Abstract
The methionine residues in the calcium (Ca2+) regulatory protein calmodulin (CaM) are structurally and functionally important. They are buried within the N- and C-domains of apo-CaM but become solvent-exposed in Ca2+-CaM, where they interact with numerous target proteins. Previous structural studies have shown that methionine substitutions to the noncoded amino acids selenomethionine, ethionine, or norleucine, or mutation to leucine do not impact the main chain structure of CaM. Here we used differential scanning calorimetry to show that these substitutions enhance the stability of both domains, with the largest increase in melting temperature (19–26°C) achieved with leucine or norleucine in the apo-C-domain. Nuclear magnetic resonance spectroscopy experiments also revealed the loss of a slow conformational exchange process in the Leu-substituted apo-C-domain. In addition, isothermal titration calorimetry experiments revealed considerable changes in the enthalpy and entropy of target binding to apo-CaM and Ca2+-CaM, but the free energy of binding was largely unaffected due to enthalpy-entropy compensation. Collectively, these results demonstrate that noncoded and coded methionine substitutions can be accommodated in CaM because of the structural plasticity of the protein. However, adjustments in side-chain packing and dynamics lead to significant differences in protein stability and the thermodynamics of target binding.
Introduction
The incorporation of noncoded amino acids into proteins is an emerging tool for investigating protein structure and function (1–3). Applications include the use of selenomethionine (SeMet) for x-ray structure determination (4), modified aromatic amino acids as spectroscopic probes (5), and proteins containing Pro analogs (such as thiaproline) as vehicles for drug delivery (6). Since many noncoded amino acids differ from their natural homologs by only a single atom or functional group, they can also be useful for studying atom-level contributions to protein folding or target interactions. Although the majority of these proteins maintain an identical three-dimensional structure and activity compared to the wild-type protein, they often have altered stabilities (6–8), indicating that noncoded substitutions impact protein flexibility and side-chain packing.
The Met residues in the small Ca2+ binding protein calmodulin (CaM) are interesting targets for modification by noncoded substitution or mutation. In the absence of Ca2+, each of the two globular domains of CaM forms a compact “closed” structure with the hydrophobic residues, including four Met residues from the N-domain (M36, M51, M71, and M72) and C-domain (M109, M124, M144, and M145) sequestered from the solvent (Fig. 1). Ca2+ binding to the four helix-loop-helix “EF-hand” motifs of CaM induces conformational changes that “open” the N- and C-domains, and exposes distinct hydrophobic target-protein binding patches, with the Met residues forming nearly 50% of the hydrophobic surface area of each patch (9,10). The “central linker” connecting the N- and C-domains is highly flexible and contains an additional Met residue, M76, that remains solvent-exposed in both Ca2+-free CaM (apo-CaM) and Ca2+-bound CaM (Ca2+-CaM) (11). The flexibility of the central linker, combined with the structural plasticity of the Met side chains in the hydrophobic patches, enables CaM to interact with more than 100 different target proteins in a sequence-independent manner, requiring only a combination of hydrophobic and basic residues in the binding region (9,12). Target protein binding by Ca2+-CaM in vivo can regulate important processes such as muscle contraction and neurotransmission, as well as cell growth, proliferation, and movement (13). In addition, the unique structural and surface features of apo-CaM enable Ca2+-independent binding of CaM to a different subset of proteins, some of which have been implicated in the targeting or localization of CaM, whereas others require apo-CaM for activity (10,14,15).
Figure 1.
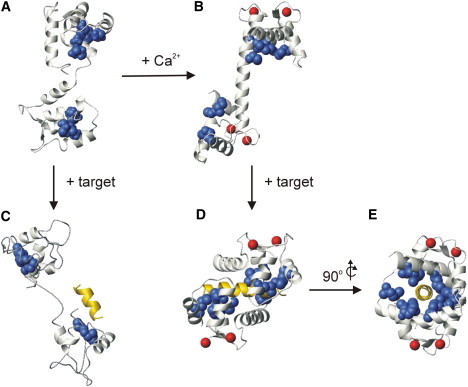
CaM structures and target interactions. (A) Apo-CaM (PDB:1DMO), (B) Ca2+-CaM (PDB:1CLL), (C) apo-CaM in complex with the SK channel CaM binding domain peptide (PDB:1QX7), (D) Ca2+-CaM in complex with the smMLCKp peptide (PDB:1CDL), and (E) 90° y-axis rotation of panel D. In each panel CaM is shown in ivory ribbon representation, the Met side chains are shown as navy blue space-fill representation, Ca2+ ions are represented by red spheres, and the bound target peptides are shown in yellow ribbon representation. CaM is oriented with the N- and C-domains on the top and the bottom, respectively, in each panel. Note that electron density was observed only for apo-CaM residues 5–72, 81–89, 97–101, and 116–146 in panel C.
To examine the effect of noncoded and coded Met substitutions on the structure and target interactions of CaM, we previously generated and studied CaM proteins with all nine Met residues substituted with selenomethionine (SeMet-CaM), norleucine (Nle-CaM), or ethionine (Eth-CaM), as well as a quadruple C-terminal M109/M124/M144/M145→Leu4 mutant (CT-CaM) and a CT-CaM variant, with the remaining five N-terminal Met residues substituted with SeMet (SeMet-CT-CaM) (16–21). Spectroscopic and biochemical studies revealed that none of these substitutions significantly impact the main chain structure of Ca2+-CaM or the Ca2+-dependent conformational changes in the protein, and they have relatively small effects on Ca2+-CaM's affinity for target proteins and peptides (16–21). However, since the SeMet, Eth, Nle, and Leu substitutions each influence side-chain properties such as structure, flexibility, polarity, and polarizability (17,22), they are expected to alter side-chain packing. This in turn should be reflected in distinct N- and C-domain stabilities and unique thermodynamic signatures for ligand binding to the various Met-substituted proteins. To directly examine these thermodynamic effects, we studied the stability and target interactions of the Met-substituted CaMs using high-sensitivity differential scanning calorimetry (DSC) and isothermal titration calorimetry (ITC), respectively. The data show that the Met substitutions result in a general, and in some cases dramatic increase in the stability of the N- and C-domains, and lead to significant changes in the thermodynamics of ligand binding.
Materials and Methods
Proteins and peptides
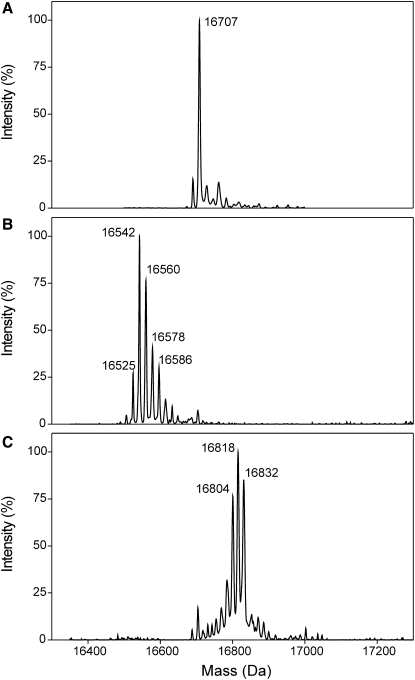
All CaM proteins (wt-CaM, SeMet-CaM, Eth-CaM, Nle-CaM, CT-CaM, and SeMet-CT-CaM) were expressed in Escherichia coli and purified to homogeneity (>95%) with the use of phenyl-Sepharose affinity chromatography as previously described (19–21). The amino acid sequences of wt-CaM and the mutant CT-CaM protein are directly encoded by their genomic sequences, resulting in proteins having 100% incorporation of Met or Leu at each respective position. Incorporation of SeMet, Eth, or Nle involves protein expression from the wt-CaM or CT-CaM genomic sequences in the Met auxotrophic E. coli strain DL-41 (21), and results in <100% substitution levels. SeMet incorporation was determined by nuclear magnetic resonance (NMR) spectroscopy and amino acid analysis to be ∼85% for SeMet-CaM and ∼95% for SeMet-CT-CaM, and incorporation was random with each protein (18,21). Nle and Eth were also randomly incorporated into CaM at levels of ∼85% in each case as determined by amino acid analysis, matrix-assisted laser desorption ionization (MALDI) mass spectrometry, and electrospray ionization mass spectrometry (19) (Fig. 2).
Figure 2.
Noncoded amino acid substitution in CaM. Electrospray ionization mass spectra of (A) wt-CaM, (B) Nle-CaM, and (C) Eth-CaM show the level and distribution of Nle or Eth incorporation in place of the nine Met residues of the wild-type CaM protein. Nle is 18 Da smaller than Met, and the Nle-CaM spectrum (panel B) shows a series of protein masses that decrease as the number of incorporated Nle residues increases. Peaks can be clearly seen for species containing from three to nine Nle substitutions, with the most abundant species at 16,542 Da representing CaM with all nine of its Met residues replaced with Nle. The extra peak at 16,525 represents a loss of NH3 from the +9 Nle species. On the other hand, Eth is 14 Da larger than Met, and the Eth-CaM spectrum (panel C) shows a series of protein masses that increase with increasing incorporation levels of Eth. The +7 through +9 species are the most abundant, with the fully Eth-substituted protein seen at 16,832 Da. Samples were analyzed using a VG Quattro (Micromass, Manchester, UK) ESI triple quadrupole mass spectrometer.
The smMLCKp peptide (Ac-ARRKWQKTGHAVRAIGRLSS-NH2) and NtMKP1b peptide (Ac-NGWSRLRRKFSSGIMK-NH2) were each commercially synthesized and shown to be >95% pure by high-pressure liquid chromatography and MALDI mass spectrometry. ANS (8-anilino-1-naphalenesulfonate) was purchased from Sigma (St. Louis, MO). Protein, peptide, and ANS concentrations were determined using the predicted molar extinction coefficients of ε276 = 2900 M−1 cm−1 for each CaM protein, ε280 = 5690 M−1 cm−1 for the smMLCKp or NtMKP1b peptides, and ε350 = 5000 M−1 cm−1 for ANS.
Differential scanning calorimetry
All DSC experiments were performed on a VP-DSC microcalorimeter (MicroCal, Northampton, MA). Samples were prepared by dissolving lyophilized protein in 20 mM HEPES, 100 mM KCl, pH 7.5 (hereafter referred to as HK-buffer) to a concentration of ∼300 μM, and samples were dialyzed in HK-buffer overnight at room temperature. Postdialysis protein samples were diluted to 180 μM (3 mg/mL) with the dialysis buffer and supplemented with either 2.5 mM EDTA + 2.5 mM EGTA for apo-CaM samples, or 2 mM CaCl2 for Ca2+-CaM samples using concentrated stock solutions of 0.5 M EDTA, 0.5 M EGTA, or 1 M CaCl2. To obtain optimal baseline reproducibility, all DSC experiments were performed in “continuous scanning mode”; the first buffer-buffer scan was discarded because of a different thermal history, as described in the MicroCal VP-DSC user's manual. Each heating scan consisted of a 15-min prescan thermostat period at 10°C, and a 10–125°C heating scan (90°/h) with a filter period of 16 s, and passive thermal compensation between the sample and reference cells. The samples were passively cooled, and the cells were refilled between 30°C and 15°C during the cooling period. As previously reported for wt-CaM (23), we found that the thermal denaturation of the apo-CaM proteins was essentially reversible (>95%) if the proteins were heated just to the end of their thermal transitions and immediately cooled. However, when apo-CaM or Ca2+-CaM proteins were heated to 125°C, which was routinely done to obtain sufficient post-transition baselines for data fitting, the thermal transitions were not fully reversible. Data analysis was performed using MicroCal Origin software, with the partial specific volume for each protein assumed to be 0.720 cm3g−1 (24). For the apo-CaM proteins, an instrument baseline was subtracted from the experimental data and then pre- and post-transition baseline segments were manually defined and connected using the “progress baseline” function. The denaturation data for each apo-CaM protein were best described by two unfolding transitions corresponding to denaturation of the N- and C-domains of the protein. Data for the Ca2+-CaM proteins were analyzed similarly to the apo-CaM data, except that the post-transition baselines were beyond the high temperature limit of the calorimeter (>125°C) and were estimated during analysis. Repeat analysis of the Ca2+-CaM protein data using different post-transition baselines gave good reproducibility in the determination of Tm (±1.0°C), but the ΔHd values for the Ca2+-CaM proteins could not be accurately determined due to the strong dependence of this parameter on the baseline position.
NMR spectroscopy
1H, 15N heteronuclear single quantum coherence (HSQC) and NMR relaxation experiments were performed at 30°C on a Bruker (Fallanden, Switzerland) AVANCE 500 MHz NMR spectrometer equipped with a triple-resonance inverse cryoprobe with single-axis z-gradient. The samples consisted of 0.8 mM 15N-wt-CaM or 15N-CT-CaM in 100 mM KCl, 2 mM EDTA, 10% D2O, 0.5 mM DSS, and 0.03% NaN3. 15N T2 values were obtained using Carr-Purcell-Meiboom-Gill (CPMG) experiments with 180° pulse train delay times of 1 ms or 5 ms, and T1ρ values were obtained by applying a 2.5 kHz spin-locking pulse during the relaxation delay time as previously described (25). Backbone resonance assignments for apo-wt-CaM were obtained using standard triple-resonance NMR spectroscopy experiments (HNCACB, HN(CA)CO, HN(CO)CACB, and HNCO) recorded on a Bruker AVANCE 700 MHz NMR spectrometer under the solution conditions described above, using samples of uniformly 13C- and 15N-labeled wt-CaM. NMR data were processed using NMRPipe/NMRDraw (26) and analyzed using NMRView software (27).
Steady-state fluorescence spectroscopy
Steady-state ANS fluorescence emission spectra were recorded on a Varian (Palo Alto, CA) Cary Eclipse spectrofluorimeter at 25°C. The samples consisted of 20 μM CaM protein and 60 μM ANS in HK-buffer containing 2 mM CaCl2 for Ca2+-CaM samples or 1 mM EDTA + 1 mM EGTA for apo-CaM samples. ANS was selectively excited at 370 nm using an excitation slit width of 5 nm, and steady-state fluorescence emission spectra were recorded from 400 to 600 nm using an emission slit width of 5 nm, and averaged over five scans.
Isothermal titration calorimetry
All ITC experiments were performed on a MicroCal VP-ITC microcalorimeter. ANS-binding experiments consisted of sequential injection of 5 mM ANS into 50 μM CaM protein in HK-buffer plus either 5 mM CaCl2 for Ca2+-CaM samples or 2 mM EDTA + 2 mM EGTA for apo-CaM samples. Peptide-binding experiments involved sequential injection of 0.4–0.5 mM smMLCKp peptide into 18–20 μM CaM in HK-buffer plus 2 mM CaCl2, or 0.6–0.65 mM NtMKP1b peptide into 30 μM CaM protein in HK-buffer plus 2 mM EDTA and 2 mM EGTA. The heat of dilution/mixing was estimated from separate control experiments or by using the average heat of injection after saturation, and these values were subtracted before curve fitting. All data (except for the Ca2+-Eth-CaM•smMLCKp titration data) were analyzed using the “one set of sites” model supplied in the MicroCal Origin software to determine the apparent stoichiometry (N), association constant (Ka), and enthalpy change (ΔH) associated with binding. When appropriate, the Gibbs free energy (ΔG), entropy (TΔS), and heat capacity (ΔCp) changes were calculated using standard thermodynamic equations (ΔG = −RT lnKa), (ΔG = ΔH − TΔS), and (ΔCp = dΔH/dT). ITC data for titrations of Ca2+-Eth-CaM with the smMLCKp peptide did not fit well with the “one set of sites” model, but were adequately described by the “two sets of sites” model supplied in the MicroCal Origin software.
Results
Thermal stability of Met-substituted CaM proteins
Previous DSC studies with wild-type CaM (wt-CaM) have shown that the N-domain is more stable (i.e., has a higher transition midpoint temperature, Tm) and has a substantially larger calorimetric enthalpy of denaturation (ΔHd) than the C-domain in the presence and absence of Ca2+ (23,24). Although the Tm for each domain can shift dramatically in response to different solution conditions, mutation, or ligand-binding, the large disparity in ΔHd is always maintained, enabling the two calorimetric unfolding transitions to be specifically assigned to the N- and C-domains of the protein (23,24,28,29).
Our DSC data agreed well with previous studies, revealing an asymmetrical unfolding profile for apo-wt-CaM with Tm values of 48°C for the C-domain and 60°C for the N-domain (Fig. 3 A; Table 1). The melting curves for each Met-substituted apo-CaM protein were also best described by two distinct transitions for the N- and C-domains, but the Tm for each domain was considerably different for each protein (Fig. 3, left column; Table 1). SeMet or Eth substitutions at all nine Met positions increased the Tm of each domain by 4–7°C, with the N-domain retaining a higher Tm than the C-domain in each case. In contrast, the Tm of the C-domain of apo-CT-CaM increased by 19°C, and the Tm of the unmodified N-domain decreased by 5°C, resulting in the N-domain unfolding before the C-domain in apo-CT-CaM. SeMet substitution at the remaining five N-terminal Met positions in CT-CaM increased the Tm of the N-domain by 5°C, but had essentially no effect on the C-domain in comparison to apo-CT-CaM. Met→Nle substitutions also dramatically increased the Tm of the C-domain (+26°C); however, the Tm of the apo-Nle-CaM N-domain was 2°C lower than that of the wild-type N-domain. Therefore, the DSC data show that the order of temperature-induced domain unfolding for the Leu- and Nle-substituted apo-CaM proteins (N-domain unfolds before C-domain), is reversed in comparison to apo-wt-CaM (N-domain unfolds before C-domain), a result that was also confirmed by NMR and fluorescence spectroscopy (see Fig. S1 in the Supporting Material).
Figure 3.

Temperature-induced denaturation of Met-substituted CaM proteins as monitored using DSC. The left column shows the temperature dependence of the excess heat capacity (solid line) and deconvolution into individual transitions for the N-domain (dotted line labeled “N”) and C-domain (dotted line labeled “C”) for (A) apo-wt-CaM, (C) apo-SeMet-CaM, (E) apo-Eth-CaM, (G) apo-Nle-CaM, (I) apo-CT-CaM, and (K) apo-SeMet-CT-CaM. The right column shows the temperature dependence of the absolute heat capacity for (B) Ca2+-wt-CaM, (D) Ca2+-SeMet-CaM, (F) Ca2+-Eth-CaM, (H) Ca2+-Nle-CaM, (J) Ca2+-CT-CaM, and (L) Ca2+-SeMet-CT-CaM, with the labels “N” and “C” corresponding to the unfolding of the N- and C-domains, respectively.
Table 1.
Thermodynamic parameters for the temperature-induced denaturation of CaM proteins as determined by DSC
| N-domain |
C-domain |
|||
|---|---|---|---|---|
| Protein | Tm (°C)∗ | ΔHd (kJ/mol) † | Tm (°C) | ΔHd (kJ/mol) |
| apo-wt-CaM | 59.5 ± 0.1 | 177 ± 3.2 | 48.3 ± 0.2 | 84 ± 3.2 |
| apo-SeMet-CaM | 66.2 ± 0.1 | 169 ± 2.7 | 54.7 ± 0.1 | 102 ± 2.7 |
| apo-Eth-CaM | 64.3 ± 0.3 | 142 ± 6.2 | 52.7 ± 0.3 | 103 ± 6.2 |
| apo-Nle-CaM‡ | 57.5 ± 0.4 | 160 ± 20 | 74.6 ± 0.9 | 86 ± 9 |
| apo-CT-CaM | 54.7 ± 0.1 | 173 ± 2.2 | 67.1 ± 0.1 | 124 ± 2.2 |
| apo-SeMet-CT-CaM | 59.9 ± 0.1 | 166 ± 4.4 | 69.2 ± 0.1 | 123 ± 4.4 |
| Ca2+-wt-CaM | 113 ± 1.0 | 93 ± 1.0 | ||
| Ca2+-SeMet-CaM | 118 ± 1.0 | 97 ± 1.0 | ||
| Ca2+-Eth-CaM | 121 ± 1.0 | 100 ± 1.0 | ||
| Ca2+-Nle-CaM | >125 ± 1.0 | 97 ± 1.0 | ||
| Ca2+-CT-CaM | 124 ± 1.0 | 85 ± 1.0 | ||
| Ca2+-SeMet-CT-CaM | >125 ± 1.0 | 92 ± 1.0 | ||
Tm is the transition midpoint temperature.
ΔHd is the calorimetric enthalpy of denaturation.
Values represent the average and standard deviation (SD) of four independent measurements. All other Ka error estimates are derived from curve-fitting uncertainties.
As with wt-CaM (23,24), Ca2+ binding dramatically increases the thermal stability of each Met-substituted CaM protein such that the melting curves are incomplete at the high temperature limit of the microcalorimeter (125°C) (Fig. 3, right column). Consequently, it was possible to obtain reasonable estimates of the Tm (±1.0°C) but not the ΔHd for the Ca2+-bound N- and C-domains (see Materials and Methods; Table 1). For each Ca2+-CaM protein, the smaller endothermic transition was found at lower temperatures followed by the larger endothermic peak, indicating that the C-domain unfolds before the N-domain in each case. Incorporation of SeMet resulted in an increase in the Tm of both domains by 4–5°C, whereas the Tm increased by 7–8°C with Eth. Incorporation of Nle increased the Tm of the Ca2+-N-domain by more than 12°C and the Ca2+-C-domain by 4°C. Interestingly, the Tm of the Leu-substituted C-domain of Ca2+-CT-CaM was 8°C lower than Ca2+-wt-CaM, and the Tm of the unmodified N-domain was increased by 11°C with respect to the wild-type domain. Incorporation of SeMet further increased the Tm of the N-domain in Ca2+-SeMet-CT-CaM and increased the Tm of the C-domain by 7°C in comparison to Ca2+-CT-CaM. The changes in Tm of unmodified CaM domains are attributed to altered interactions between the two domains during the unfolding process, as described in detail in the Discussion section.
Leu substitutions eliminate slow conformational exchange in the apo-C-domain
In addition to having different stabilities, the N- and C-domains of apo-wt-CaM exhibit different conformational exchange behaviors, with the apo-N-domain having a relatively rigid structure, and the apo-C-domain undergoing exchange between major (>90%) and minor (<10%) conformations on the timescale of several hundred microseconds (25). This exchange is clearly visible in the weak signal intensity of many C-domain resonances in the 1H, 15N HSQC NMR spectrum of apo-15N-wt-CaM (Fig. 4, A and C, and Fig. S2) (30). Conformational exchange in the apo-C-domain can also be observed by comparing NMR-obtained 15N T1ρ data with T2 data, or by comparing 15N T2 data obtained using different pulse train delays (τcp) for the backbone amide resonances (25,31). In these experiments the larger T1ρ/T2 ratios or larger ΔR2 values (where ΔR2 = R2(τcp = 5 ms) − R2(τcp = 1 ms), and R2 = 1/T2) identify many residues throughout the apo-C-domain of wt-CaM with signals that are influenced by slow conformational exchange, whereas T1ρ/T2 ratios close to one and smaller ΔR2 values demonstrate a lack of such exchange in the apo-N-domain (Fig. 4, E and F).
Figure 4.
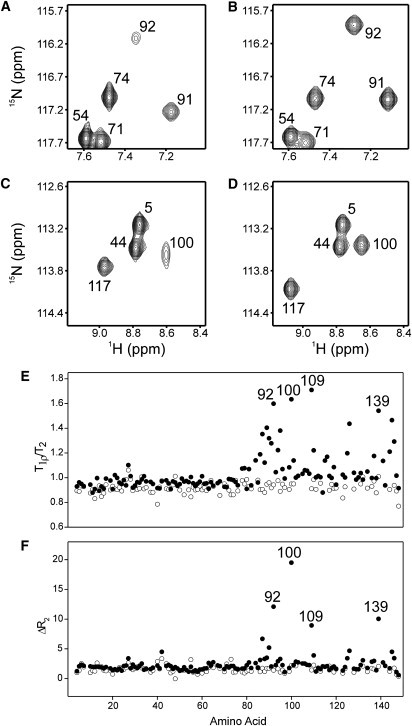
NMR investigation of slow conformational exchange in apo-wt-CaM and apo-CT-CaM. Panels A and C show selected regions of the 1H, 15N HSQC NMR spectrum of apo-15N-wt-CaM, and panels B and D show the same regions of the apo-15N-CT-CaM spectrum. (E) Ratio of the 15N T1ρ and T2 values plotted versus amino acid residue for apo-wt-CaM (•) and apo-CT-CaM (○). (F) ΔR2 values (where ΔR2 = R2(τcp = 5 ms) – R2(τcp = 1 ms), and R2 = 1/T2), plotted versus amino acid residue for apo-wt-CaM (•) and apo-CT-CaM (○).
The dramatically enhanced stability of the Leu- and Nle-substituted apo-C-domains suggests that these aliphatic substitutions might also influence conformational exchange in this domain. Since the mutant CT-CaM protein is amenable to uniform 15N-labeling, we produced apo-15N-CT-CaM and subjected the protein to a similar set of NMR experiments as described above for apo-15N-wt-CaM. The near-perfect HSQC signal overlap for the N-domain residues of apo-CT-CaM and apo-wt-CaM support our previous conclusion that the C-terminal Met→Leu mutations do not influence the structure of the folded N-domain of CaM (20). As expected, the four Met→Leu mutations induced chemical shift changes throughout the C-domain, but overall these shifts were small enough to assign and analyze ∼70% of the nonoverlapped backbone signals in this region based on the spectrum of apo-wt-CaM (Fig. S2). As shown in Fig. 4, the Met→Leu mutations dramatically improved the signal intensities for the C-domain residues of apo-CT-CaM and resulted in T1ρ/T2 ratios and ΔR2 values similar to those for the N-domain. These NMR data indicate that the Met→Leu mutations greatly reduce the slow conformational exchange behavior in the C-domain of apo-CaM.
Thermodynamics of ANS binding to Met-substituted CaM proteins
To investigate the influence of the Met substitutions on the thermodynamics of ligand binding to CaM, we first studied the interaction with the small hydrophobic and fluorescent molecule ANS, which is commonly used to probe exposed hydrophobic protein surfaces. ITC and steady-state fluorescence spectroscopy experiments revealed no ANS binding to any of the apo-CaM proteins, presumably due to a lack of hydrophobic surface area and an electrostatic repulsion between the negatively charged sulfonate of ANS and the acidic surfaces of the apo-N- and -C-domains. In contrast, there were dramatic fluorescence enhancements, as well as large exothermic signals generated in ITC titrations of each Ca2+-CaM protein with ANS, indicating that there is an enthalpically favorable interaction between ANS and the exposed hydrophobic patches of each protein (Fig. 5, Table 2). The gradual and incomplete saturation of each ANS-binding isotherm indicates that ANS binds with relatively low affinity to each protein, with c-values (c = Ka[CaM]) well below the range of 1–1000, which is necessary for complete thermodynamic characterization by ITC (32). Consequently, the simplest best fit for each ITC data set was obtained using a “one set of sites” model, with the apparent association constant (Ka) values representing the overall low-affinity interaction with multiple ANS molecules. The apparent Ka values determined in this manner were very reproducible, as shown in replica experiments with Ca2+-wt-CaM (Ka = 4.1 ± 0.1 × 103 M−1 at 25°C), and the Ka values decreased by a small amount with increasing temperature (Table 2). Although the enthalpy of binding (ΔH) could not be accurately determined because of the low c-value, it was clear that the magnitude of the exothermic signals were only slightly smaller at lower temperatures (Fig. 5 B), implying that the change in heat capacity (ΔCp = dΔH/dT) associated with the binding of ANS to Ca2+-wt-CaM is small and negative.
Figure 5.
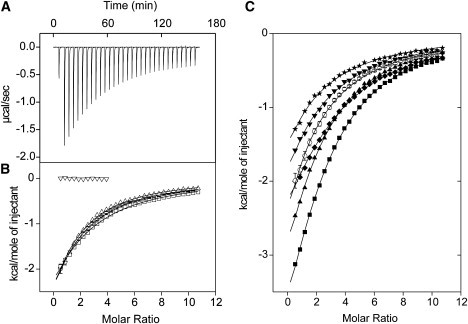
ANS binding to Met-substituted CaM proteins as monitored using ITC. (A) Baseline-corrected raw calorimetric data for the sequential injection of 5 mM ANS into 50 μM Ca2+-wt-CaM at 25°C. (B) Integrated heat signals (corrected for heat of dilution effects) are plotted as a function of molar ratio for ANS titrations of apo-wt-CaM at 25°C (▿), or Ca2+-wt-CaM at 20°C (▵), 25°C (○), or 30°C (□). (C) Integrated heat signals plotted as a function of molar ratio for ANS titrations of Ca2+-wt-CaM (○), Ca2+-SeMet-CaM (▾), Ca2+-Eth-CaM (★), Ca2+-Nle-CaM (♦), Ca2+-CT-CaM (▪), and Ca2+-SeMet-CT-CaM (▴), each performed at 25°C. The solid lines through the data represent the best fit using the “one set of sites” model supplied in the MicroCal Origin software. The data for Ca2+-wt-CaM at 25°C in panels B and C are shown as the average, with SD error bars derived from three independent titrations.
Table 2.
Thermodynamic and fluorescence parameters for ANS-binding to Ca2+-CaM proteins
| Protein | Temperature (°C) | Ka (M−1) | λmax (nm) | Fluorescence intensity at λmax (a.u.) |
|---|---|---|---|---|
| Ca2+-wt-CaM | 20 | 4.8 ± 0.3 × 103 | ||
| Ca2+-wt-CaM | 25 | 4.1 ± 0.1 × 103∗ | 481 | 166 |
| Ca2+-wt-CaM | 30 | 3.7 ± 0.2 × 103 | ||
| Ca2+-SeMet-CaM | 25 | 3.7 ± 0.3 × 103 | 485 | 134 |
| Ca2+-Eth-CaM | 25 | 3.2 ± 0.3 × 103 | 483 | 110 |
| Ca2+-Nle-CaM | 25 | 5.0 ± 0.3 × 103 | 479 | 285 |
| Ca2+-CT-CaM | 25 | 7.1 ± 0.2 × 103 | 474 | 332 |
| Ca2+-SeMet-CT-CaM | 25 | 6.2 ± 0.2 × 103 | 476 | 312 |
Apparent association constant (Ka) values were determined by ITC, and the steady-state fluorescence emission wavelength maximum (λmax) and fluorescence intensity at λmax (in arbitrary units, a.u.) were obtained from steady-state fluorescence emission spectra.
Value represents the average and SD of three independent measurements. All other Ka error estimates are derived from curve-fitting uncertainties.
Each of the Met substitutions clearly influenced the enthalpy of ANS binding to CaM, as evidenced by the large difference in heat released with each protein (Fig. 5 C). The fluorescence properties of ANS were also distinct when bound to each protein, with considerably larger fluorescence enhancements observed for the Nle- and Leu-substituted proteins, and somewhat smaller enhancements observed with Ca2+-SeMet-CaM and Ca2+-Eth-CaM (Table 2). Of interest, these changes were accompanied by only small differences in ANS affinity among the Ca2+-CaM proteins, with all Ka values in the range of 3.2–7.1 × 103 M−1. However, there was a distinct correlation between the Ka values, the magnitude of the exothermic signals, and the fluorescence enhancements, each increasing in the order of Eth < SeMet < Met < Nle < Leu as the substituted residues (Table 2).
Thermodynamics of smMLCKp peptide binding to Met-substituted Ca2+-CaM proteins
The complex of Ca2+-CaM with the CaM-binding domain (CaMBD) of smooth muscle myosin light chain kinase (smMLCKp peptide) is a model system for the canonical “wrap around” binding mode involving peptide interactions with both the N- and C-domain hydrophobic patches of Ca2+-CaM (33,34) (see Fig. 1, D and E). ITC experiments showed that smMLCKp peptide binding to each Met-substituted protein is of high affinity, with an exothermic, sigmoidal-shaped binding isotherm that saturates near a 1:1 stoichiometric ratio (Fig. 6, A and B). With the exception of Ca2+-Eth-CaM (discussed below), the ITC data for each protein were best described by a “one set of sites” binding model with Ka values of 107–108 M−1. Because these high Ka values are near the upper accuracy limit of the ITC technique (c-value: 200–5000), the experimental uncertainty in Ka is relatively large for the smMLCKp peptide measurements (e.g., Ka = 3.6 ± 2.6 × 107 M−1 for Ca2+-Nle-CaM at 25°C; Table 3). However, since the overall range of Ka values in Table 3 corresponds to a small variation in the ΔG (−39.2 to −45.5 kJ/mol), and because ΔH is accurately determined in situations of strong binding, we were able to obtain good estimates for the entropy (TΔS) and the ΔCp for smMLCKp peptide binding to the Ca2+-CaM proteins (34,35).
Figure 6.
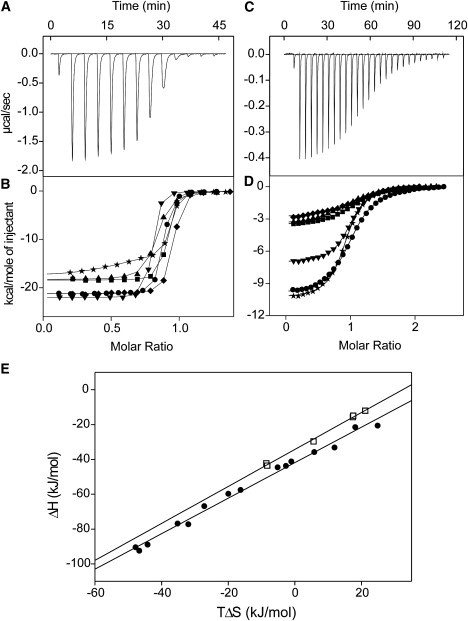
Peptide binding to CaM proteins as monitored by ITC. Baseline-corrected raw calorimetric data for the titration of (A) 18 μM Ca2+-Nle-CaM with 0.4 mM smMLCKp peptide at 25°C, or (C) 30 μM apo-CT-CaM titrated with 0.63 mM NtMKP1b peptide at 25°C. Integrated heat signals (corrected for heat of dilution effects) are plotted as a function of molar ratio for (B) smMLCKp peptide titrations or (D) NtMKP1b peptide titrations of wt-CaM (•), SeMet-CaM (▾), Eth-CaM (★), Nle-CaM (♦), CT-CaM (▪), and SeMet-CT-CaM (▴). The solid lines through the data represent the best fit using the “one set of sites” model supplied in the MicroCal Origin software, except for the Ca2+-Eth-CaM data, which were best fitted using the “two sets of sites” model. (E) Enthalpy (ΔH) versus entropy (TΔS) plot for smMLCKp peptide (•) binding to Ca2+-CaM proteins, or NtMKP1b peptide (□) binding to apo-CaM proteins. The solid lines were obtained by linear regression.
Table 3.
Thermodynamic parameters for peptide binding to CaM proteins as determined by ITC
| Protein | Conditions∗ | N | Ka (M−1) | ΔH (kJ/mol) | TΔS (kJ/mol) | ΔCp (kJ/mol•K) |
|---|---|---|---|---|---|---|
| smMLCKp peptide | ||||||
| Ca2+-wt-CaM | 25.0°C | 0.8 ± 0.0 | 6.7 ± 0.2 × 107 | −88.9 ± 0.1 | −44.2 | |
| Ca2+-wt-CaM | 17.6°C | 0.9 ± 0.0 | 1.3 ± 0.3 × 107 | −59.7 ± 0.7 | −20.0 | −3.6 |
| Ca2+-wt-CaM | 10.1°C | 0.8 ± 0.0 | 4.6 ± 1.1 × 107 | −35.8 ± 0.3 | 5.8 | |
| Ca2+-SeMet-CaM | 25.0°C | 0.8 ± 0.0 | 9.7 ± 0.6 × 107 | −92.4 ± 0.2 | −46.7 | |
| Ca2+-SeMet-CaM | 17.6°C | 0.8 ± 0.0 | 1.3 ± 0.2 × 107 | −66.8 ± 0.9 | −27.3 | −3.4 |
| Ca2+-SeMet-CaM | 10.2°C | 0.8 ± 0.0 | 2.5 ± 0.4 × 107 | −41.2 ± 0.3 | −1.1 | |
| Ca2+-Eth-CaM | 25.0°C | −74.3 ± 2.2 | ||||
| Ca2+-Eth-CaM | 17.6°C | −49.1 ± 0.7 | −3.4 | |||
| Ca2+-Eth-CaM | 10.2°C | −23.8 ± 0.3 | ||||
| Ca2+-Nle-CaM† | 25.0°C | 0.9 ± 0.1 | 3.6 ± 2.6 × 107 | −90.4 ± 3.7 | −47.8 ± 3.3 | |
| Ca2+-Nle-CaM | 17.6°C | 1.0 ± 0.0 | 2.5 ± 0.5 × 107 | −57.5 ± 0.5 | −16.3 | −3.9 |
| Ca2+-Nle-CaM | 10.2°C | 0.9 ± 0.0 | 2.0 ± 1.2 × 108 | −33.2 ± 0.3 | 11.9 | |
| Ca2+-CT-CaM | 25.0°C | 0.9 ± 0.0 | 8.4 ± 0.7 × 107 | −77.2 ± 0.2 | −32.0 | |
| Ca2+-CT-CaM | 17.6°C | 1.0 ± 0.0 | 2.3 ± 0.9 × 107 | −43.7 ± 0.9 | −2.7 | −3.8 |
| Ca2+-CT-CaM | 10.2°C | 1.0 ± 0.0 | 2.4 ± 0.9 × 108 | −20.6 ± 0.1 | 24.8 | |
| Ca2+-SeMet-CT-CaM | 25.0°C | 0.8 ± 0.0 | 1.9 ± 0.2 × 107 | −76.8 ± 0.5 | −35.2 | |
| Ca2+-SeMet-CT-CaM | 17.6°C | 0.9 ± 0.0 | 1.1 ± 0.2 × 107 | −44.5 ± 0.7 | −5.2 | −3.7 |
| Ca2+-SeMet-CT-CaM | 10.2°C | 0.9 ± 0.0 | 1.9 ± 1.0 × 107 | −21.5 ± 0.7 | 18.1 | |
| NtMKP1b peptide | ||||||
| apo-wt-CaM | 0 mM KCl | 1.0 ± 0.0 | 8.0 ± 0.3 × 105 | −42.3 ± 0.2 | −8.6 | |
| apo-wt-CaM | 100 mM KCl | 0.9 ± 0.2 | 1.9 ± 0.3 × 104 | −32.4 ± 7.5 | −8.0 | |
| apo-SeMet-CaM | 0 mM KCl | 1.0 ± 0.0 | 1.5 ± 0.1 × 106 | −29.7 ± 0.1 | 5.6 | |
| apo-Eth-CaM | 0 mM KCl | 0.9 ± 0.0 | 1.5 ± 0.1 × 106 | −43.6 ± 0.3 | −8.3 | |
| apo-Nle-CaM | 0 mM KCl | 1.1 ± 0.0 | 6.1 ± 0.8 × 105 | −12.0 ± 0.2 | 21.1 | |
| apo-CT-CaM† | 0 mM KCl | 1.1 ± 0.0 | 6.2 ± 0.8 × 105 | −15.7 ± 0.6 | 17.4 ± 0.9 | |
| apo-CT-CaM | 100 mM KCl | 0.8 ± 0.2 | 4.3 ± 1.9 × 104 | −7.0 ± 2.9 | 19.5 | |
| apo-SeMet-CT-CaM | 0 mM KCl | 0.9 ± 0.0 | 4.5 ± 0.3 × 105 | −14.9 ± 0.2 | 17.5 | |
All smMLCKp peptide experiments were performed in 100 mM KCl, and all NtMKP1b peptide experiments were performed at 25°C.
Value represents the average and SD of three independent measurements. All other error estimates are derived from curve-fitting uncertainties.
Titration curves for Ca2+-Eth-CaM were also exothermic but were not perfectly sigmoidal, and they were better fitted to models that assumed the presence of two or more binding events, each with similar affinity to the other CaM proteins (Ka values 107–108 M−1) but with different ΔH values (Fig. 6 B). Considering that Eth incorporation into CaM is random but incomplete (see Materials and Methods and Fig. 2), the ITC data suggest that there are detectable thermodynamic differences in smMLCKp peptide binding to some of the partially substituted Eth-CaM subpopulations. Since these thermodynamic differences could not be accurately resolved in the ITC data, only the ΔH for the initial binding event is reported for Ca2+-Eth-CaM (Table 3).
SmMLCKp peptide binding to each Ca2+-CaM protein was associated with large negative changes in ΔH and TΔS at 25°C, and both ΔH and TΔS become more positive at lower temperatures, which is typical for Ca2+-CaM-peptide interactions (35,36) (Table 3). Of interest, the thermodynamic parameters showed a distinct correlation with side-chain structure, with ΔH and TΔS differing by no more than 8 kJ/mol for proteins with the geometrically similar Met, SeMet, and Nle residues, but ΔH and TΔS were 10–20 kJ/mol smaller in magnitude at all temperatures for proteins with the longer Eth or shorter Leu residues. Despite these thermodynamic differences, a plot of ΔH versus TΔS for all of the smMLCKp experiments yields a straight line (R = 0.99) with a slope equal to unity (1.02 ± 0.03) (Fig. 6 E). This distinct enthalpy-entropy compensation is evidence that the mechanism of smMLCKp peptide binding is the same with each Ca2+-CaM protein at all temperatures. The ΔCp values were all large and negative (ΔCp = −3.4 to −3.9 kJ/mol K), consistent with a substantial burial of hydrophobic surface area in each complex. Since each domain of Ca2+-CaM contributes ∼−1.6 kJ/mol to the ΔCp of binding (35), these ΔCp values provide further evidence that each protein binds the peptide using both the N- and C-domains.
Thermodynamics of NtMKP1b peptide binding to Met-substituted apo-CaM proteins
We recently showed that the CaMBD of Nicotiana tabacum (tobacco) mitogen-activated protein kinase phosphatase (NtMKP1) binds to the C-domain of several CaM isoforms in the absence of Ca2+ in a manner resembling the complex of apo-wt-CaM with the CaMBD of the small conductance Ca2+-activated potassium channel (Fig. 1 C) (37,38). A peptide derived from this region (NtMKP1b) also bound to each Met-substituted apo-CaM protein in an exothermic, stoichiometric manner, similar to that previously reported with apo-wt-CaM (Fig. 6, C and D). The affinity for the NtMKP1b peptide was salt-dependent, with 100 mM KCl weakening the interaction with both apo-wt-CaM and apo-CT-CaM by more than 10-fold. Consequently, NtMKP1b peptide binding to the apo-CaM proteins was examined primarily in the absence of salt, where the c-value is in the optimal range for ITC (c-value: 14–45; Table 3).
Similarly to the ANS and smMLCKp peptide binding experiments, ITC experiments revealed very small effects of the Met substitutions on the overall affinity of apo-CaM for the NtMKP1b peptide. More specifically, the peptide bound with <2-fold lower affinity to the Nle- and Leu-substituted proteins, and <2-fold higher affinity to the SeMet- and Eth-substituted proteins in comparison to apo-wt-CaM (Table 3). However, there were substantial differences in ΔH and TΔS of NtMKP1b peptide binding, with both values being significantly more positive with the Nle- and Leu-substituted proteins, comparable for apo-wt-CaM and apo-Eth-CaM, and intermediate for apo-SeMet-CaM. The thermodynamics of NtMKP1b peptide binding to apo-CT-CaM and apo-SeMet-CT-CaM were almost identical, consistent with the peptide binding exclusively to the C-domain. As with the smMLCKp peptide, a ΔH versus TΔS plot yielded a linear compensatory relationship with R = 0.99, and a slope of 1.06 ± 0.03, suggesting that the mechanism of NtMKP1b peptide binding is the same for each apo-CaM protein (Fig. 6 E).
Discussion
Studies with several small globular proteins have shown that “conservative” Met substitutions to SeMet, Eth, Nle, Leu, Ile, or Val generally have very minor effects on protein structure, and only a small influence on protein stability that can be either stabilizing or destabilizing depending on the location and number of substituted residues (7,8,39–43). Stabilizing effects are typically attributed to the increased hydrophobicity of the SeMet, Eth, Nle, Leu, Ile, and Val side chains in comparison to Met, whereas destabilizing effects are predominantly attributed to the disruption of specific van der Waals packing interactions with neighboring side chains.
SeMet, Eth, Nle, and Leu substitutions also had a very small effect on the global structures of apo-CaM or Ca2+-CaM, as also shown in our previous work (17–21). However, the changes in stability with CaM were typically larger than those seen with other proteins, and nearly each substitution type increased the Tm of the N- and C-domains of the protein (Table 1). In general, SeMet and Eth had similar effects, increasing the Tm of each domain by 4–8°C, whereas the more hydrophobic Nle and Leu residues generally had larger effects, especially in the apo-C-domain, which had an increased Tm by as much as 26°C. With apo-CT-CaM, this increased stability was also accompanied by a loss of conformational exchange in the C-domain of the protein (Fig. 4).
The general stability enhancement of the Met-substituted CaM proteins suggests that the side chains of CaM can reorganize to largely compensate for geometric restraints and restricted degrees of freedom introduced by the different Met analogs. The ability to accommodate four additional methylene groups within the interior of both the N- and C-domains of apo-Eth-CaM is particularly noteworthy, since larger substitutions at buried sites are almost always destabilizing (44,45). X-ray crystal structures of Ca2+-Eth-CaM and Ca2+-CT-CaM show how Eth and Leu can be accommodated in the hydrophobic patches without influencing the main chain structure of the protein (46). For example, the α-carbon backbone root mean-square deviations between the structures of Ca2+-wt-CaM and Ca2+-CT-CaM or Ca2+-Eth-CaM are 0.324 Å and 0.314 Å, respectively (46). The structure of CaM has also been shown to be very tolerant of even “nonconservative” Met mutations such as Met→Gln or Met→Arg (20,47). Moreover, NMR spectroscopy experiments have demonstrated that the side chains of CaM are unusually dynamic in solution (48). We suggest that this structural plasticity distinguishes CaM from other, more rigid proteins that are only modestly stabilized or destabilized by similar Met substitutions. Although this plasticity is evident in both the N- and C-domains of apo- and Ca2+-CaM, we note that Nle had a much larger stabilizing effect on the apo-C-domain in comparison to the apo-N-domain, and we were unable to produce a quadruple M36/M51/M71/M72→Leu4 N-domain mutant of CaM (20). This suggests that the more rigid structure of the apo-N-domain is less accommodating of side chains such as Nle and Leu, which are less flexible than Met, SeMet, or Eth.
Exceptions to the general stabilizing trend of the Met substitutions were the lower Tm values for the mutated C-domain of Ca2+-CT-CaM (−8°C) as well as for the unmodified N-domain of apo-CT-CaM (−5°C). It is also noteworthy that the unmodified N-domain of Ca2+-CT-CaM was stabilized by 11°C in comparison to Ca2+-wt-CaM (Table 1). These stability changes are attributed to altered interactions between the folded and unfolded N- and C-domains of the mutant CaM protein during the unfolding process. Bayley and co-workers have shown that even though the folded N- and C-domains of wt-CaM behave independently in solution, they interact with each other during the unfolding process, and these interactions destabilize the less stable domain and stabilize the more stable domain with respect to the isolated domains of the protein (49,50). Since the denaturation pathway for apo-CT-CaM is reversed (N-domain unfolds before C-domain) in comparison to apo-wt-CaM (C-domain unfolds before N-domain), the N-domain of apo-CT-CaM is no longer stabilized by interactions with the unfolded C-domain, and consequently the Tm value for the apo-N-domain is reduced by ∼5°C. An increased strength of the interaction between the folded N-domain and unfolded C-domain at elevated temperatures could also explain the larger separation of Tm values for the two domains of Ca2+-CT-CaM in comparison to Ca2+-wt-CaM. These strengthened interactions could occur, for example, through binding of the N-domain hydrophobic patch to Leu residues in the unfolded C-domain of Ca2+-CT-CaM, since the wild-type Ca2+-N-domain shows specificity for Leu “anchor” residues in target proteins but rarely binds to Met residues (9,51). Although these interdomain interactions do not influence the order in which the N- and C-domains denature, the interactions likely have an effect on the magnitude of the observed Tm changes for the other apo- and Ca2+-CaM proteins as well.
ITC studies revealed that the Met substitutions also had a considerable impact on the ΔH and TΔS of ANS or peptide binding to the CaM proteins. The ΔH values represent the change in noncovalent bond energy for these interactions, including hydrogen bonds, van der Waals interactions, and salt bridges. These enthalpies are generally quite large (for example, 20 kJ/mol for each hydrogen bond), and the CaM-target complexes involve a multitude of these noncovalent interactions. Thus, when the binding surfaces of CaM are modified by Met substitution, large differences in the ΔH of binding are to be expected. In solution, however, many hydrogen bonds must be broken with solvent water before target binding. In addition, the entropic penalty associated with bond formation (increasing order in the system) is nearly equal to the favorable enthalpy of bonding at typical experimental temperatures (52). Consequently, even the relatively large variations in enthalpy that are observed for the different CaM-target interactions are roughly balanced by equal but opposite changes in entropy, resulting in very small differences in the free energy of binding ΔG—a phenomenon known as enthalpy-entropy compensation (52–54). Although the role of solvent water on biomolecular binding energetics remains poorly understood (55), the differences in ΔH and TΔS can provide some clues as to how the Met substitutions impact side-chain packing and dynamics.
In the case of ANS binding to CaM, there was a clear correlation between the exothermic signals, the fluorescence λmax blue shifts, and the fluorescence intensity values, each of which increased in the order of Eth < SeMet < Met < Nle < Leu (Table 2). Since larger blue shifts (and to some extent increased fluorescence intensity) typically indicate a reduction in fluorophore solvent exposure, and larger enthalpy is generally associated with increased noncovalent interactions, the data suggest that ANS binds more deeply into the hydrophobic patches of the Leu- and Nle-substituted proteins, and more on the surface of the SeMet- and Eth-substituted proteins in comparison to Ca2+-wt-CaM. Indeed, the shorter Leu residues widen the hydrophobic cleft between L109 and L145 (46), which is the primary ANS binding site in the C-domain of Ca2+-CaM (56). Nle is also somewhat shorter than Met due to smaller C–C–C bond lengths and bond angles in comparison to C–S–C, which would open the hydrophobic patches of both the N- and C-domains. On the other hand, the ∼3 Å longer Eth side chains narrow the hydrophobic patches of Ca2+-Eth-CaM (46), which would limit the depth of ANS binding. SeMet could have a similar, albeit smaller, narrowing effect due to the increased size of selenium in comparison to sulfur and the increased C–Se bond length in comparison to the C–S bond.
The effects of the Met substitutions on the ΔH and TΔS of peptide binding are summarized in Fig. 7. With the smMLCKp peptide, the changes in ΔH (ΔΔH) and TΔS (TΔΔS) were considerably larger for the Eth- or Leu-substituted proteins, suggesting that these structurally distinct side chains perturb the close and specific packing interactions with the peptide more than SeMet or Nle, whose side-chain structures are more similar to that of Met. The heterogeneous binding thermodynamics with Ca2+-Eth-CaM also indicate that incorporation of Eth has unique disruptive effects at the different Met positions (Fig. 6 B). Binding heterogeneity was not observed with Ca2+-CT-CaM due to the 100% incorporation of Leu in the C-domain of the mutant protein. The positive ΔΔH and TΔΔS values may reflect increased flexibility in the protein-peptide complex in the case of Ca2+-Eth-CaM (17). On the other hand, the more rigid Leu side chains should decrease flexibility in the hydrophobic patch of the peptide-free C-domain, resulting in less ordering of these side chains upon peptide binding in comparison to the Met side chains of Ca2+-wt-CaM. Of interest, the ΔΔH and TΔΔS values for smMLCKp peptide binding to Ca2+-SeMet-CaM were negative at all temperatures, possibly indicating that there are stronger London dispersion forces between the peptide and the very polarizable Se atoms of SeMet (Fig. 7).
Figure 7.
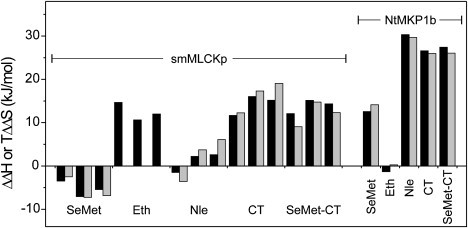
Effect of Met substitutions on the thermodynamics of peptide binding to CaM. ΔΔH (black bars) or TΔΔS (gray bars) values, in comparison to wt-CaM, are shown for Met-substituted CaM proteins binding to the smMLCKp or NtMKP1b peptides. For each protein binding to the smMLCKp peptide, values are shown for experiments performed at 25°C, 17.6°C, and 10.2°C (viewed left to right).
In contrast to the smMLCKp peptide, Eth did not significantly affect the thermodynamics of NtMKP1b peptide binding. This is consistent with the predicted structure of the complex, where the peptide binds on the surface of the closed apo-C-lobe with limited “lock and key” packing interactions with the Met or Eth side chains (Fig. 1 C) (37,38). With this peptide we found large positive ΔΔH and TΔΔS values for the Leu- and Nle-substituted proteins. We attributed this predominantly to less peptide-induced order in the C-domains of each protein, which are already significantly more stable than apo-wt-CaM in the absence of peptide (Table 1). Of importance, however, the ΔΔH and TΔΔS values for apo-SeMet-CaM were also large and positive, but the C-domain was only marginally stabilized by SeMet, suggesting that other factors, such as solvent reorganization, also make an important contribution to the thermodynamics of NtMKP1b peptide binding.
Overall, this study shows that the Met→SeMet, →Eth, →Nle, or →Leu substitutions generally enhance the stability of CaM, likely due to the increased hydrophobicity of each Met analog and a minimization of steric constraint due to the structural plasticity in the N- and C-domains. The Met substitutions also cause changes in side-chain packing and dynamics that are reflected in distinct ΔH and TΔS values for ANS or peptide binding to each protein, but the overall target affinity is largely unchanged due to enthalpy-entropy compensation. These results support the general view of CaM as an especially dynamic and structurally adaptable protein, and clearly show that noncoded and coded Met substitutions can have large effects on protein stability and the thermodynamics of ligand interactions. Our data with apo-CT-CaM show that the natural selection of Met rather than Leu results in dramatically lower stability and slow conformational exchange in the apo-C-domain, which could be functionally important because these properties result in the apo-C-domain being susceptible to oxidative regulation of protein turnover and target interactions (57,58). In addition, the enhanced stability of the apo-C-domain should impact its affinity for Ca2+, which would alter the range of Ca2+-stimuli to which the protein can respond in vivo (59). Considering that the Met substitutions do not appreciably perturb target binding affinity, the enhanced stabilities and potentially altered Ca2+ affinities of these Met-substituted CaMs could be utilized to engineer novel CaM-based “cameleon” Ca2+-sensors (60), or expand the stability range and longevity of CaM-affinity columns (61) for future research or industrial applications.
Acknowledgments
This research was supported by an operating grant from the Canadian Institutes for Health Research. The calorimetry equipment used was purchased through grants provided by the Canada Foundation for Innovation and the Alberta Science and Research Authority. H. J. Vogel holds a Scientist award from the Alberta Heritage Foundation for Medical Research.
Supporting Material
References
- 1.Budisa N. Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew. Chem. Int. Ed. Engl. 2004;43:6426–6463. doi: 10.1002/anie.200300646. [DOI] [PubMed] [Google Scholar]
- 2.Hendrickson T.L., Crecy-Lagard V., Schimmel P. Incorporation of nonnatural amino acids into proteins. Annu. Rev. Biochem. 2004;73:147–176. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 3.Link A.J., Mock M.L., Tirrell D.A. Non-canonical amino acids in protein engineering. Curr. Opin. Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson W.A., Horton J.R., LeMaster D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bann J.G., Pinkner J., Hultgren S.J., Frieden C. Real-time and equilibrium (19)F-NMR studies reveal the role of domain-domain interactions in the folding of the chaperone PapD. Proc. Natl. Acad. Sci. USA. 2002;99:709–714. doi: 10.1073/pnas.022649599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budisa N., Minks C., Medrano F.J., Lutz J., Huber R. Residue-specific bioincorporation of non-natural, biologically active amino acids into proteins as possible drug carriers: structure and stability of the per-thiaproline mutant of annexin V. Proc. Natl. Acad. Sci. USA. 1998;95:455–459. doi: 10.1073/pnas.95.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budisa N., Huber R., Golbik R., Minks C., Weyher E. Atomic mutations in annexin V–thermodynamic studies of isomorphous protein variants. Eur. J. Biochem. 1998;253:1–9. doi: 10.1046/j.1432-1327.1998.2530001.x. [DOI] [PubMed] [Google Scholar]
- 8.Ratnaparkhi G.S., Varadarajan R. Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics. Biochemistry. 2000;39:12365–12374. doi: 10.1021/bi000775k. [DOI] [PubMed] [Google Scholar]
- 9.Ishida H., Vogel H.J. Protein-peptide interaction studies demonstrate the versatility of calmodulin target protein binding. Protein Pept. Lett. 2006;13:455–465. doi: 10.2174/092986606776819600. [DOI] [PubMed] [Google Scholar]
- 10.Yamniuk A.P., Vogel H.J. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 11.Yuan T., Ouyang H., Vogel H.J. Surface exposure of the methionine side chains of calmodulin in solution. A nitroxide spin label and two-dimensional NMR study. J. Biol. Chem. 1999;274:8411–8420. doi: 10.1074/jbc.274.13.8411. [DOI] [PubMed] [Google Scholar]
- 12.Ikura M., Ames J.B. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc. Natl. Acad. Sci. USA. 2006;103:1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 14.Jurado L.A., Chockalingam P.S., Jarrett H.W. Apocalmodulin. Physiol. Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- 15.Ishida H., Borman M.A., Ostrander J., Vogel H.J., Macdonald J.A. Solution structure of the calponin homology (CH)-domain from the smoothelin-like 1 protein: a unique Apo-calmodulin binding mode and the possible role of the C-terminal type 2 CH-domain in smooth muscle relaxation. J. Biol. Chem. 2008;283:20569–20578. doi: 10.1074/jbc.M800627200. [DOI] [PubMed] [Google Scholar]
- 16.Edwards R.A., Walsh M.P., Sutherland C., Vogel H.J. Activation of calcineurin and smooth muscle myosin light chain kinase by Met-to-Leu mutants of calmodulin. Biochem. J. 1998;331:149–152. doi: 10.1042/bj3310149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weljie A.M., Vogel H.J. Tryptophan fluorescence of calmodulin binding domain peptides interacting with calmodulin containing unnatural methionine analogues. Protein Eng. 2000;13:59–66. doi: 10.1093/protein/13.1.59. [DOI] [PubMed] [Google Scholar]
- 18.Yuan T., Weljie A.M., Vogel H.J. Tryptophan fluorescence quenching by methionine and selenomethionine residues of calmodulin: orientation of peptide and protein binding. Biochemistry. 1998;37:3187–3195. doi: 10.1021/bi9716579. [DOI] [PubMed] [Google Scholar]
- 19.Yuan T., Vogel H.J. Substitution of the methionine residues of calmodulin with the unnatural amino acid analogs ethionine and norleucine: biochemical and spectroscopic studies. Protein Sci. 1999;8:113–121. doi: 10.1110/ps.8.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M., Li M., Wang J.H., Vogel H.J. The effect of Met→Leu mutations on calmodulin's ability to activate cyclic nucleotide phosphodiesterase. J. Biol. Chem. 1994;269:15546–15552. [PubMed] [Google Scholar]
- 21.Zhang M., Vogel H.J. Two-dimensional NMR studies of selenomethionyl calmodulin. J. Mol. Biol. 1994;239:545–554. doi: 10.1006/jmbi.1994.1393. [DOI] [PubMed] [Google Scholar]
- 22.Gellman S.H. On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry. 1991;30:6633–6636. doi: 10.1021/bi00241a001. [DOI] [PubMed] [Google Scholar]
- 23.Protasevich I., Ranjbar B., Lobachov V., Makarov A., Gilli R. Conformation and thermal denaturation of apocalmodulin: role of electrostatic mutations. Biochemistry. 1997;36:2017–2024. doi: 10.1021/bi962538g. [DOI] [PubMed] [Google Scholar]
- 24.Tsalkova T.N., Privalov P.L. Thermodynamic study of domain organization in troponin C and calmodulin. J. Mol. Biol. 1985;181:533–544. doi: 10.1016/0022-2836(85)90425-5. [DOI] [PubMed] [Google Scholar]
- 25.Tjandra N., Kuboniwa H., Ren H., Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur. J. Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson B.A., Blevins R.A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Lafitte D., Tsvetkov P.O., Devred F., Toci R., Barras F. Cation binding mode of fully oxidised calmodulin explained by the unfolding of the apostate. Biochim. Biophys. Acta. 2002;1600:105–110. doi: 10.1016/s1570-9639(02)00450-8. [DOI] [PubMed] [Google Scholar]
- 29.Tsvetkov P.O., Protasevich I.I., Gilli R., Lafitte D., Lobachov V.M. Apocalmodulin binds to the myosin light chain kinase calmodulin target site. J. Biol. Chem. 1999;274:18161–18164. doi: 10.1074/jbc.274.26.18161. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 31.Wang C., Palmer A.G. Solution NMR methods for quantitative identification of chemical exchange in 15N-labeled proteins. Magn. Reson. Chem. 2003;41:866–876. [Google Scholar]
- 32.Wiseman T., Williston S., Brandts J.F., Lin L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 33.Meador W.E., Means A.R., Quiocho F.A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 34.Wintrode P.L., Privalov P.L. Energetics of target peptide recognition by calmodulin: a calorimetric study. J. Mol. Biol. 1997;266:1050–1062. doi: 10.1006/jmbi.1996.0785. [DOI] [PubMed] [Google Scholar]
- 35.Brokx R.D., Lopez M.M., Vogel H.J., Makhatadze G.I. Energetics of target peptide binding by calmodulin reveals different modes of binding. J. Biol. Chem. 2001;276:14083–14091. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 36.Yamniuk A.P., Vogel H.J. Structural investigation into the differential target enzyme regulation displayed by plant calmodulin isoforms. Biochemistry. 2005;44:3101–3111. doi: 10.1021/bi047770y. [DOI] [PubMed] [Google Scholar]
- 37.Rainaldi M., Yamniuk A.P., Murase T., Vogel H.J. Calcium-dependent and -independent binding of soybean calmodulin isoforms to the calmodulin binding domain of tobacco MAPK phosphatase-1. J. Biol. Chem. 2007;282:6031–6042. doi: 10.1074/jbc.M608970200. [DOI] [PubMed] [Google Scholar]
- 38.Schumacher M.A., Crum M., Miller M.C. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12:849–860. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Budisa N., Steipe B., Demange P., Eckerskorn C., Kellermann J. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem. 1995;230:788–796. doi: 10.1111/j.1432-1033.1995.tb20622.x. [DOI] [PubMed] [Google Scholar]
- 40.Gassner N.C., Baase W.A., Hausrath A.C., Matthews B.W. Substitution with selenomethionine can enhance the stability of methionine-rich proteins. J. Mol. Biol. 1999;294:17–20. doi: 10.1006/jmbi.1999.3220. [DOI] [PubMed] [Google Scholar]
- 41.Holder J.B., Bennett A.F., Chen J., Spencer D.S., Byrne M.P. Energetics of side chain packing in staphylococcal nuclease assessed by exchange of valines, isoleucines, and leucines. Biochemistry. 2001;40:13998–14003. doi: 10.1021/bi011267t. [DOI] [PubMed] [Google Scholar]
- 42.Lipscomb L.A., Gassner N.C., Snow S.D., Eldridge A.M., Baase W.A. Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme. Protein Sci. 1998;7:765–773. doi: 10.1002/pro.5560070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohmura T., Ueda T., Hashimoto Y., Imoto T. Tolerance of point substitution of methionine for isoleucine in hen egg white lysozyme. Protein Eng. 2001;14:421–425. doi: 10.1093/protein/14.6.421. [DOI] [PubMed] [Google Scholar]
- 44.Karpusas M., Baase W.A., Matsumura M., Matthews B.W. Hydrophobic packing in T4 lysozyme probed by cavity-filling mutants. Proc. Natl. Acad. Sci. USA. 1989;86:8237–8241. doi: 10.1073/pnas.86.21.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandberg W.S., Terwilliger T.C. Influence of interior packing and hydrophobicity on the stability of a protein. Science. 1989;245:54–57. doi: 10.1126/science.2787053. [DOI] [PubMed] [Google Scholar]
- 46.Skene R.J. University of Calgary; Calgary, Alberta, Canada: 2001. Crystallographic Studies of Calcium-Binding Proteins: Aeromonas salmonicida Surface Array Protein and Calmodulin. [Google Scholar]
- 47.Chin D., Means A.R. Methionine to glutamine substitutions in the C-terminal domain of calmodulin impair the activation of three protein kinases. J. Biol. Chem. 1996;271:30465–30471. doi: 10.1074/jbc.271.48.30465. [DOI] [PubMed] [Google Scholar]
- 48.Lee A.L., Kinnear S.A., Wand A.J. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat. Struct. Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 49.Biekofsky R.R., Martin S.R., McCormick J.E., Masino L., Fefeu S. Thermal stability of calmodulin and mutants studied by (1)H-(15)N HSQC NMR measurements of selectively labeled [(15)N]Ile proteins. Biochemistry. 2002;41:6850–6859. doi: 10.1021/bi012187s. [DOI] [PubMed] [Google Scholar]
- 50.Masino L., Martin S.R., Bayley P.M. Ligand binding and thermodynamic stability of a multidomain protein, calmodulin. Protein Sci. 2000;9:1519–1529. doi: 10.1110/ps.9.8.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap K.L., Kim J., Truong K., Sherman M., Yuan T. Calmodulin target database. J. Struct. Funct. Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 52.Dunitz J.D. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem. Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 53.Cooper A., Johnson C.M., Lakey J.H., Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys. Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 54.Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 55.Whitesides G.M., Krishnamurthy V.M. Designing ligands to bind proteins. Q. Rev. Biophys. 2005;38:385–395. doi: 10.1017/S0033583506004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steiner R.F. Location of a binding site for 1-anilinonaphthalene-8-sulfonate on calmodulin. Arch. Biochem. Biophys. 1984;228:105–112. doi: 10.1016/0003-9861(84)90051-1. [DOI] [PubMed] [Google Scholar]
- 57.Ferrington D.A., Sun H., Murray K.K., Costa J., Williams T.D. Selective degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 2001;276:937–943. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 58.Tsvetkov P.O., Ezraty B., Mitchell J.K., Devred F., Peyrot V. Calorimetry and mass spectrometry study of oxidized calmodulin interaction with target and differential repair by methionine sulfoxide reductases. Biochimie. 2005;87:473–480. doi: 10.1016/j.biochi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 59.Gifford J.L., Walsh M.P., Vogel H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 60.Miyawaki A., Llopis J., Heim R., McCaffery J.M., Adams J.A. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 61.Berggard T., Arrigoni G., Olsson O., Fex M., Linse S. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J. Proteome Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.