and U24 snoRNAs on Northern blots of RNA isolated from Rlp7p-depleted cells. No 5′ extended U18 or U24 snoRNAs were detectable (data not shown). Similarly, levels of other small RNAs (U1, U3, U4, U5, U6) were not affected (data not shown). Thus, the potential cooperation between Rlp7p and Rat1p/Xrn1p does not extend to processing of snoRNA precursors.
Recently, it has been found that Rlp7p is complexed with Nop7p and a number of other protein components and may therefore be part of a large processing complex committed to large ribosomal subunit biogenesis (P. Harnpicharnchai, E. Horsey, J. Jakovljevic, J. Roman, M. Unlu, M. Rout, and J. Woolford, Jr., personal communication). In separate studies, a snoRNA and trans-acting protein factors may also be involved with Rlp7p in pre-rRNA processing on the basis of genetic disruption of their functions. Depletion of the essential snR30 RNA, a member of the box H/ACA class of snoRNAs, leads to accumulation of the 27SA3 pre-rRNA (6). To investigate whether Rlp7p was a component of the snR30 ribonucleoprotein, we performed immunoprecipitations of HA-tagged Rlp7p followed by direct labeling of the resulting associated small RNAs. These analyses revealed no reproducible Rlp7p-snoRNA association (data not shown). Likewise, accumulation of 27SA3 is visualized in a strain that harbors a temperature-sensitive mutation in the Ebp2 protein, which is also required for large rRNA biogenesis (30). Depletion of Rpf1p also leads to accumulation of the 27SA3 precursor and a defect in biogenesis of the 5.8S and 25S rRNAs (K. A. Wehner, S.J.L., and S.J.B., unpublished results). In the two examples (Ebp2 and Rpf1p) where it has been studied, the 27SBL to 27SBS precursor ratios were unchanged, which is distinct from what we have observed with Rlp7p. A third protein, Rai1p, is a nonessential protein that binds to Rat1p and, when disrupted, also leads to accumulation of 27SA3 (31). Therefore, Ebp2p, Rpf1p, and Rai1p may interact with Rlp7p to carry out processing of the 27SA rRNA precursors.
Imp3p and Rlp7p are required for processing of rRNA precursors of the small and large ribosomal subunits, respectively. Imp3p has homology to a small subunit protein and is required for pre-18S rRNA processing, whereas Rlp7p has homology to a large subunit protein and is required for pre-5.8S and pre-25S processing. Both bear conserved RNA-binding domains, although as of yet neither has been shown to bind directly to RNA. It is tempting to speculate that perhaps each is acting as a placeholder for its homologous ribosomal protein on the nascent pre-rRNA.
Acknowledgments
We thank John Aris for the anti-Nop1 monoclonal antibody, Jesus de la Cruz, University of Sevilla, Spain, for the protocol for [5,6-3H] uracil labeling, and Jim Jamieson for the use of his microscope. We are grateful to Baserga lab members Karen Wehner and Jen Gallagher, who provided insightful comments on the manuscript and help in preparation of the figures. F.D. was supported by the Anna Fuller Fund for Molecular Oncology of the Yale Cancer Center. S.J.B. is a member of the Yale Cancer Center.
Abbreviations
- pre-rRNA
precursor rRNA
- snoRNA
small nucleolar RNA
- snoRNP
small nucleolar ribonucleo-protein
- ITS
internal transcribed spacer
- YPD
yeast extract/peptone/dextrose
References
- 1.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 2.Warner J R. Trends Biochem Sci. 1999;24:437–439. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 3.Volarevic S, Stewart M J, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma S C, Thomas G. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 4.Venema J, Tollervey D. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 5.Kressler D, Linder P, de la Cruz J. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 8.van Hoof A, Lennertz P, Parker R. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geerlings, T. H., Vos, J. C. & Raue, H. A. (2000) RNA, in press. [DOI] [PMC free article] [PubMed]
- 10.Johnson A W. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larimer F W, Hsu C L, Maupin M K, Stevens A. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 12.Petfalski E, Dandekar T, Henry Y, Tollervey D. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu L H, Henras A, Lu Y J, Zhou H, Zhou W X, Zhu Y Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S J, Baserga S J. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravind L, Koonin E V. J Mol Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- 16.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancer A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, et al. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuta K, Hashimoto T, Otaka E. Curr Genet. 1995;28:19–25. doi: 10.1007/BF00311877. [DOI] [PubMed] [Google Scholar]
- 18.Lalo D, Mariotte S, Thuriaux P. Yeast. 1993;9:1085–1091. doi: 10.1002/yea.320091007. [DOI] [PubMed] [Google Scholar]
- 19.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobel S G, Synder M. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 24.Aris J P, Blobel G. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. New York 275, 28764–28773: Wiley; 1995. [Google Scholar]
- 26.de la Cruz J, Kressler D, Tollervey D, Linder P. EMBO J. 1997;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Mickecz A, Neu E, Krawinkel U, Hemmerich P. Biochem Biophys Res Commun. 1999;258:530–536. doi: 10.1006/bbrc.1999.0682. [DOI] [PubMed] [Google Scholar]
- 28.Henriquez R G, Blobel G, Aris J P. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 29.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber, M. D., Dworet, J. H., Shire, K., Frappier, L. & McAlear, M. A. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 31.Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson A W. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]