Abstract
The genome of thylakoidless cyanobacterium Gloeobacter violaceus encodes a fast-cycling rhodopsin capable of light-driven proton transport. We characterize the dark state, the photocycle, and the proton translocation pathway of GR spectroscopically. The dark state of GR contains predominantly all-trans-retinal and, similar to proteorhodopsin, does not show the light/dark adaptation. We found an unusually strong coupling between the conformation of the retinal and the site of Glu132, the homolog of Asp96 of BR. Although the photocycle of GR is similar to that of proteorhodopsin in general, it differs in accumulating two intermediates typical for BR, the L-like and the N-like states. The latter state has a deprotonated cytoplasmic proton donor and is spectrally distinct from the strongly red-shifted N intermediate known for proteorhodopsin. The proton uptake precedes the release and occurs during the transition to the O intermediate. The proton translocation pathway of GR is similar to those of other proton-pumping rhodopsins, involving homologs of BR Schiff base proton acceptor and donor Asp85 and Asp96 (Asp121 and Glu132). We assigned a pair of FTIR bands (positive at 1749 cm−1 and negative at 1734 cm−1) to the protonation and deprotonation, respectively, of these carboxylic acids.
Abbreviations used: BR, bacteriorhodopsin; PR, green-absorbing proteorhodopsin; GR, Gloeobacter rhodopsin; ASR, sensory rhodopsin from Anabaena; XR, xanthorhodopsin from Salinibacter ruber; DM, n-dodecyl-β-D-maltoside; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DMPA, 1,2-dimyristoyl-sn-glycero-3-phosphate; FTIR, Fourier-transform infrared
Introduction
Microbial rhodopsins are ubiquitous retinal-binding membrane proteins with ion-transporting and photosensory functions. The archetypal well-studied microbial rhodopsins are of haloarchaeal origin and are best represented by the light-driven proton pump BR. PR is a eubacterial homolog of BR originally found in uncultured marine γ-proteobacteria (1). Similar to BR, it binds all-trans-retinal covalently and performs transmembrane light-driven proton translocation, implying that it can be used as a supplementary source of energy (1–3). The exact contribution of the PR-dependent bioenergetic pathway is still controversial and may vary with the taxonomy of the host (4–7). In the past few years it was realized that there are thousands of different PR species, with great taxonomic, ecological, structural, spectral, and, possibly, functional diversity (8–14). The last is suggested not only by the analysis of the primary structures (11) but also by the presence of both fast (typical for ion pumps) and slow (typical for photosensors) photocycles and corresponding photoelectric signals (15,16). Genes encoding PR-like proteins are found not only in α-, β-, and γ-proteobacteria but also in Bacteroidetes, Actinobacteria, Planctomycetes, Firmicutes, and Cyanobacteria (8,12,17,18) as well as in marine archaea and some dinoflagellates, which probably obtained them by lateral gene transfer (19,20).
Most of the biophysical studies done on the original green-absorbing PR (1) expressed in Escherichia coli imply the BR-like nature of the light-driven proton translocation. Under physiological conditions (mildly alkaline pH), PR transports protons light-dependently in the extracellular direction in liposomes, oocytes, and E. coli cells (16,21,22). Accordingly, it shows light-induced spectral changes in the infrared region of strongly bound water characteristic for transport rhodopsins (23). The photocycle of PR is relatively fast, and it showed most of the photointermediates known for BR, with the apparent exception of the L intermediate (21,24,25). Finally, a combination of site-directed mutagenesis and time-resolved spectroscopy showed that the homologs of the primary proton acceptor (Asp85, present as Asp97 in PR) and donor (Asp96, present as Glu108 in PR) of the Schiff base of BR are involved in proton translocation in PR as well (24). On the other hand, there are many notable differences between PR and BR, the most interesting one being the unusually high pKa of Asp97 (7.1–7.6), which may be related to the alkaline character of the marine environment (21,24). This can originate from the structural differences in the extracellular half of PR, such as the absence of the pair of proton-releasing glutamates (Glu194 and Glu204 of BR), weaker coupling of the Schiff base counterion to Arg94 (Arg82 of BR), its unique interaction with Asn230 and His75, and a relatively strong hydration of Asp227 (Asp212 of BR) (26–29).
Among approximately a dozen sequenced cyanobacterial genomes, only two rhodopsin species have been found. The first one is a unique photosensory rhodopsin from Anabaena (Nostoc) sp. PCC7120 (ASR) (30), possibly serving as a receptor for phycobilin biosynthesis. The second rhodopsin was found in a thylakoidless unicellular cyanobacterium Gloeobacter violaceus PCC 7421 (31). Its primary structure resembles that of PR, and because it has a glutamate residue at the position of the Schiff base proton donor, it was suggested to have a proton-pumping function as well (32). Indeed, when expressed in E. coli, GR demonstrates light-driven proton transport and correspondingly fast photocycle (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished). It should be noted that the exact physiological role of such proton-pumping rhodopsin in the presence of the proton gradient-generating chlorophyll-based photosynthetic apparatus is puzzling. Even though the amino acid sequence of GR is closer to that of PR than of BR, it forms a separate cluster together with XR, the carotenoid-associated proton pump from Salinibacter ruber (33,34), and a few other sequences from proteobacteria, actinobacteria, and Chloroflexii (8,12). The recently obtained crystal structure of XR (35) may give more hints toward general structural features of this group of rhodopsins, in particular, participation of a His side chain in the extracellular hydrogen-bonded network and unusual connectivity of the cytoplasmic Glu (homolog of Asp96 of BR) to the Schiff base-forming Lys.
In this work, we tried to define the pathway of proton translocation in the photocycle of GR using the combination of site-directed mutagenesis, heterologous expression, and three kinds of spectroscopy, including Raman, time-resolved FTIR, and time-resolved visible. We compared characteristics of the photocycle and the dark state of GR with those of PR and BR and concluded that overall path of proton transport involves the same carboxylic residues (homologs of BR's Asp85 and Asp96). At the same time, we found that the photocycle and the dark state of GR are different from those of both PR and BR in several respects. In addition to substantially lower pKa of the Schiff base counterion Asp121 (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished), the most interesting features of GR are an apparent accumulation of the BR-like N intermediate with the deprotonated cytoplasmic proton donor Glu132 and unusually strong coupling between this proton donor and the retinal.
Materials and Methods
Heterologous expression of the wild-type and mutant GR
The heterologous expression of wild-type GR in E. coli as well as production of GR mutants are described in detail elsewhere (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished). Briefly, genomic DNA of Gloeobacter violaceous was used to amplify the gene encoding GR by standard PCR using nondegenerate primers 5′ATGTTGATGACCGTATTTTCTTCTGC3′ and 5′CTAGGAGATAAGACTGCC-TCCCCG3′. E. coli strain DH5α was used for the cloning, and transformants were grown in LB medium in the presence of ampicillin (50 μg/mL) at 35°C. The amplification product was inserted under the lacUV5 promoter into the pKJ900 plasmid, which also carried a gene for β-carotene dioxygenase under pBAD promoter (36). GR was expressed in E. coli strain β/UT, which was constructed by transforming plasmid pORANGE into UT5600, resulting in the ability to synthesize β-carotene (36). β/UT cells transformed with pKJ900 were able to produce retinal and GR endogenously after being induced with 1 mM IPTG and 0.2% L-(+)-arabinose for 24 h at 30°C. The pKJ900 plasmid encoding wild-type gloeoopsin was used as a template for site-directed mutagenesis. Site-directed mutagenesis was carried out with the two-step megaprimer PCR method with Pfu polymerase (36).
Isolation, purification, and reconstitution of GR
The typical protein yields were in the range of 2–5 mg of GR per liter of culture, which produces E. coli membranes with an intense color and negligible cytochrome bands (around 410 nm). Isolation of the GR-containing E. coli membranes, their treatment with DM and incorporation into polyacrylamide gels was performed as described for ASR (37). For vibrational spectroscopy and proton kinetics, further purification was performed via solubilization in DM, Ni2+-NTA resin binding, and reconstitution into DMPC/DMPA liposomes as described earlier (37). The notable difference between GR and other His-tagged microbial rhodopsins is its relatively fast rate of hydrolysis of the C-terminus, so that His-tag purification had to be performed using freshly isolated membranes.
Spectroscopy in the visible and infrared ranges
Static visible spectra were collected using a Cary 50 spectrophotometer (Varian, Palo Alto, CA). Time-resolved laser difference spectroscopy in the visible range and global multiexponential fitting of the data were performed as before (37,38). Static Raman spectra were collected using either a Renishaw (Toronto, Ontario, Canada) Raman Imaging Microscope, System 2000, with excitation at 785 nm, or Bruker (Billerica, MA) FRA 106/s accessory to the IFS66vs spectrometer, with excitation at 1024 nm, at 2 cm−1 resolution, using concentrated suspensions of GR liposomes. Time-resolved rapid-scan FTIR difference spectra were measured in transmission mode using Bruker IFS66vs spectrometer as described elsewhere (37,38), using fast return and 4 cm−1 spectral resolution. The interferogram acquisition time was 12 ms.
Results and Discussion
Characterization of the dark state of GR by Raman spectroscopy
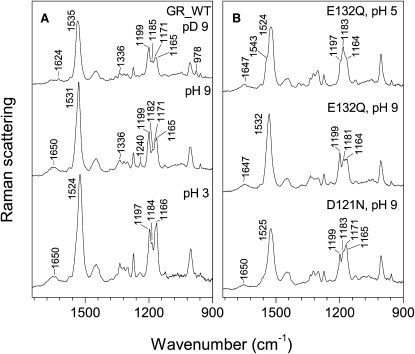
Similar to many other microbial rhodopsins, GR exists in two spectral forms in a pH-dependent equilibrium, involving the bluish acidic (λmax ≈ 550 nm) and reddish alkaline (λmax ≈ 543 nm) pigments. The transition between the two forms was found to be rather broad, with the midpoint at pH ≈ 5 (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished), which is higher than that for BR but lower than for PR and XR (24,39). For this reason, we chose to study the alkaline (proton-pumping) form of GR at pH 9 to avoid contamination by the acidic form. First, we characterized the chromophore of GR by preresonance Raman spectroscopy (Fig. 1 A), using GR reconstituted into DMPC/DMPA liposomes to increase the protein concentration and purity. The basic features of the spectra in the fingerprint region are consistent with the retinal chromophore being in the all-trans-configuration (40), with predominant C-C stretches at 1199 and 1165/1171 cm−1, almost the same as for PR (24). Because the accumulation of the Raman spectra can proceed for many hours, thermal isomerization of the chromophore (dark adaptation) should be considered. We checked for the presence of the dark adaptation using both visible and Raman spectroscopy and have not detected appreciable blue shifts in the visible absorption maximum (not shown) or the appearance of the 1185 cm−1 band characteristic of 13-cis-retinal (41) in 8 h. The lack of the appreciable dark adaptation is reminiscent of that of PR as studied by Raman and FTIR spectroscopies (24) as well as by solid-state NMR (42).
Figure 1.
Preresonance Raman spectra (excited at 1024 nm, Bruker FRA 106/s) of DMPC/DMPA liposomes containing GR variants at room temperature. (A) Wild-type at pD 9 and pH 9 (0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M CHES), and at pH 3 (0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M sodium citrate); (B) E132Q at pH 5 (0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M MES) and pH 9, and D121N at pH 9.
The observed position of the C=N stretching vibration at ∼1650 cm−1 is typical for the protonated Schiff base of retinal (43) and is higher than that for BR but lower than for PR (27,28). The actual position of this band may be slightly different because of the distortion given by minor contribution of the protein amide I vibrations at 1660 cm−1. On deuteration, this Schiff base vibration downshifts to 1624 cm−1 (Fig. 1 A), which indicates strong counterion, similar to that of PR (27,28,42). Other H/D-exchange dependent bands of interest identifiable from our data include in-plane N-H/N-D bending vibrations of the Schiff base at 1336/978 cm−1 and combination bands of the retinal C12-C13 stretch with the Schiff base lysine rocks at 1240 and 1165 cm−1, which lose their intensity on deuteration (40,44,45). Interestingly, the main ethylenic stretch upshifts by 4 cm−1 and broadens on the H/D exchange (also observed by FTIR, see Fig. 6), which is untypical for all-trans-BR (40). It is possible that such upshift and broadening represent higher 13-cis-retinal content of the deuterated sample, as also suggested by the stronger 1185 cm−1 C-C stretching vibration band of the chromophore.
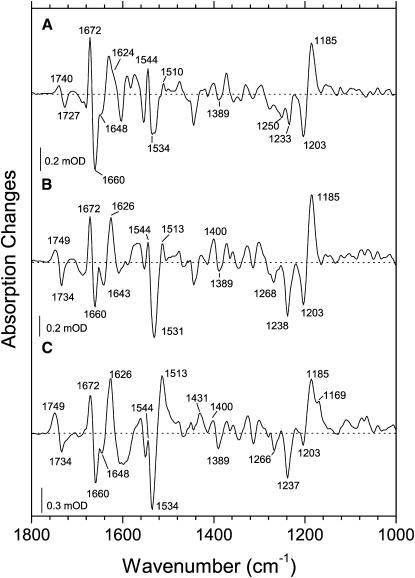
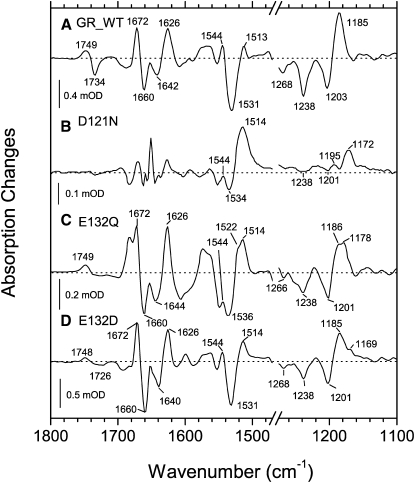
Figure 6.
Light-induced rapid-scan FTIR difference spectra of DMPC/DMPA liposome films with the wild-type GR measured at 2°C, hydrated with 0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M CHES in H2O or D2O. (A) pD 9, 1 ms delay; (B) pH 9, 1 ms delay; and (C) pH 9, 50 ms delay, but note the 12-ms interferogram acquisition time.
The major ethylenic (C=C) stretching band shifts from 1531 to 1524 cm−1 on transition to the acidic form of GR at pH 3, presumably accompanied by protonation of the Schiff base counterion (Fig. 1 A), consistent with the correlation of the position of this band with the maximum in the visible spectral range (46). The fingerprint region of the acidic form indicates that it also contains predominantly all-trans-retinal, unlike in BR, where a significant amount of 13-cis-species is present (47). It was found that when the putative Schiff base counterion Asp121 (the homolog of BR's Asp85) was replaced, the D121N mutant existed as a bluish pigment (λmax ≈ 560 nm), similar to the acidic form of the wild-type, but in the wide pH range (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished), as expected from the neutralization of the counterion. Accordingly, the Raman spectrum of the D121N mutant (Fig. 1 B) looks similar to that of the acidic species of GR (Fig. 1 A) with the main ethylenic stretch at 1525 cm−1, but with a higher proportion of the 13-cis species as evident from a shoulder at 1183 cm−1. Similar to the acidic form of the wild-type but much more pronounced, the 1171 cm−1 C-C stretch is strongly enhanced, reminiscent of the homologous D97N mutant and the acidic form of PR (24) and the O intermediate of BR (48). This enhancement may be a general feature of the protonated all-trans-retinal Schiff bases with the protonated carboxylic counterion.
Unexpectedly, the GR mutant, in which the homolog of BR's cytoplasmic proton donor of the Schiff base (Asp96) is replaced (E132Q), turned into the bluish pigment on purification at neutral pH. The titration of the membranes containing E132Q showed that the pKa of the transition between acidic and alkaline forms (presumably the pKa of Asp121) is upshifted by at least 2 pH units (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished), which was not observed in the homologous E108Q mutant of PR (24). This would indicate unusually strong coupling of the Glu132 site with the retinal Schiff base vicinity, much stronger than that observed for BR and fungal rhodopsins (49,50), but consistent with a recent structure of XR (35). We tested the perturbation of retinal configuration in the E132Q mutant by measuring Raman spectra of its acidic (pH 5, higher than for the wild-type) and alkaline (pH 9) forms (Fig. 1 B). The positions of the ethylenic stretches are similar to those observed for the corresponding forms of the wild-type GR (Fig. 1 A), but the bands look broader, especially at pH 5, implying the presence of the extra species. Indeed, the fingerprint regions indicate major contribution of 13-cis species as obvious from the 1181 cm−1 peak for the alkaline form, which is much stronger than in the wild-type. The effect is even more dramatic for the acidic form of the E132Q mutant, where the dominant 1183 cm−1 C-C stretch and the strong 1543 cm−1 shoulder of the 1524 cm−1 ethylenic C=C stretch are observed. Splits of the ethylenic stretch of similar magnitude were observed for the 13-cis-15-anti-protonated retinal Schiff bases, or the N-like states (51,52), possibly implying a more cytoplasmically open conformation of this mutant at low pH. The dramatic increase of the 13-cis content in the blue species on a mutation in the cytoplasmic portion is similar to that observed for D85N/F42C mutant of BR (52), where it was proposed to originate from the negative charge of Asp96 leading to the formation of the N-like state. This argument is not applicable to the situation in the E132Q mutant, as the homologous residue is neutral, but points at the strong coupling of this site with the retinal. Additional evidence of the perturbation of the retinal by the E132Q replacement comes from the downshifted position of the Schiff base C=N stretching vibration (1647 cm−1 vs. 1650 cm−1 in the wild-type), consistent with the higher 13-cis content.
Proton translocation steps in the photocycle of GR studied by time-resolved spectroscopy in the visible range
At neutral pH, the photocycle of GR is relatively fast, with a characteristic time of turnover <100 ms, characteristic for proton pumps (not shown). We studied the details of the photocycle at pH 9 (to monitor pure alkaline form), where it is somewhat slower than at neutral pH but similar in other respects. The last time constant of the GR photocycle is ∼140 ms (Figs. 2 and 3), comparable to the turnover of the photocycle of PR under similar conditions (24). An overall character of the photocycle of GR (Fig. 2) is similar to that of the alkaline form of PR under the same conditions (24,25) as well as of solubilized XR (39). Two red-shifted intermediates (faster K-like and slower O-like) dominate the absorption changes. Even less M intermediate than an already low amount observed for PR accumulates in the blue spectral region, probably due to its extremely fast (submillisecond) decay. On the other hand, the detailed global multiexponential analysis (Fig. 3) reveals significant differences between the photocycles of GR and PR. We detect six statistically valid exponential processes, which correspond to the transitions between mixtures of the intermediates (21,53), with the characteristic times τ1 11 μs, τ2 110 μs, τ3 640 μs, τ4 3.2 ms, τ5 22 ms, and τ6 140 ms. The initial spectrum in Fig. 3 represents the K-GR difference spectrum, and from the spectral shapes of the kinetic components (where negative absorption means formation of intermediate(s), whereas the positive absorption corresponds to their disappearance), it can be assumed that τ1 and τ2 correspond to the transition of the K-like intermediate to the L/M mixture, τ3 to the transition of the L/M mixture to the N/O mixture, and τ4 to the transition of N to O during the reequilibration of the N/O mixture (probably involving a substate of the O intermediate). The last two kinetic components τ5 and τ6 correspond to the return of the final mixture to the parent state, possibly via an additional intermediate spectrally similar to the ground state, as was postulated for PR (25).
Figure 2.
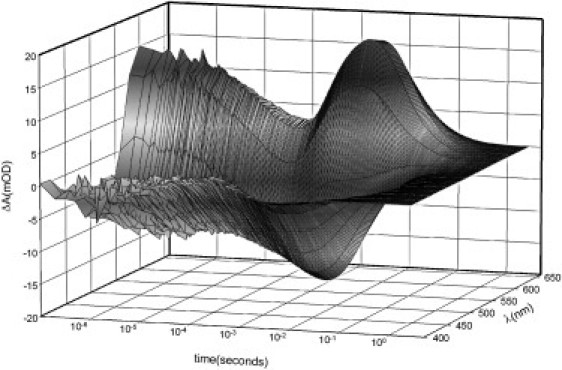
Kinetics of the light-induced absorption changes in the photocycle of wild-type GR in DM-treated E. coli membranes encased in polyacrylamide gel and incubated at pH 9, 100 mM NaCl, 50 mM KH2PO4, 50 mM CHES, room temperature. The kinetics was measured every 20 nm.
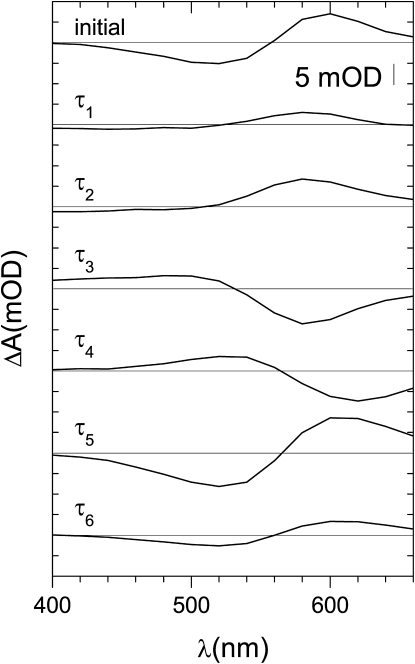
Figure 3.
Spectra of the kinetic components of the photocycle extracted by global multiexponential analysis of the data in Fig. 2. The spectrum labeled “initial” represents extrapolation of the fit to zero time point. Characteristic times of the spectral components are τ1 11 μs, τ2 110 μs, τ3 640 μs, τ4 3.2 ms, τ5 22 ms, and τ6 140 ms. The negative absorption means formation of intermediate(s), whereas the positive absorption corresponds to their disappearance. See text for details.
The most striking difference between the photocycles of GR and PR is the apparent presence of the L-like and the N-like intermediates (Fig. 3). The L-like intermediate was not detected for PR either in similar membrane preparations (24,25) or in liposomes (21) but can exist in different lipid compositions (25) (S. Pierobon, L. Shi, and L. S. Brown, unpublished). From the broad shallow short-wavelength feature in the spectra of the three fastest components in Fig. 3, it appears that the L intermediate of GR exists in extremely fast equilibrium with the M state (as indicated by the simultaneous presence of the spectral features of the same sign at both 400 nm and 500 nm). The extremely shallow character of the negative features in the first two spectra can probably be explained by combination of the relatively low extinction coefficients of the L and M states, spectral overlap of L and K, and the simultaneous presence of the L and M intermediates forming from a single K state. It should be noted that in DMPC/DMPA liposomes of GR, the kinetics of absorption changes at 457 nm hint at the accumulation (peaked at ∼50 μs) of the L intermediate as well (Fig. 4). The N-like intermediate was detected in PR before (21,24,25), but in the membrane preparations similar to our GR samples it was postulated to be represented by a strongly red-shifted, O-like species, having a 13-cis configuration of the retinal. On the other hand, only slightly red-shifted N species were found in PR liposomes in a complex mixture with strongly red-shifted O-like species (21) (S. Pierobon, L. Shi, and L. S. Brown, unpublished). What we observe in GR-containing E. coli membranes (Fig. 3, especially obvious from the spectra of the processes with τ3 and τ4) is more similar to the situation in BR, where the N intermediate with a spectrum close to that of the parent state (slightly blue-shifted for BR, slightly red-shifted for GR) clearly precedes the strongly red-shifted O intermediate. This interpretation is strongly supported by our time-resolved FTIR analysis presented below (see Fig. 6). We believe that the true (not strongly red-shifted) N intermediate exists in PR as well but can be easily observed only in GR and some PR preparations in artificial lipids (S. Pierobon, L. Shi, and L. S. Brown, unpublished) because of the much faster decay of the M intermediate, which merges with the O intermediate otherwise, preventing accumulation of the distinct N intermediate. In principle, according to the molecular definition of the N intermediate, its spectral maximum should be blue-shifted compared to the parent state, as it has a relaxed 13-cis chromophore. It should be noted that this basic assumption can be challenged because the N spectrum can be affected by the changed protein environment, as may be the case in PR and in some BR mutants. Our findings show that GR is “normal” (wild-type BR-like) in this respect.
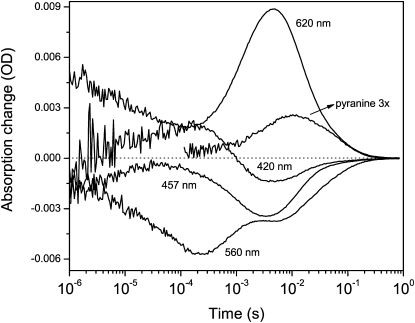
Figure 4.
Kinetics of the light-induced proton release and uptake measured by the absorption changes of 100 μM pyranine in the unbuffered suspension of DMPC/DMPA liposomes of the wild-type GR in 5 mM NaCl, pH 7.4, at room temperature. The upward deflection of the pyranine signal at 457 nm (with the photocycle signal subtracted) corresponds to the proton uptake.
We studied kinetics of the proton release and uptake in the photocyle of GR using the pH-sensitive dye pyranine (54) at pH 7.4, well above the pKa of the counterion and still within the range of sensitivity of the dye. We had to use DMPC/DMPA liposomes, as the proton signal was hard to detect in the membranes because of their high buffering capacity and relatively low concentration of GR. The obtained proton kinetics shows the proton uptake (around 10 ms time) preceding the release at the end of the photocycle (Fig. 4). Such proton kinetics is very similar to that of PR in E. coli membranes (24) but not in micelles (55) and is consistent with the absence of the extracellular glutamates (homologs of BR's Glu194 and Glu204) responsible for the fast proton release (56,57). It should be noted that the kinetics of the proton uptake lags behind the rise of the signal at 620 nm, reflecting the reequilibration of the N/O mixture (Fig. 4). Such kinetic uncoupling implies that the proton uptake by Glu132 occurs between the N and O intermediates (or between the two substates of the O intermediate), which reequilibrate on the timescale of a few milliseconds (see the spectrum of the process with τ4, Fig. 3). This is also confirmed by FTIR (see Fig. 6 below), where reprotonation of the cytoplasmic carboxylate is observed in the same time domain.
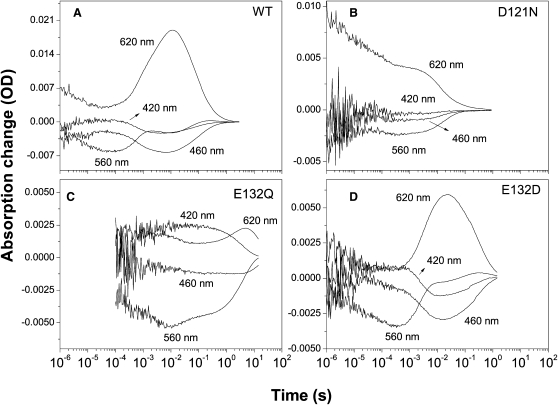
To learn the nature of the primary proton donor and acceptor of the Schiff base, we compared the photocycle of the wild-type GR with those of several mutants at pH 9 (Fig. 5). From the Raman spectra shown above (Fig. 1), it can be expected that, in analogy to BR and PR, Asp121 (homolog of Asp85 of BR) serves not only as a counterion but also as a primary proton acceptor of the Schiff base. Thus, one can expect to see no appreciable Schiff base deprotonation (M intermediate) in the photocycle of the D121N mutant of GR. This is indeed what was observed, as both the positive absorption change at 420 nm (M) and absorption changes of the later intermediates produced from M (at 620 nm) disappear in the D121N mutant's photocycle (Fig. 5 B). The observed photocycle is very similar to that in the acidic form of PR and its homologous D97N mutant (21,24), suggesting that Asp121 is indeed the primary proton acceptor for the retinal Schiff base.
Figure 5.
Kinetics of the light-induced absorption changes in the photocycles of GR variants in DM-treated E. coli membranes encased in polyacrylamide gels. (A) Wild-type; (B) D121N; (C) E132Q; and (D) E132D. Conditions: pH 9 (0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M CHES), room temperature. The differences in the amplitudes and noise levels reflect the differences in the expression, the early part of the 132Q kinetics is removed because of the high noise. Relatively high noise of the earlier parts of the traces is caused by the smoothing of the later parts during linear-to-logarithmic conversion.
If Glu132 (the homolog of Asp96 of BR) serves as a proton donor for the Schiff base of GR, one has to expect that the E132Q mutant will have a very slow decay of the M intermediate corresponding to the reprotonation of the Schiff base from the bulk, as was shown for homologous E108Q mutant of PR (24). The situation is complicated by the fact that the dark state of the E132Q mutant is substantially contaminated by the 13-cis species (see above, Fig. 1 B). We found that both the replacement of Glu132 and the presence of 13-cis-retinal are reflected in the photocycle of E132Q (Fig. 5 C). First, the M decay becomes very slow (several seconds at pH 9), reflecting inefficient reprotonation of the Schiff base in the absence of the internal proton donor. Second, slow-decaying red-shifted intermediates typical for the 13-cis cycle (and, possibly, slow O intermediate of the trans-cycle) are observed as a strong positive signal at 620 nm, coexisting with the M intermediate. Because the E132Q mutation produces such dramatic global conformational effects, it is not possible to make a clear-cut conclusion on the proton-donating role of Glu132 just based on this mutant phenotype. Thus, a second, less perturbing mutation E132D was introduced at this site. The photocycle of the E132D mutant (Fig. 5 D) is not very different from that of the wild-type, but the decay of M is delayed ∼10-fold, consistent with the proton-donating role of Glu132, as Asp132 may have somewhat higher pKa than Glu. Additionally, the photocycle turnover in this mutant is several times slower than in the wild-type. Even though the phenotypes of E132Q and E132D mutants are suggestive, an additional verification of the nature of the proton donor of the Schiff base had to be done using FTIR spectroscopy (see below).
Verification of the proton translocation pathway by time-resolved FTIR spectroscopy
To verify the proton translocation pathway suggested by the results from the visible spectroscopy (see above) and to gain more detailed molecular information on the late intermediates of the photocycle, we performed rapid-scan FTIR measurements on the DMPC/DMPA liposomes of GR in the tens of milliseconds time domain (Fig. 6). The measurements were performed at 2°C to slow down the photocycle to observe the pure N intermediate, which equilibrates with the O state in a few milliseconds at room temperature (Fig. 3). Comparison of the spectra for the first and the second time points (Fig. 6, B and C, so-called first and second slices, at 1 ms and 50 ms delays after the laser flash, but note the 12-ms interferogram acquisition time) confirms our conclusions on the BR-like character of the last part of the photocycle, in which the N-like (not strongly red-shifted) intermediate precedes the O-like (strongly red-shifted) one. Moreover, the FTIR spectra give molecular characteristics of these two states, which are also BR-like rather than PR-like, using the following criteria. First, the main ethylenic stretch at 1513 cm−1 (corresponding to red-shifted intermediates) has a low amplitude in the first but not in the second slice, unlike in PR (24), which is consistent with the formation of the O-like product on this timescale. Accordingly, in the first slice, we observe a positive ethylenic stretch (with a possible contribution from the amide-II) at 1544 cm−1, which decreases in amplitude in the second slice. This could be interpreted as a decay of a classic, not strongly red-shifted, N-like intermediate (which has a double ethylenic stretch in BR (58)). Second, in the fingerprints region, a strong negative C-C stretch at 1203 cm−1 with the corresponding positive 1185 cm−1 band indicative of 13-cis-retinal (41) in the first slice is replaced with very weak negative band at 1203 cm−1 and a strong 1169 cm−1 shoulder on the 1185 cm−1 band (Fig. 6 C), which shows the accumulation of a product with distorted all-trans-retinal (45,48). Additionally, the strong D2O-sensitive 1400 cm−1 band (presumably, coupled N-H and C15-H in-plane bending) in the first slice, which is characteristic for the N intermediate, is replaced by the strong 1431 cm−1 band (possibly, retinal methyl deformation) typical for the O intermediate of BR (45,58) in the second slice. Taken together, these results strongly suggest that in GR a classic, not strongly red-shifted, N intermediate with 13-cis-retinal and BR-like characteristics equilibrates with rather typical O intermediate with twisted all-trans-retinal during a few milliseconds after the photoexcitation. On the other hand, despite the BR-like character of the retinal changes in the photocycle, the protein changes are quite different. For example, strong amide I bands present in both slices (negative 1660 cm−1 and positive 1672 cm−1 and 1626 cm−1) are not typical for BR, but the former two are close to those observed for PR (21). An additional negative band at 1643 cm−1 can be assigned to the C=N stretch of the Schiff base, as it shifts to around 1624 cm−1 (the exact position is hard to define because of the overlap with a stronger positive amide I band) in D2O (Fig. 6), consistent with the results of Raman spectroscopy (Fig. 1).
Similarly, the positions of the C=O stretches of protonated carboxylic acids (positive at 1749 cm−1 and negative at 1734 cm−1, downshifting to 1740/1727 cm−1 in D2O, Fig. 6 A) are closer to those in PR (21,24) than in BR. This should not be surprising considering the absence of the extracellular dyad of glutamates, the presence of glutamate as a homolog of Asp96 of BR, and the absence of the homolog of Asp115 (which usually contributes to this spectral region) in both PR and GR. The positive 1749 cm−1 band can be tentatively assigned to the protonation of the primary proton acceptor, Asp121 (1755 cm−1 for the homologs in PR and BR (21,24,45)), and the corresponding negative band of the deprotonated species may be located at 1389 cm−1 (for the homologs, 1384 cm−1 in PR (21) and 1385 cm−1 in BR (59); the strong negative band at 1734 cm−1, which decreases its amplitude in the second slice, should be assigned to the deprotonation of the primary proton donor, Glu132 (for the homologs, 1728 cm−1 in PR (21,24), 1742 cm−1 in BR (58), and 1720 cm−1 in the D96E mutant of BR (60)). It should be noted that the homologous negative band in PR does not develop appreciably (21,24), especially in E. coli membranes, consistent with the weak accumulation of the N intermediate. The noted decrease of the amplitude of this negative band concomitant with the N→O transition is consistent with the reprotonation of Glu132 detected as a proton uptake on the same timescale (Fig. 4). Thus, we can conclude that, unlike in PR in E. coli membranes (24) but similar to BR, we observe the true N state with the deprotonated cytoplasmic proton donor, 13-cis-retinal, and a visible spectrum similar to that of the parent state (so-called N1 state (61)).
To confirm the tentative assignments of the bands for carboxylic acids and thus define the proton translocation pathway, we measured light-induced FTIR difference spectra of several GR mutants (Fig. 7). First, if Asp121 is the proton acceptor of the Schiff base as suggested by the visible spectroscopy (Fig. 5), one should not see signals of protonation of Asp121 and deprotonation of Glu132 in the D121N mutant, as its photocycle does not accumulate the late intermediates (M, N, O). Indeed, we do not observe any signals from protonated carboxylic acids in this mutant (Fig. 7 B, done at pH 7 to avoid the titration of the Schiff base), whereas the retinal bands (1514 cm−1 ethylenic, 1172 cm−1 C-C stretch) suggest the presence of all-trans red-shifted late intermediate(s). Second, if Glu132 is indeed the cytoplasmic proton donor for the Schiff base, one expects to see the signal of protonation of Asp121 but not the negative carboxylic band at 1734 cm−1 in the E132Q mutant. Because the photocycle of E132Q is extremely slow (Fig. 5), we used static light-dark difference FTIR spectroscopy of the photostationary mixture, which is supposed to be dominated by the M intermediate because of its slow decay. Unfortunately, the emerging picture is much more complex because of the presence of even slower red-shifted intermediate(s), some of which belong to the 13-cis cycle (Figs. 1 and 5). As expected from this, the FTIR spectrum of the photostationary mixture of intermediates of the E132Q photocycle (Fig. 7 C) shows the retinal bands associated with the red-shifted intermediates with protonated Schiff base (1186 cm−1 and 1178 cm−1 C-C stretches, and 1514 cm−1 and 1522 cm−1 C=C stretches), but the exact interpretation of these bands is difficult because of the possibility of secondary photochemistry. More importantly, the carboxylic stretch region of E132Q shows the band at 1749 cm−1 caused by protonation of Asp121 but not the sharp negative band at 1734 cm−1 tentatively assigned to Glu132. Thus, it appears that Glu132 serves as a Schiff base proton donor, and its deprotonation in the N intermediate is manifested by the appearance of the negative 1734 cm−1 band. At the same time, it may be dangerous to base such conclusion using the mutant as globally perturbed as E132Q. We double-checked our assignment using another, less perturbing, mutant E132D (Fig. 7 D). In a homologous mutant of PR (E108D), the 1728 cm−1 band of Glu108 is upshifted to 1744 cm−1 for Asp108 (24). If we expect a shift of the same magnitude in GR, the new band of Asp132 in the E132D mutant may overlap with the positive band of Asp121 and disappear as a result of the cancellation. Indeed, although we do not observe a strong negative band at 1734 cm−1 in E132D, we can not detect its new, upshifted, position. There is only a minor negative band at a lower frequency (1726 cm−1), but it is almost not D2O-sensitive (not shown), probably coming from lipid esters. Although all of this would be consistent with the expected band overlap for Asp132 and Asp121, the relative amplitude of the positive Asp121 band does not decrease as much as expected. This can be explained by the lower amplitude of the negative band of Asp132, which is expected from lower accumulation of the N1 state because of the higher pKa of Asp132. This is supported by the much slower rate of the Schiff base reprotonation in the E132D mutant shown above (Fig. 5). To sum up, the FTIR measurements of the GR mutants support the BR-like proton translocation pathway, where the homologs of BR's Asp85 and Asp96 act as proton acceptor and donor for the retinal Schiff base, respectively.
Figure 7.
Light-induced rapid-scan FTIR difference spectra of DMPC/DMPA liposome films with the GR variants: (A) wild-type, 1 ms delay at 2°C, pH 9 (0.1 M NaCl + 0.05 M KH2PO4 + 0.05 M CHES); (B) D121N, 1 ms delay at 7°C, pH 7 (0.1 M NaCl + 0.05 M KH2PO4); (C) E132Q, photostationary (>540 nm illumination) light-dark difference at 20°C, pH 9; and (D) E132D, 1 ms delay at 2°C, pH 9, but note the 12-ms interferogram acquisition time.
Conclusions
In this work, we provided biophysical characterization of novel cyanobacterial light-driven proton pump from Gloeobacter violaceus PCC 7421 (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished), GR. We characterized the dark state, the photochemical reaction cycle, and the proton translocation pathway of GR using a combination of spectroscopic methods (in the infrared and visible ranges) with site-directed mutagenesis. The general conclusion is that the photochemistry and proton translocation pathway of GR are proteorhodopsin-like, but with interesting differences, which, in some respects, make GR look more BR-like. The differences between GR and PR may reflect adaptation to the freshwater environment (pH ≈ 7.2 and low salt) of Gloeobacter as opposed to the marine environment (pH ≈ 8.2 and high salt) of PR-harboring bacteria. The main biospectroscopic features of GR can be summarized as follows.
First, the dark state of GR contains primarily all-trans-retinal both in its alkaline and acidic forms and does not show appreciable signs of the dark or light adaptation. This, along with the strong Schiff base counterion, makes it similar to PR (24,27,42). Unexpectedly, the replacement of Glu132, the putative cytoplasmic Schiff base proton donor (the homolog of Asp96 of BR), led to a dramatic increase in the 13-cis content of the dark state of GR in addition to the shift of the equilibrium between the alkaline and acidic forms described elsewhere (A. R. Choi, L. Shi, L. S. Brown, and K.-H. Jung, unpublished). Although the coupling of the retinal to the cytoplasmic carboxylic acid was shown for mutants of BR and a fungal rhodopsin (49,50,52), this is probably the strongest case of such coupling observed. Interestingly, such a phenomenon was not detected in the homologous E108Q mutant of PR (24). At the same time, the observed strong coupling is consistent with the recent structure of homologous XR, where connectivity of the Schiff base-forming Lys and cytoplasmic Glu is established in the dark state (35). A possible significance of such coupling for GR may be reflected in the observed greatly increased rate of reprotonation of the Schiff base and a shift of the M→N equilibrium toward N. This may create the much needed unidirectional step in the photocycle, in the absence of the early proton release ensuring unidirectionality of the M1→M2 reaction in BR.
Second, the photocycle of GR is, in general, similar to that of PR (25), having low accumulation of the M intermediate and being dominated by the two red-shifted intermediates (K-like and O-like). Similar to PR (24), the proton uptake precedes the proton release. On the other hand, we observed two striking BR-like features in the GR's photocycle, which were not observed for PR under similar conditions. We detected the accumulation of the L-like and N-like intermediates, and, importantly and differently from PR (24), the N-like intermediate is not strongly red-shifted and has a deprotonated cytoplasmic proton donor.
Finally, we established that the main participants in the proton translocation pathway are the same in GR, PR, and BR, with the primary proton acceptor of the Schiff base being homologous to Asp85 and the cytoplasmic proton donor to Asp96 of BR. We showed that the homolog of Asp85 of BR (Asp121 in GR) serves as a Schiff base counterion and a proton acceptor, and its protonation in the late intermediates of the photocycle produces a distinct positive vibrational band at 1749 cm−1. The homolog of Asp96 of BR (Glu132 in GR) deprotonates in the N intermediate, producing negative vibrational band at 1734 cm−1, and reprotonates during the transition to the O intermediate, concomitant with the proton uptake from the cytoplasmic bulk.
Acknowledgments
We thank Prof. Jacek Lipkowski for use of the Raman spectrometer and Samantha Veitch for help in collection of the spectroscopic data in the visible range.
The research was supported by a Korea Research Foundation Grant (KRF 2004-042-C00113 to K.H.J.), the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science & Technology, Korea to K.H.J., the second stage of Brain Korea 21 graduate Fellowship Program for A.R.C., and grants from the Canada Foundation for Innovation/Ontario Innovation Trust, the Natural Sciences and Engineering Research Council of Canada, the Premier's Research Excellence Award, Research Corporation, and Advanced Foods and Materials Network to L.S.B.
References
- 1.Béjà O., Aravind L., Koonin E.V., Suzuki M.T., Hadd A. Bacterial rhodopsin: evidence for a new type of phototrophy in the Sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 2.Walter J.M., Greenfield D., Bustamante C., Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc. Natl. Acad. Sci. USA. 2007;104:2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez A., Bradley A.S., Waldbauer J.R., Summons R.E., DeLong E.F. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. USA. 2007;104:5590–5595. doi: 10.1073/pnas.0611470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwalbach M.S., Brown M., Fuhrman J.A. Impact of light on marine bacterioplankton community structure. Aquat. Microb. Ecol. 2005;39:235–245. [Google Scholar]
- 5.Stingl U., Desiderio R.A., Cho J.C., Vergin K.L., Giovannoni S.J. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 2007;73:2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Consarnau L., Gonzalez J.M., Coll-Llado M., Gourdon P., Pascher T. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 7.Campbell B.J., Waidner L.A., Cottrell M.T., Kirchman D.L. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ. Microbiol. 2008;10:99–109. doi: 10.1111/j.1462-2920.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- 8.McCarren J., DeLong E.F. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ. Microbiol. 2007;9:846–858. doi: 10.1111/j.1462-2920.2006.01203.x. [DOI] [PubMed] [Google Scholar]
- 9.Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabehi G., Kirkup B.C., Rosenberg M., Stambler N., Polz M.F. Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 2007;1:48–55. doi: 10.1038/ismej.2007.10. [DOI] [PubMed] [Google Scholar]
- 11.Spudich J.L. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Sharma A.K., Zhaxybayeva O., Papke R.T., Doolittle W.F. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ. Microbiol. 2008;10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 13.Jung J.Y., Choi A.R., Lee Y.K., Lee H.K., Jung K.-H. Spectroscopic and photochemical analysis of proteorhodopsin variants from the surface of the Arctic Ocean. FEBS Lett. 2008;582:1679–1684. doi: 10.1016/j.febslet.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Atamna-Ismaeel N., Sabehi G., Sharon I., Witzel K.P., Labrenz M. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2008;2:656–662. doi: 10.1038/ismej.2008.27. [DOI] [PubMed] [Google Scholar]
- 15.Wang W.-W., Sineshchekov O.A., Spudich E.N., Spudich J.L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 16.Sineshchekov O.A., Spudich J.L. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem. Photobiol. Sci. 2004;3:548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]
- 17.Brown L.S., Jung K.-H. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem. Photobiol. Sci. 2006;5:538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- 18.Sabehi G., Loy A., Jung K.-H., Partha R., Spudich J.L. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3:e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frigaard N.U., Martinez A., Mincer T.J., DeLong E.F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 20.Sharma A.K., Spudich J.L., Doolittle W.F. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich T., Geibel S., Kalmbach R., Chizhov I., Ataka K. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 2002;321:821–838. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 22.Dioumaev A.K., Wang J.M., Balint Z., Váró G., Lanyi J.K. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry. 2003;42:6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 23.Furutani Y., Ikeda D., Shibata M., Kandori H. Strongly hydrogen-bonded water molecule is observed only in the alkaline form of proteorhodopsin. Chem. Phys. 2006;324:705–708. [Google Scholar]
- 24.Dioumaev A.K., Brown L.S., Shih J., Spudich E.N., Spudich J.L. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 25.Váró G., Brown L.S., Lakatos M., Lanyi J.K. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 2003;84:1202–1207. doi: 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partha R., Krebs R., Caterino T.L., Braiman M.S. Weakened coupling of conserved arginine to the proteorhodopsin chromophore and its counterion implies structural differences from bacteriorhodopsin. Biochim. Biophys. Acta. 2005;1708:6–12. doi: 10.1016/j.bbabio.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Bergo V., Amsden J.J., Spudich E.N., Spudich J.L., Rothschild K.J. Structural changes in the photoactive site of proteorhodopsin during the primary photoreaction. Biochemistry. 2004;43:9075–9083. doi: 10.1021/bi0361968. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda D., Furutani Y., Kandori H. FTIR study of the retinal Schiff base and internal water molecules of proteorhodopsin. Biochemistry. 2007;46:5365–5373. doi: 10.1021/bi700143g. [DOI] [PubMed] [Google Scholar]
- 29.Rangarajan R., Galan J.F., Whited G., Birge R.R. Mechanism of spectral tuning in green-absorbing proteorhodopsin. Biochemistry. 2007;46:12679–12686. doi: 10.1021/bi700955x. [DOI] [PubMed] [Google Scholar]
- 30.Jung K.H., Trivedi V.D., Spudich J.L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y., Kaneko T., Sato S., Mimuro M., Miyashita H. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 32.Spudich J.L., Jung K.-H. Microbial rhodopsins: phylogenetic and functional diversity. In: Briggs W.R., Spudich J.L., editors. Handbook of Photosensory Receptors. 1st ed. Wiley-VCH; Weinheim, Germany: 2005. pp. 1–24. [Google Scholar]
- 33.Mongodin E.F., Nelson K.E., Daugherty S., Deboy R.T., Wister J. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balashov S.P., Imasheva E.S., Boichenko V.A., Anton J., Wang J.M. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luecke H., Schobert B., Stagno J., Imasheva E.S., Wang J.M. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl. Acad. Sci. USA. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S.Y., Waschuk S.A., Brown L.S., Jung K.-H. Screening and characterization of proteorhodopsin color-tuning mutations in Escherichia coli with endogenous retinal synthesis. Biochim. Biophys. Acta Bioenergetics. 2008;1777:504–513. doi: 10.1016/j.bbabio.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Shi L., Yoon S.R., Bezerra A.G., Jr., Jung K.-H., Brown L.S. Cytoplasmic shuttling of protons in Anabaena sensory rhodopsin: implications for signaling mechanism. J. Mol. Biol. 2006;358:686–700. doi: 10.1016/j.jmb.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Waschuk S.A., Bezerra A.G., Jr., Shi L., Brown L.S. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eucaryote. Proc. Natl. Acad. Sci. USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imasheva E.S., Balashov S.P., Wang J.M., Lanyi J.K. pH dependent transitions in xanthorhodopsin. Photochem. Photobiol. 2006;82:1406–1413. doi: 10.1562/2006-01-15-RA-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith S.O., Braiman M.S., Myers A.B., Pardoen J.A., Courtin J.M.L. Vibrational analysis of the all-trans-retinal chromophore in light-adapted bacteriorhodopsin. J. Am. Chem. Soc. 1987;109:3108–3125. [Google Scholar]
- 41.Smith S.O., Pardoen J.A., Lugtenburg J., Mathies R.A. Vibrational analysis of the 13-cis-retinal chromophore in dark-adapted bacteriorhodopsin. J. Phys. Chem. 1987;91:804–819. [Google Scholar]
- 42.Pfleger N., Lorch M., Woerner A.C., Shastri S., Glaubitz C. Characterisation of Schiff base and chromophore in green proteorhodopsin by solid-state NMR. J. Biomol. NMR. 2008;40:15–21. doi: 10.1007/s10858-007-9203-5. [DOI] [PubMed] [Google Scholar]
- 43.Aton B., Doukas A.G., Callender R.H., Becher B., Ebrey T.G. Resonance Raman studies of the purple membrane. Biochemistry. 1977;16:2995–2999. doi: 10.1021/bi00632a029. [DOI] [PubMed] [Google Scholar]
- 44.Maeda A., Sasaki J., Pfefferlé J.M., Shichida Y., Yoshizawa T. Fourier transform infrared spectral studies on the Schiff base mode of all-trans bacteriorhodopsin and its photointermediates, K and L. Photochem. Photobiol. 1991;54:911–921. [Google Scholar]
- 45.Zscherp C., Heberle J. Infrared difference spectra of the intermediates L, M, N, and O of the bacteriorhodopsin photoreaction obtained by time-resolved attenuated total reflection spectroscopy. J. Phys. Chem. B. 1997;101:10542–10547. [Google Scholar]
- 46.Kakitani H., Kakitani T., Rodman H., Honig B., Callender R. Correlation of vibrational frequencies with absorption maxima in polyenes, rhodopsin, bacteriorhodopsin and retinal analogues. J. Phys. Chem. 1983;87:3620–3628. [Google Scholar]
- 47.Pande C., Callender R.H., Chang C.-H., Ebrey T.G. Resonance Raman spectra of the “blue” and the regenerated “purple” membranes of Halobacterium halobium. Photochem. Photobiol. 1985;42:549–552. doi: 10.1111/j.1751-1097.1985.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 48.Smith S.O., Pardoen J.A., Mulder P.P.J., Curry B., Lugtenburg J. Chromophore structure in bacteriorhodopsin's O640 photointermediate. Biochemistry. 1983;22:6141–6148. [Google Scholar]
- 49.Furutani Y., Sumii M., Fan Y., Shi L., Waschuk S.A. Conformational coupling between the cytoplasmic carboxylic acid and the retinal in a fungal light-driven proton pump. Biochemistry. 2006;45:15349–15358. doi: 10.1021/bi061864l. [DOI] [PubMed] [Google Scholar]
- 50.Lanyi J.K., Schobert B. Propagating structural perturbation inside bacteriorhodopsin: crystal structures of the M state and the D96A and T46V mutants. Biochemistry. 2006;45:12003–12010. doi: 10.1021/bi061310i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fodor S.P., Ames J.B., Gebhard R., van den Berg E.M., Stoeckenius W. Chromophore structure in bacteriorhodopsin's N intermediate: implications for the proton-pumping mechanism. Biochemistry. 1988;27:7097–7101. doi: 10.1021/bi00418a064. [DOI] [PubMed] [Google Scholar]
- 52.Dioumaev A.K., Brown L.S., Needleman R., Lanyi J.K. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry. 1998;37:9889–9893. doi: 10.1021/bi980934o. [DOI] [PubMed] [Google Scholar]
- 53.Dioumaev A.K. Evaluation of intrinsic chemical kinetics and transient product spectra from time-resolved spectroscopic data. Biophys. Chem. 1997;67:1–25. doi: 10.1016/s0301-4622(96)02268-5. [DOI] [PubMed] [Google Scholar]
- 54.Grzesiek S., Dencher N.A. Time course and stoichiometry of light-induced proton release and uptake during the photocycle of bacteriorhodopsin. FEBS Lett. 1986;208:337–342. [Google Scholar]
- 55.Krebs R., Alexiev U., Partha R., DeVita A.M., Braiman M. Detection of fast light-activated H+ release and M intermediate formation from proteorhodopsin. BMC Physiol. 2002;2:5. doi: 10.1186/1472-6793-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balashov S.P., Imasheva E.S., Ebrey T.G., Chen N., Menick D.R. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 57.Brown L.S., Sasaki J., Kandori H., Maeda A., Needleman R. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J. Biol. Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 58.Pfefferlé J.M., Maeda A., Sasaki J., Yoshizawa T. Fourier transform infrared study of the N intermediate of bacteriorhodopsin. Biochemistry. 1991;30:6548–6556. doi: 10.1021/bi00240a027. [DOI] [PubMed] [Google Scholar]
- 59.Maeda A. Application of FTIR spectroscopy to the structural study on the function of bacteriorhodopsin. Isr. J. Chem. 1995;35:387–400. [Google Scholar]
- 60.Bousché O., Braiman M., He Y.-W., Marti T., Khorana H.G. Vibrational spectroscopy of bacteriorhodopsin mutants. Evidence that Asp-96 deprotonates during the M-N transition. J. Biol. Chem. 1991;266:11063–11067. [PubMed] [Google Scholar]
- 61.Dioumaev A.K., Brown L.S., Needleman R., Lanyi J.K. Coupling of the reisomerization of the retinal, proton uptake, and reprotonation of Asp-96 in the N photointermediate of bacteriorhodopsin. Biochemistry. 2001;40:11308–11317. doi: 10.1021/bi011027d. [DOI] [PubMed] [Google Scholar]