Summary
The MYST family of lysine acetyltransferases has been intensely studied because of its broad conservation and biological significance. In humans, there are multiple correlations between the enzymes and development and disease. In model organisms, genetic and biochemical studies have been particularly productive because of mechanistic insights they provide in defining substrate specificity, the complexes through which the enzymes function, and the sites of their activity within the genome. Established and emerging data from yeast reveal roles for the three MYST enzymes in diverse chromosomal functions. In particular, recent studies help explain how MYST complexes coordinate with other modifiers, the histone variant H2A.Z, and remodeling complexes to demarcate silent and active chromosomal domains, facilitate transcription, and enable repair of DNA damage.
Keywords: SAS, Esa1, NuA4, SAS3, NuA3
Overview
The DNA coding sequence of nearly every cell in an organism is identical. However, in each cell, DNA is packaged with histones and other proteins to form chromatin, which is anything but uniform or static. Covalent modifications of chromatin provide a dynamic mechanism for controlling key cellular processes as diverse as transcription, recombination, replication, repair of DNA damage, and apoptosis. Notably, these modifications leave the underlying genomic DNA sequence intact. Among the variety of chromatin protein modifications characterized, lysine acetylation and deacetylation have been particularly well studied, initially through definition of the enzymes that control these reactions.
Among the many lysine acetyltransferases (KATs) that have been identified, the GNAT and MYST families have been intensely studied because of their broad conservation, diverse functions, and associations with metazoan development and disease (reviewed in [1-3*]). The MYST gene family was first reported in 1996, defined by its founding members in yeast and humans, MOZ, YBF2/SAS3, SAS2 and TIP60 [4,5].
MOZ was identified as the recurrent translocation partner in an acute myeloid leukemia, the yeast SAS2 and SAS3 genes were found to influence transcriptional silencing, and TIP60 was shown to encode a human protein that interacts with the HIV-1 Tat trans-activating factor. These MYST family members were initially hypothesized to be histone KATs, based on limited sequence similarity to the GNATs Gcn5 and Hat1, whose enzymatic activity had been very recently defined [6-8], and to other acetyltransferases with non-histone substrates. Since these first reports, remarkable progress has been made.
Recent comprehensive reviews of yeast, fly and vertebrate MYST enzymes provide details of their substrate specificity and structure, the complexes through which they act, their biological functions, and connections to development and disease [9*-13*]. In this review, the focus will be to extend this foundation to highlight several newly emerging areas that reveal links between MYST targeting and other chromatin-modulating activities. In particular interactions between MYSTs and histone methylation and remodeling activities provide increasing mechanistic insight into how chromosomal transactions may be integrated. The recent data considered here come primarily from analysis in yeast and underscore important areas that are worthy of even broader study.
MYST Complexes and Substrate Specificity
Many of the first MYST genes were found through mutant analyses. Sequences of the cloned genes underlying the mutations allowed probing of growing genomic sequence databases and the recognition that most eukaryotes had multiple MYST representatives. These ranged from 2-3 genes in yeast species to at least 5 in vertebrates, along with splice variants whose numbers are expected to grow with further study. Early phenotypic data revealed roles for MYSTs in transcriptional silencing and sex-specific dosage compensation, and that MYST KAT activity did not merely fine-tune chromatin function, but could actually be essential for life.
In parallel with the genetic studies, biochemical purification provided an increasingly clear view of the complexes through which the MYSTs act and first glimpses of how their distinct substrate specificities influence functional output. Comprehensive structural and functional analyses of each yeast MYST complex have been reported (reviewed in [11*,14*-16*]).
Sas2 is the catalytic subunit of the simplest complex, SAS, which targets histone H4-K16 and includes the Sas4 and Sas5 subunits and targets nucleosomal histone H3. The Sas3 enzyme resides in the NuA3 complex that has at least four additional subunits. By contrast, the only essential yeast MYST protein is Esa1, and it is found in two complexes: NuA4 includes at least thirteen subunits, whereas Piccolo is a distinct sub-complex with only three subunits, all of which are shared with NuA4. Although both Piccolo and NuA4 target multiple residues of nucleosomal H4 and H2A, Piccolo is distinguished by its extremely robust activity. Details of complex purifications and information about the noncatalytic subunits of the yeast complexes are summarized in [11*,14*,17,18*].
Much of what has been learned of the yeast MYST complexes has distinct parallels in recent studies of the human and Drosophila complexes. Indeed, striking similarities are noted in subunit composition between the yeast and human NuA4 complexes, in which Tip60 is the catalytic subunit, Piccolo and the human HBO1 complex, and NuA3 and human MOZ/MORF complexes [9*]. Likewise, the human MSL complex with hMOF as its catalytic subunit is similar to the Drosophila MOF-containing dosage compensation complex, although hMOF is also found in additional complexes and is responsible for the majority of H4-K16 acetylation in the cell [19].
Beyond the biochemical purification of complexes, X-ray crystal studies of Esa1 demonstrate that its catalytic core domain and CoA interactions are nearly superimposable with those of the GNAT Gcn5, even though the two enzymes have only limited amino acid sequence similarity [10] and distinct substrate specificities. Of note is that together, Esa1 and Gcn5 appear responsible for the majority of global H4 and H3 acetylation and are generally found upstream of actively transcribed genes [20,21].
Targeting Complexes to Chromatin
Binding and reconstitution studies reveal that Esa1 targeting to substrates is mediated by the noncatalytic Epl1 and Yng2 complex subunits, which also serve to stabilize the enzyme [22-24*]. It is the histone-fold domain (HFD) regions of histones, rather than the tails themselves, that appear responsible for tight substrate binding and efficient tail acetylation [24]. Tail-independent binding was observed earlier for MOF [25] and interactions with HFDs and nucleosomal basic patches may also be relevant for Sas2 and its functions in establishing chromatin boundaries ([26*] and see below).
Many KATs, such as Esa1, must be directed to nucleosomal targets through partner proteins, yet a recent study demonstrates that is not be the case for all MYSTs. X-ray crystal structural analysis of human MOZ reveals that whereas it has a central region that is very similar to Esa1, divergent N- and C-terminal regions can strongly bind DNA [27*]. Binding is mediated through both a TFIIA-type zinc finger and a separate helix-turn-helix motif, and mutational analysis demonstrates that KAT activity and DNA binding are not mutually interdependent. It will be important to determine in future studies if specificity of MOZ targeting in vivo is influenced by other complex members or is controlled by the MYST domain.
In vivo Analysis of Genomic Targets
Details of residue-specific targets of MYST activity were initially defined using recombinant proteins and purified complexes. More complete understanding of genomic targets has come through directed chromatin immunoprecipitation (ChIP) and global microarray studies to monitor gene expression and genomic patterns of binding and activity in vivo. It is clear that each of the yeast MYSTs have distinct genomic patterns of activity, a principle likely to be shared by metazoan MYSTs as studies are expanded in those organisms.
SAS-directed acetylation of H4-K16 is important in defining boundaries between transcriptionally active and transcriptionally silenced regions of the genome. Early studies demonstrated this role in subtelomeric regions, where sas2Δ mutants suffered a spread of hypoacetylated histone H4 and the Sirtuin deacetylase Sir2 [28,29], which in concert with other silent chromatin components can ultimately extend the reach of heterochromatin beyond its normal limits. SAS-mediated boundary functions are also observed at other silenced regions of the genome [30,31] and increased Sas2 expression can restrict the spread of silencing by preventing binding of the Sir3 silencing protein that is ordinarily targeted to deacetylated H4K-16[26*].
NuA3's acetylation of H3 has been studied using sas3Δ mutants, histone mutants, epitope-tagged Sas3, and isoform specific antibodies. Results from these studies together point to a role for NuA3 in contributing to transcriptional activation of many target genes. It has recently been shown that this is a role shared by Gcn5-directed acetylation for many of the same targets [32]. Such overlapping activity is consistent with the observation that either single mutant has only modest phenotypes, whereas simultaneous loss of Gcn5 and Sas3 activity results in cell death [33], presumably due to cumulative loss of expression of genes required for cell growth and cell cycle progress. Two independent studies point to additional roles for NuA3 in chromatin boundaries [31,34], although the molecular details have not yet been as thoroughly explored as for SAS.
The essential yeast MYST Esa1 has multiple roles in vivo, supporting the idea that it may also have many critical genomic targets. Early analysis revealed that ribosomal protein genes, whose expression is rate-limiting for growth, were important targets for Esa1 activation [35]. Subsequent directed and genome-wide ChIP experiments support a role for Piccolo and NuA4 in global and targeted H4 acetylation [20-22,36,37]. This acetylation contributes not only to activation, but also transcriptional silencing and repair of DNA damage [38-41*].
Less is known about the significance of Esa1-directed H2A acetylation. Genome-wide analyses suggest that in general, H2A-K7 acetylation is not associated with transcriptional activation, although Esa1-dependent acetylation is observed at the promoters of several genes examined individually [37,42]. Notably, the bromodomain protein Bdf1 binds hyperacetylated histones, including acetylated H2A-K7 in intergenic regions, and deletion of BDF1 is lethal in an esa1 conditional mutant background [42,43]. Bdf1 also links the acetylation state of nucleosomes modified by NuA4 to Pre-Initiation Complex (PIC) assembly and interacts with the SWR complex, which incorporates the H2A variant H2A.Z into chromatin [44*].
In fact, important recent studies define H2A.Z (encoded by the HTZ1 gene) as an additional substrate of Esa1 [45*-47*]. H2A.Z's SWR assembly complex shares 4 subunits with NuA4 not found in Piccolo [48-50]. Mutational analysis demonstrates that H2A.Z contributes to barrier functions by contributing to restricting the spread of silencing proteins [31,45,51] and ChIP analysis demonstrates that H2A.Z influences localization of genes to the nuclear periphery and kinetics of their reactivation after repression [52*]. Several independent genomic maps of H2A.Z occupancy have been completed [47*,53-56] [41*,44*,57*]. Because H2A.Z-containing nucleosomes are found throughout the genome, understanding the regulation of their assembly and modification by Esa1 and other KATs is an area deserving further attention (see below).
Interactions between Modifications and Modifiers
The significance of one chromatin modification inhibiting or enhancing modification at another site is increasingly recognized. This concept, sometimes referred to as ‘cross-talk’ between modifications has been recently reviewed [3*, 58*]. Specific modifications can also be influenced by ATP-dependent chromatin remodeling complexes and assembly processes. Several examples of ‘cross-chromatin’ modification have been recently reported for yeast MYSTs and provide new insights into their regulation and targeting (Figure 1).
An overview of the functions and sites of action of MYST complexes throughout the yeast genome.
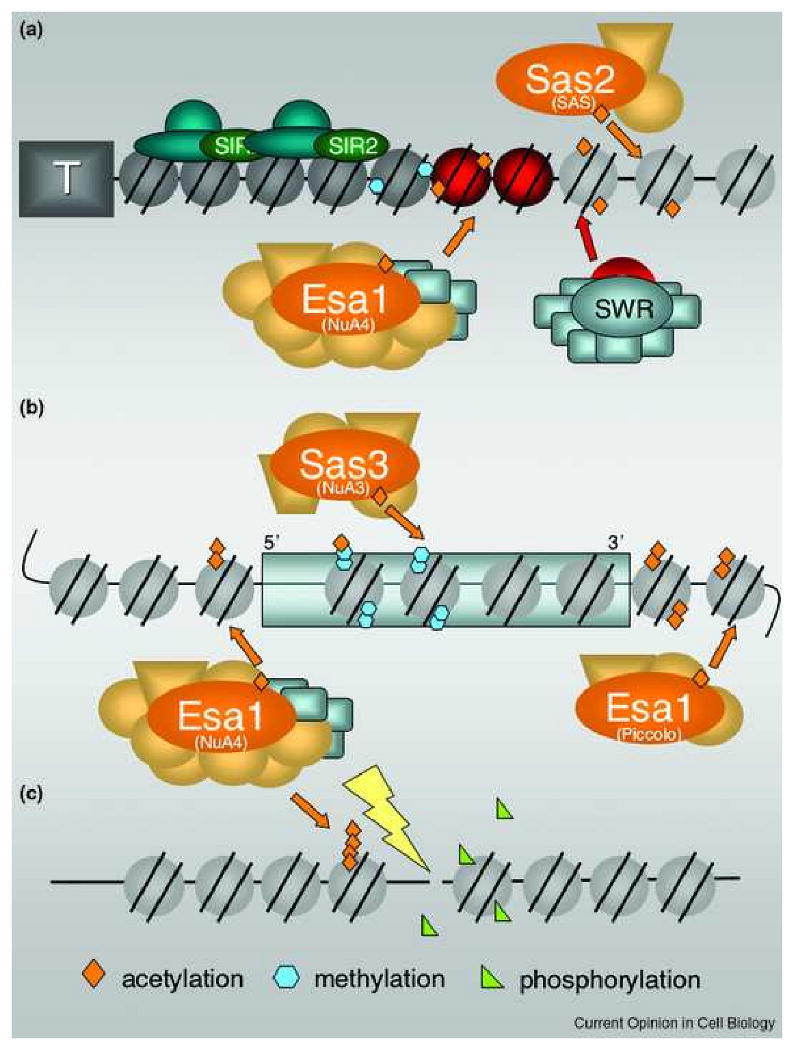
A. Boundary Functions. At subtelomeric regions, the Sas2 MYST of the SAS complex acetylates H4-K16, thereby facilitating the SWR complex's assembly of H2A.Z-containing nucleosomes (red). H2A.Z is then acetylated by the Esa1 MYST, shown here in the context of the large NuA4 complex, which has four of the same subunits found in SWR. These events facilitate formation of heterochromatin (dark nucleosomes): euchromatin (light nucleosomes) boundaries and prevent the spread of silencing proteins such as the SIR complex (blue and green), that inchudes the Sir2 deacetylase (green). Methylation of H3 by Dot1 and Set1 (complexes not shown) is also important in boundary formation. Not depicted here, but recent studies demonstrate that H2A.Z and Set1 act in concert throughout the genome, not just at subtelomeric regions, to restrict the spread of silent chromatin (see text). B. Transcriptional Control. Acetylation by Sas3 and Esa1 occurs throughout the genome. As part of NuA4, Esa1 targets 5′ regions of genes, often sites of decreased nucleosome density. This acetylation can be important for their transcriptional activation. NuA3 also mediates transcription and targets many of the same target genes as Gcn5 (not shown). For Sas3 to acetylate H3-K14, methylation by Set1 and Set2 (not shown) must occur to facilitate targeting of NuA3 by its PHD finger component Yng1. Esa1 also acetylates much of the genome through its very active, smaller Piccolo complex. C. DNA Damage Repair. At sites of DNA damage, acetylation by Esa1 and NuA4 are required for repair to occur. Cycles of phosphorylation and dephosphorylation of H4 and H2A, along with the INO remodeling complex (not shown) are also important in mediating repair and recruitment of NuA4.
SAS-directed acetylation of H4-K16 is required for efficient H2A.Z incorporation into subtelomeric chromatin [59*]. As noted above, Esa1 acetylates H2A.Z, and increased gene dosage of SAS2 can displace the Sir3 silencing protein in telomeric regions and promote Dot1 mediated methylation of H3-K79 [26*], which functions to limit further binding and spreading of Sir3. The H3-K4 methyltransferase Set1 is also important for telomeric silencing and contributes to the restriction of Sir3p binding [60,61]. In fact, acetylated H2A.Z and Set1 act in cooperation throughout the genome, not just near telomeres, to mediate an anti-silencing protective mechanism that prevents the spread of silent chromatin into normally active regions [47*, 57*]. Thus, two MYSTs acting in concert with two methyltransferases and a variant histone assembly/remodeling complex are important on both a local and genome-wide scale for restricting inappropriate silencing (Figure 1A).
To be acetylated by NuA3, histone H3 must be methylated at H3-K4 and H3-K36 [62*]. It is the PHD finger of the Yng1 component of NuA3 that binds H3K4 after it has been trimethylated and then promotes H3-K14 acetylation and subsequent transcriptional activation [62*-64*]. The mechanistic contribution of H3-K36 methylation to NuA3's KAT activity remains unknown, although it also appears linked to NuA4. In this case, deletion of the genes encoding Set2 and Set1, the H3-K36 and H3-K4 methyltransferases, respectively, are associated with decreased Esa1-dependent H4-K8 acetylation [65] (Figure 1B).
Interactions between NuA4 and SWR have also been documented with the ISW1 chromatin remodeling complex. In particular, both expression microarrays and mutant analyses indicated that these three complexes act in parallel to maintain normal growth, mediated through both activation of genes with TATA-containing promoters and repression of stress-induced genes [41*]. Another important remodeling complex, the essential RSC complex, is also functionally linked to NuA4 and SAGA complexes, both of which can stimulate SWR's effects on transcriptional elongation. In this case, it appears that RSC is recruited to nucleosomes acetylated by Esa1 and Gcn5 thereby facilitating the movement of RNA Pol II through chromatin [66*].
Finally, DNA repair and maintenance of genomic integrity are intimately linked to chromatin modification and remodeling activities (reviewed in [67*-70*]). The connection with MYST proteins was first found for Esa1 when it was discovered that the four H4 residues that it targets are critical for repair of double strand breaks [38]. NuA4 and deacetylase complexes are recruited to sites of damage [38,71,72] and dynamic (de)acetylation and (de)phosphorylation of H4 and H2A act in concert with the INO remodeling complex to effect repair [39,72,73] (Fig. 1C).
Conclusions and Outstanding Questions
This brief review has focused on yeast MYSTs, and as noted above, important progress in analysis of metazoan MYSTs has also recently been reviewed [9*-13*]. Since the discovery of the enzyme family, a broad view of MYST function through multimeric complexes has been achieved. Although we now know many of the individual histone residues on which each yeast MYST protein may act, and where in the genome, and for which cellular processes each is critical, many important questions remain to be addressed. Chief among these include defining mechanisms for regulating MYST activity, how complex assembly and disassembly may be regulated, and which additional non-histone targets may be most important biologically. Some clues to answering these questions already exist.
The form of MYST regulation that has been most deeply explored is complex targeting, with some examples as discussed above. However, it is likely that regulation of activity of the complexes may also be mediated through posttranslational modification of the catalytic subunits, or other members of the complex. Already, some instances are known, notably the UV-induced sumoylation of human Tip60 [74*] that appears to enhance its catalytic activity.
Change in expression of MYST complex components is also a potential point for regulation, particularly for the components that are known to exist in more than one modifying or remodeling complex. Searching publicly available expression array datasets available for yeast (http://www.yeastgenome.org/) reveals environmental and genetic conditions under which MYST complex component expression is enhanced or inhibited. It may prove fruitful to pursue the effects of these conditions on both complex levels and activity.
Little is yet known about complex assembly and disassembly and whether this may be a point of regulation. One study has shown that nuclear import of SAS complex subunits is mediated through different karyopherins, demonstrating that there is likely to be no cytoplasmic pre-assembly of active complex [75]. Understanding assembly and whether subunit exchange between complexes drives activity should answer many interesting questions. For example, what dictates Piccolo vs. NuA4 assembly? Because addition of NuA4 subunits to the Piccolo core appears to decrease Esa1's KAT activity, yet enable its targeting to sites of DNA damage, it will be important to know if pools of the complexes are sensitive to damage induction.
The initial focus defining MYST substrates was on the amino terminal tails of histones, however increasingly sensitive mass spectrometry has shown that histones are modified at many additional positions (reviewed in [76*]). Because the enzymes responsible for many of these additional sites have not yet been identified, it is possible that some or all of the MYSTs may prove to act on residues beyond those defined to date. Indeed, lysine acetylation is a prevalent posttranslational modification and new affinity based proteomic studies are being developed to define the complete spectrum of acetylated proteins [77*]. Expansion of this and parallel approaches, particularly assisted by mutant analysis should be informative. Already, candidate protein analysis has revealed that both ATM and p53 can be acetylated by Tip60 and that this acetylation is important for growth control and DNA damage response (reviewed in [78*,79*]). It has been observed that SAS complex members [80*] and NuA4 are important for mediating human p53's activation function when expressed in yeast whereas, NuA3 appears to interfere [81,82]. Thus, it appears that not only do MYST proteins likely have multiple non-histone substrates, but that further studies in yeast may be useful in dissecting complex regulatory pathways in which human MYST proteins function.
Acknowledgments
Thanks to S. Jacobson, E. Scott, C. Chang and M. Koch for discussion and critical reading of this manuscript. Work in the lab has been funded by the National Institutes of Health GM-56469.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allis CD, Berger SL, Côté J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]; * A proposal summarizing a new nomencature for lysine methyltransferases (KMTs), demethylases (KDTs), and acetyltransferases (KATs). Includes tables summarizing enzymes found in human, D. melanogaster, S. cerevisiae, and S. pombe along with their substrate specificity.
- 3.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–925. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]; * A comprehensive review of KAT complexes including useful tables summarizing GNAT and MYST complexes and subunits, discussions of in vivo functions and interplay between modifications. Also see [58].
- 4.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dube I, Frischauf AM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 5.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 6.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–25677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 8.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 9.Avvakumov N, Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]; * One of a series of reviews covering the biology, chemistry and genetics of KATs from a special review issue of Oncogene. See [*10-13].
- 10.Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]; * See [9].
- 11.Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–5384. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]; * See [9].
- 12.Rea S, Xouri G, Akhtar A. Males absent on the first (MOF): from flies to humans. Oncogene. 2007;26:5385–539494. doi: 10.1038/sj.onc.1210607. [DOI] [PubMed] [Google Scholar]; * See [9].
- 13.Yang XJ, Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–5419. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]; * See [9].
- 14.Utley RT, Côté J. The MYST family of histone acetyltransferases. Curr Top Microbiol Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 15.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avvakumov N, Côté J. Functions of myst family histone acetyltransferases and their link to disease. Subcell Biochem. 2007;41:295–317. [PubMed] [Google Scholar]; * Another useful MYST review focusing on complex components and their associations with disease.
- 17.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Barrios A, Selleck W, Hnatkovich B, Kramer R, Sermwittayawong D, Tan S. Expression and purification of recombinant yeast Ada2/Ada3/Gcn5 and Piccolo NuA4 histone acetyltransferase complexes. Methods. 2007;41:271–277. doi: 10.1016/j.ymeth.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The most recent overview of methods to purify KAT complexes that builds on earlier affinity studies useful for recombinant and cellular purifications (for example [22]).
- 19.Smith ER, Cayrou C, Huang R, Lane WS, Côté J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry. 2007;46:2091–2099. doi: 10.1021/bi602366n. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A study demonstrating that histone tails are not required for Piccolo's binding to nucleosomes or free histones. Instead, it is the HFD regions (in H4 residues 21-52) that are critical for binding and activity, and the authors propose that binding on this relatively open surface allows the enzyme to be tethered and participate in cycles of acetylation of tail residues.
- 25.Akhtar A, Becker PB. The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep. 2001;2:113–118. doi: 10.1093/embo-reports/kve022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Côté J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In contrast to Piccolo [24], the KMT Dot1 requires the H4 tail for binding and methylation of H3-K79. In telomeric regions, Dot1 competes with the silencing protein Sir3 for this binding site, and over-expression of Sas2 results in both increased H4-K16 acetylation and H3-K79 methylation, correlated with loss of the SIR silencing complex.
- 27.Holbert MA, Sikorski T, Carten J, Snowflack D, Hodawadekar S, Marmorstein R. The Human Monocytic Leukemia Zinc Finger Histone Acetyltransferase Domain Contains DNA-binding Activity Implicated in Chromatin Targeting. J Biol Chem. 2007;282:36603–36613. doi: 10.1074/jbc.M705812200. [DOI] [PubMed] [Google Scholar]; * A demonstration of direct DNA binding of a MYST protein. The MOZ protein has a structure similar to Esa1 in the KAT domain, but its divergent N-terminal TFII-type zinc finger and C-terminal helix-turn-helix motifs facilitate strong binding to DNA. Of note is that MOZ, unlike other MYSTs tested, has equally strong activity for both nucleosomes and free histones, presumably because it does not require auxiliary factors to mediate nucleosome binding.
- 28.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 29.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 30.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. Embo J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosaleny LE, Ruiz-Garcia AB, Garcia-Martinez J, Perez-Ortin JE, Tordera V. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 2007;8:R119. doi: 10.1186/gb-2007-8-6-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 36.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 37.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 38.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 39.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Specific roles for Esa1 are reported for silencing rDNA and telomeric reported genes. Esa1 binds within the rDNA and influences acetylation levels, as does the sirtuin deacetylase Sir2. Of note, increased gene dosage of either Esa1 or Sir2 can reciprocally suppress silencing defects of the other mutant.
- 41.Lindstrom KC, Vary JC, Jr, Parthun MR, Delrow J, Tsukiyama T. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol. 2006;26:6117–6129. doi: 10.1128/MCB.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A genetic interaction network is defined between the ATPase-dependent ISW1 remodeling complex, SWR and NuA4. Microarray expression studies also reveal parallel functions for TATA-controlled genes and the normal repression of stress-responsive genes.
- 42.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 44.Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol Cell Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study links PIC assembly, as defined by TFIID occupancy to SWR-directed chromatin remodeling through NuA4 acetylation followed by binding of the Bdf1 bromodomain protein. This role appears restricted to NuA4, rather than being shared by SAGA, although both H3 and H4 tails are bound by Bdf1.
- 45.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]; * One of several reports (also see 46-47; 52; 57] defining H2A.Z as a substrate for Esa1 and defining this acetylation as important for its function in transcriptional regulation and boundary formation. These reports build on earlier genetic and genomic studies of HTZ1 and H2A.Z [48-51; 53-56] and further demonstrate a role for H2A.Z in nuclear localization and coordination with Set1 to mediate functional genomic boundaries.
- 46.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [45].
- 47.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [45]
- 48.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Côté J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 52.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [45].
- 53.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [45].
- 58.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]; * An in-depth consideration of examples and principles of ‘cross-talk’ between different histone modifications to fine-tune chromatin regulation. Also see [3].
- 59.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Mutantional analysis and ChIP experiments demonstrate that acetylation of H4-K16 is required for optimal recruitment of H2A.Z at subtelomeric areas proximal to silenced regions.
- 60.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 62.Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Together with [63-65], it is demonstrated that methylation of H3 by Set1 and Set2 is required to recruit NuA3 to chromatin, via its PHD-containing Yng1 component. Methylation thereby facilitates H3 acetylation and transcriptional regulation of target genes.
- 63.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [62].
- 64.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [62].
- 65.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 66.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; * An in vitro assay of RNA Pol II stalled on a nucleosome was used to demonstrate that acetylation by NuA4 and SAGA could promote the RSC remodeling complex's recruitment to facilitate elongation.
- 67.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]; * Along with [68-70], this is one of a series of reviews summarizing current understanding of chromatin and its modification and remodeling to DNA repair and genomic stability.
- 68.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]; * See [67].
- 69.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; * See [67].
- 70.Wurtele H, Verreault A. Histone post-translational modifications and the response to DNA double-strand breaks. Curr Opin Cell Biol. 2006;18:137–144. doi: 10.1016/j.ceb.2006.02.008. [DOI] [PubMed] [Google Scholar]; * See [67].
- 71.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Côté J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng Z, Ke Y, Ding X, Wang F, Wang H, Ahmed K, Liu Z, Xu Y, Aikhionbare F, Yan H, et al. Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene. 2007 doi: 10.1038/sj.onc.1210710. in press. [DOI] [PubMed] [Google Scholar]; * Demonstration that the human Tip60 MYST is sumoylated in response to UV-irradiation and that this modification enhances its KAT activity.
- 75.Schaper S, Franke J, Meijsing SH, Ehrenhofer-Murray AE. Nuclear import of the histone acetyltransferase complex SAS-I in Saccharomyces cerevisiae. J Cell Sci. 2005;118:1473–1484. doi: 10.1242/jcs.01739. [DOI] [PubMed] [Google Scholar]
- 76.Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A review of progress made in defining and understanding non-tail modifications of histones.
- 77.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]; * This study combined immunoaffinity of acetylated lysine peptides with nano-HPLC/MS/MS analysis to identify acetylation targets from HeLa cells and mouse mitochondria. Many new KAT substrates have been identified.
- 78.Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]; * Along with [79], a current overview of diverse roles for the Tip60 MYST.
- 79.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]; * See [78].
- 80.Yousef AF, Xu GW, Mendez M, Brandl CJ, Mymryk JS. Coactivator requirements for p53-dependent transcription in the yeast Saccharomyces cerevisiae. Int J Cancer. 2008;122:942–946. doi: 10.1002/ijc.23174. [DOI] [PubMed] [Google Scholar]; * An exploration of the contributions of chromatin regulators on full length human p53's activation functions in yeast A number of effects are reported, including that activation is strongly reduced upon deletion of SAS2 and the genes encoding YEATS domain components found in SAS, NuA4 and SWR, and INO complexes.
- 81.Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Côté J. Role of an ING1 Growth Regulator in Transcriptional Activation and Targeted Histone Acetylation by the NuA4 Complex. Mol Cell Biol. 2001;21:7629–7640. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nourani A, Howe L, Pray-Grant MG, Workman JL, Grant PA, Côté J. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J Biol Chem. 2003 doi: 10.1074/jbc.C300036200. [DOI] [PubMed] [Google Scholar]


