Abstract
We demonstrate a method for simultaneous structure and function determination of integral membrane proteins. Electrical impedance spectroscopy shows that Staphylococcus aureus α-hemolysin channels in membranes tethered to gold have the same properties as those formed in free-standing bilayer lipid membranes. Neutron reflectometry provides high-resolution structural information on the interaction between the channel and the disordered membrane, validating predictions based on the channel's x-ray crystal structure. The robust nature of the membrane enabled the precise localization of the protein within 1.1 Å. The channel's extramembranous cap domain affects the lipid headgroup region and the alkyl chains in the outer membrane leaflet and significantly dehydrates the headgroups. The results suggest that this technique could be used to elucidate molecular details of the association of other proteins with membranes and may provide structural information on domain organization and stimuli-responsive reorganization for transmembrane proteins in membrane mimics.
Introduction
The primary goal of structural biology is to understand the structure-function relationship of proteins, which constitute the machinery of life. Despite the stunning achievements in determining protein structures using electron microscopy (1) and x-ray crystallography (2), the number of solved membrane protein structures significantly lags that of soluble proteins (3), in part because of the inherent difficulty of crystallizing proteins whose native environment is a disordered fluid membrane. Furthermore, these methods do not demonstrate whether the structural models are functionally accurate. NMR provides dynamic information about protein structure (4) but the technique is still limited to the study of relatively small proteins (i.e., with molecular mass <50 kDa) (5). Neutron reflectometry can provide complementary information to NMR and crystallography if a suitable interface for membrane protein reconstitution can be developed. The challenge lies in fabricating a biomimetic interface that retains the fluidity of a cell wall while providing long-term stability that is needed for structural characterization methods.
Tethered bilayer lipid membranes (tBLMs) combine the fluidity of a lipid bilayer with a stable platform for analysis with analytical techniques, including electrochemistry (6,7) and surface-sensitive scattering (8). Synthetic chemistry has recently been developed (8,9) that facilitates the formation of tBLMs in which the membranes are intrinsically disordered, thus permitting the reconstitution of proteins such as ion channels (10). Such membrane platforms are sufficiently resilient to be long-term stable (10,11) and can be characterized with neutron reflection at high resolution (8,12) by taking advantage of the possibility, offered by tBLM systems, to vary isotopic contrast and incorporate proteins in situ. This enables the acquisition of data sets before and after protein reconstitution, and under different isotopic labeling schemes, from one physical sample.
Although α-hemolysin (α-HL) spontaneously forms ion channels in free-standing bilayer lipid membranes (BLMs) (13), it is more difficult to reconstitute it into surface-bound membranes, presumably because of the rigidity of the membranes' inner leaflets (14). With a novel synthetic approach to tBLM synthesis, successful reconstitution of the membrane channels was recently demonstrated (10). In this work, we prepared a mixed self-assembled monolayer (SAM) of a thiahexa(ethylene oxide)-substituted lipid analog, 20-tetradecyloxy-3,6,9,12,15,18,22-heptaoxahexatricontane-1-thiol (WC14) (8) and β-mercaptoethanol (β-ME), then completed the bilayer with phospholipid via rapid solvent exchange (see Methods). Electrochemical impedance spectroscopy (EIS) and neutron reflection (NR) show that the resultant tBLMs are impermeable to ions and contain water in the submembrane space (8), which should facilitate protein reconstitution (7,15).
Materials and Methods
Materials
WC14 was synthesized, purified, and characterized in house, as reported previously in the Supplementary Material of McGillivray et al. (8). 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (DPhyPC) and pyridyl–dithiopropionate-poly(ethylene glycol)-distearoyl-sn-glycerophosphatidyl-ethanolamine (PDP-PEG2000-DSPE) were from Avanti Polar Lipids (Birmingham, AL). β-ME (Sigma-Aldrich, St. Louis, MO) was distilled before use. Poly(ethylene glycol) (PEG) and D2O (99.9% isotope purity) were obtained from Fluka (Ronkonkoma, NY) and Cambridge Isotopes Laboratory (Andover, MA), respectively. H2O was from a Millipore (Billerica, MA) UHQ reagent-grade water purification system.
Conductance measurements of α-HL in free-standing bilayer membranes
Freestanding BLMs (16) were formed from DPhyPC on ∼100 μm diameter holes in a thin Teflon partition separating two identical Teflon chambers (17) that each held ∼2 mL of electrolyte solution (0.1 M KCl, 5 mM 3-(N-morpholino) propanesulfonic acid (MOPS), 5 mM 2-(N-morpholino) ethanesulfonic acid (MES) at either pH 7.5 or 4.5). A small voltage (∼2 mV) was applied across the membrane via two matched Ag/AgCl electrodes, and the ionic current was converted to voltage and amplified with an Axon Instruments 200B patch clamp amplifier (Molecular Devices, Sunnyvale, CA) in the voltage clamp mode. α-HL channels were formed by adding the toxin to one chamber while briefly stirring the solution.
Surface preparation
The chemicals used are described in the Supporting Material. SAMs were formed on thin gold layers (∼100 Å for neutron reflectivity NR, ∼2000 Å for EIS), deposited by direct current magnetron sputtering (Auto A306; BOC Edwards, Crawley, West Sussex, UK) on [100]-cut Si wafers precoated with an ∼20 Å thick Cr adhesion layer. The gold films typically had an RMS surface roughness of ∼5 Å, as measured by x-ray reflectometry (Bruker AXS, Madison, WI), and a uniformity of thickness across the surface of ± 3% or better, as determined by ellipsometry.
We demonstrated in recent work that the dilution of tether points in tBLMs, for example with β-ME, is essential for the formation of fluid bilayers in tBLMs that incorporate a ∼2 nm thick hydrated cushion between the membrane and the solid support (8). Mixed SAMs were prepared by exposing magnetron-sputtered Au films to solutions of WC14: β-ME (3:7 mol/mol, 0.2 mM total concentration) in 99.5% ethanol for >12 h. Because of the short hexa(ethylene oxide) tether, these SAMs and completed tBLMs incorporate hydrated submembrane layers that are only 15 Å thick (8,18,19). The tBLMs were subsequently completed by incubation of the moderately hydrophobic surface with a concentrated (∼10 mM) DPhyPC solution that was rapidly, within ∼5 s, replaced by vigorous injection of aqueous buffer (0.1 M NaCl, 0.005 M NaH2PO4/Na2HPO4, pH 7.5) into the sample cell. This rapid solvent exchange procedure leads to the formation of complete and electrically insulating bilayers (8). The tBLMs used here had residual specific conductances <3 μS cm−2. Because of this low background conductance, the tBLMs were well suited to detect toxin-induced membrane conductance changes. tBLMs that incorporated longer tethers, providing a thicker hydrated submembrane layer, were prepared by chemisorption of SAMs from PDP-PEG2000-DSPE without β-ME, followed by solvent exchange. Solutions with different pH values used in EIS were of the same buffer composition. However, the solvent exchange was always carried out at pH 7.5.
Electrochemical impedance spectroscopy
EIS measurements were performed using an electrochemical impedance system (Solartron Analytical, Farnborough, Hampshire, UK, model 1287 potentiostat, model 1252A frequency response analyzer and software). The spectra were obtained for frequencies between 1 and 6.5 × 104 Hz with 10 logarithmically distributed measurements per decade. Six electrochemical cells were assembled on a 20 × 40 mm large gold-coated silicon wafer used as the working electrode that had the tBLMs attached to its surface. Each of these six parallel cells had an effective real surface area of 0.44 cm2. The reference was a saturated silver-silver chloride (Ag/AgCl/NaCl(aq,sat)) microelectrode (M-401F, Microelectrodes, Inc., Bedford, NH), and the auxiliary electrode was a 0.5 mm diameter platinum wire (Aldrich, St. Louis, MO, 99.99% purity) coiled around the barrel of the reference electrode. All measurements were carried out at 0 V direct current bias versus reference electrode at T = (21 ± 1)°C.
Neutron reflectivity measurements and data modeling
NR was measured on the Advanced Neutron Diffractometer/Reflectometer (12) at the National Institute of Standards and Technology Center for Neutron Research. tBLMs were sufficiently robust to permit the injection of aqueous buffers of various isotopic compositions into the sample chamber and sequentially collect NR spectra that each take ∼6 h. After the bare tBLM was characterized at different solvent contrasts, ∼1 μM α-HL (final concentration) was injected into the sample cell in D2O-based buffer and equilibrated with the membrane for 6 h. Finally, the protein-reconstituted tBLM was examined in protein-free buffers of various isotopic compositions. The total experiment time exceeded 36 h, during which the tBLM was stable.
Data analysis was performed in terms of homogeneous layer models, or “box” models (20), using the ga_refl software developed at the NIST Center for Neutron Research (21). The reflectivity due to a model neutron scattering length density (nSLD) profile was computed using an optical matrix based on Parratt's recursion algorithm (22). Isostructural samples with distinct isotopic contrast were fitted simultaneously by consistently refining the corresponding nSLD profiles that were parameterized in terms of the underlying molecular structures (23,24). The robustness of the tBLMs permitted the characterization of the membrane at various solvent contrasts followed by protein reconstitution and further NR characterization of the reconstituted tBLM with the same physical sample without removal from the reflectometer. With the substrate and membrane structures thus established, the structural changes due to protein incorporation could be studied in full detail. Although the data at the highest momentum transfer (Qz > 0.2 Å−1) do not appear to contribute information about distinctive structural features of the tBLM with and without the protein (see Fig. 3 below and the Supporting Material), these data were included in the error-weighted fitting because they contribute information about the overall sample structure.
Figure 3.
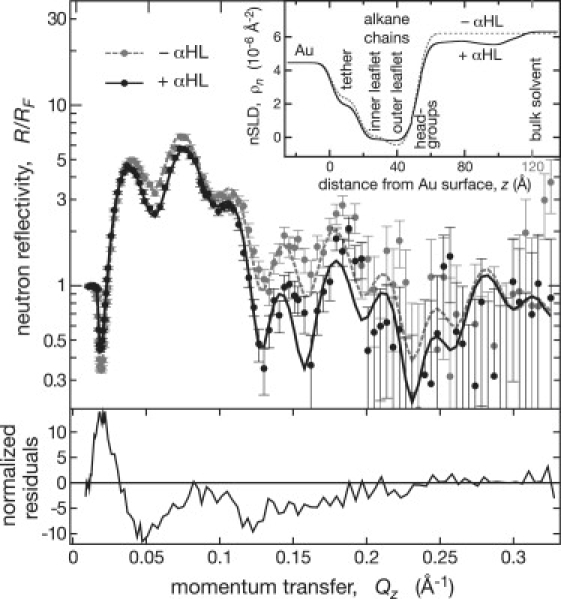
NR of protein-free and protein-reconstituted tBLMs. Before exposure to α-HL, the tBLM was characterized at two solvent contrasts (for data and modeled nSLD profiles, see Figs. S3 and S4, respectively, in the Supporting Material). Nine-hundred nanomoles of α-HL were subsequently injected into the sample cell in D2O-based buffer and equilibrated with the membrane for 6 h. The protein-reconstituted tBLM was finally examined in protein-free buffers of various isotopic compositions. (Main panel) Fresnel-normalized NR of the tBLM in D2O-based buffer before and after α-HL reconstitution. (Bottom panel) Because all data sets derive from the same physical sample, an error-weighted residuals plot shows exactly the protein contributions to the reflectivity. (Inset) nSLD profiles derived for the data in the main panel from a simultaneous fit to five data sets. Only the outer 30 Å of the inorganic surface structure of the solid substrate are shown. The nSLD profiles in the inset yield the reflectivity models shown as continuous lines in the main panel.
For a refined data analysis, the crystal structure of the α-HL channel (Protein Data Bank accession code: 7AHL) (25) was incorporated into the model by assuming that the barrel of the protein was aligned perpendicular to the membrane (z direction) and the crystal structure was undisturbed. The contribution of the protein to the nSLD profile along z was then divided into 0.5 Å thick slices in which the sums of the atomic scattering lengths, divided by the solvent-excluded volumes, were calculated. The solvent-excluded volume was determined by the method of Connolly (26), using a 1.4 Å radius probe to represent a water molecule. In the fitting to the data, the resultant protein nSLD contribution was allowed to shift freely along z relative to the bilayer membrane. The lateral protein density at/in the membrane was accounted for as a fitting parameter. Protein contributions in the aqueous volume phase outside the membrane slabs were integrated into six layers. Measurements were typically made in three solvent contrasts (D2O, H2O, and a D2O/H2O mixture with an nSLD of 4 × 10−6 Å−2, “CM4”). All solvent contrasts were simultaneously fitted for the tBLM with and without α-HL (21).
Results and Discussion
Electrochemical investigation of biomimetic interface and of protein functionality
The addition of α-HL to the aqueous phase caused significant changes in the magnitude of the tBLM impedance, |Z|, and the phase angle (Fig. 1). These EIS features are associated with ion flux induced by membrane ionophores (6). Quantitative information was obtained by modeling the impedance spectra with the equivalent circuit shown in Fig. 1 A (for details of the fitting, see the Supporting Material). The membrane is characterized by a constant phase element (CPEtBLM) (27), with α = 0.986 ± 0.001 (n = 22 samples), which suggests a nearly ideal capacitive behavior of the electrically insulating dielectric bilayer (14). The CPEtBLM coefficient, (0.62 ± 0.01) μF × cm−2 s(α-1) (n = 22 samples), agrees with the specific capacitance values for solvent-free BLMs (28). Upon incubation with α-HL, CPEtBLM increases in a protein concentration- and time-dependent manner, which suggests a net increase in the bilayer dielectric constant upon insertion of water-filled ion channels into the membrane (29). In addition, the reconstitution of ion channels creates parallel conductance pathways that bypass the membrane's high resistance. These pathways are represented by another constant phase element, CPEpores, in series with a resistor, Rpores. The nearly ohmic open channel conductance determines Rpores, whereas CPEpores is determined by a complex combination of the conductance of the aqueous reservoir between the Au surface and the inner membrane and the differential capacitance of the Au/reservoir interface.
Figure 1.
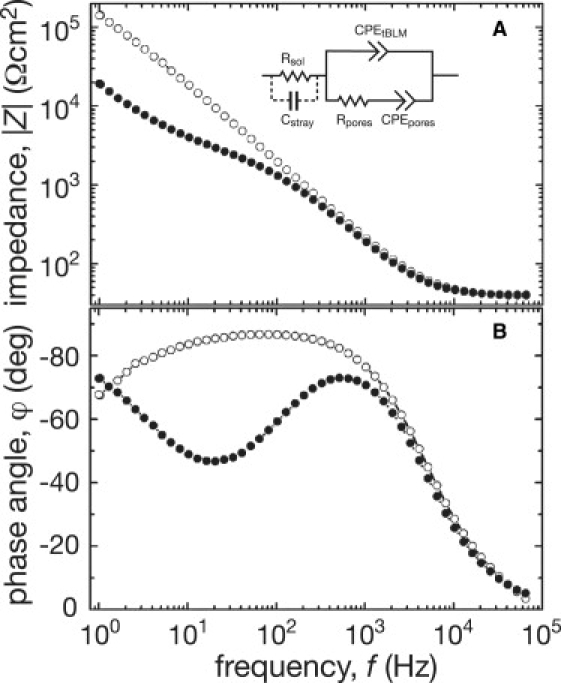
EIS spectra of a tBLM before and after α-HL reconstitution. The Bode plots (A) and phase angles (B) were measured with tBLMs in 0.1 M NaCl, 5 mM phosphate buffer at pH 7.5 before (open circles) and 2 h after the addition of α-HL to a final concentration of 50 nM (solid circles). The temperature was (21 ± 1)°C. The data were normalized with respect to the membrane surface area. (Inset) Equivalent model circuit for the tBLM.
Several criteria were used to demonstrate that the α-HL channels in the tBLMs were equivalent to those in free-standing BLMs. First, the toxin increased the tBLM conductance in proportion to both the bulk electrolyte conductivity and the ion (i.e., the conductance is greater in a KCl than in an NaCl solution), as it does in BLMs (13) (data not shown). Second, the rates at which α-HL increased the conductance of tBLMs correlated with those of free-standing BLMs at the same pH values (Fig. 2 A). Third, the conductance increased superlinearly with the bulk α-HL concentration in both membrane types (data not shown). In general, the specific conductance increases induced by α-HL in tBLMs were within an order of magnitude (usually within a factor of 2 to 7) of those in free-standing BLMs.
Figure 2.
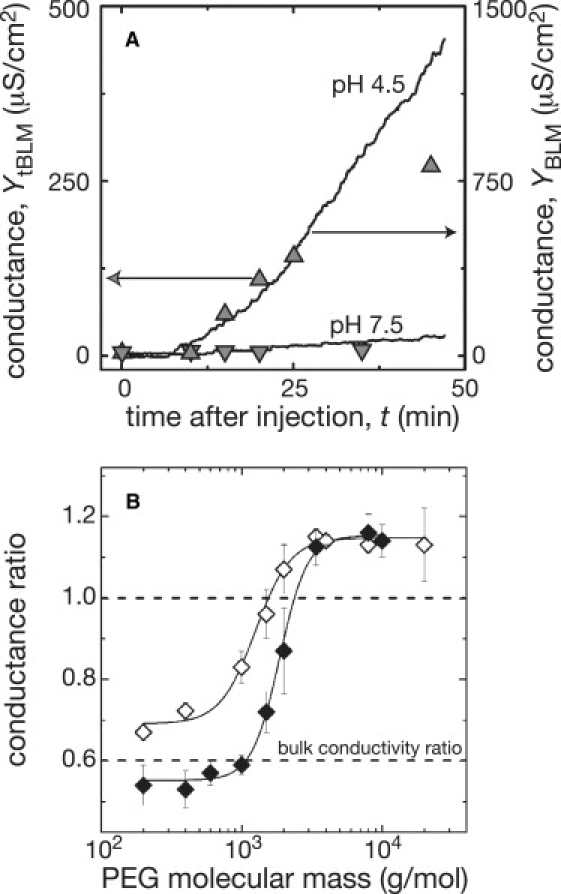
(A) Kinetics of α-HL channel formation in free-standing and tethered bilayer lipid membranes. The rate at which α-HL increased the specific conductance of tBLMs was greater at pH 4.5 (up triangles) than at pH 7.5 (downward triangles). Similar results were obtained with free-standing BLMs (solid lines). The applied potential was 10 mV alternating current (tBLM) and 2 mV direct current (BLM). In the BLM experiments, α-HL was added to the solution bathing one side of the membrane. In both experiments, the bulk concentration of α-HL was 50 nM. For the EIS data, the model-derived conductance, Yα-HL = Rpores−1, was normalized with respect to the surface area. (B) Effect of size-selected PEGs on α-HL channel conductance. Polymers with molecular mass ≤2000 g/mol decreased the conductance of tBLMs containing many α-HL channels (open diamonds) and of single α-HL channels in a free-standing BLM (solid diamonds) (30). The concentration of PEG in each experiment was 15% (w:w). The curves are drawn to guide the eye. The bottom dashed line indicates the ratio of the bulk conductivities, σ(+PEG)/σ(−PEG).
The diameter of the α-HL channel in free-standing BLMs was estimated previously from the effect of PEG on the channel conductance (30). The conductance decreases when the PEGs are small enough to partition into the pore. For α-HL, this limit is ∼2250 g/mol. Fig. 2 B shows that the effect of different molecular mass PEGs on the conductance of α-HL-doped tBLMs (open symbols) is similar to that on single α-HL channels in free-standing BLMs (solid symbols). The difference between the two partitioning plots might be due to restricted diffusion of PEG in the region adjacent to the Au electrode on the solid surface. Nevertheless, the results suggest that the values of the conductance, Yα-HL = Rpores−1, derived from the EIS model are directly related to the intrinsic conductance of α-HL.
The correspondence between the results in BLMs and tBLMs demonstrates that α-HL channels are functional in both systems. Thus, other techniques such as NR can be used to determine the structural characteristics of the α-HL-membrane complex, taking advantage of the tBLM system's high stability.
Neutron reflectometry of the biomimetic interface
The sample preparation led to a robust film that was stable for days and survived solvent exchanges for isomorphic contrast adjustments (12). The multiple solvent contrasts allow for unambiguous determination of model parameters. For details of the technique, see Majkrzak et al. (31) and references therein. Fig. 3 shows NR results for a tBLM with and without α-HL in D2O (for full data sets, see the Supporting Material). Changes in the scattering after exposure to α-HL are shown as error-weighted residuals of reflectivity data against the best-fit reflectivity model of the protein-free tBLM. Deviations from the baseline are most pronounced at low momentum transfer (e.g., around Qz = 0.05 Å−1, where the coordinate z is normal to the membrane plane and originates at the gold/thiol interface). This indicates significant structural changes of the bilayer upon α-HL reconstitution on the length scale, 2π/Qz ≈ 120 Å, the approximate height of the α-HL channel along its symmetry axis (25). Although less pronounced than those around Qz = 0.05 Å−1, significant deviations persist to 0.2 Å−1. However, more information can be extracted from these data sets by using more detailed fitting of the data, with additional details gleaned from the use of multiple solvent contrasts.
Simultaneously fitting the data in three solvent contrasts to a slab model with a single extramembranous layer accounts for the most prominent feature of the channel: its large cap domain exterior to the membrane (25). Structural details were derived from a composition-space model (23,24) of the membrane-inserted α-HL array. This modeling procedure assumes the protein inserts into the membrane according to its known x-ray structure (25) and suggests that the nanopore spans the tBLM with its β-barrel stem end flush with the lipid headgroup region of the inner bilayer leaflet (Fig. 4, tabulated details can be found in the Supporting Material). Although this is not obvious from the nSLD profile (Fig. 4 A), the optimized parameter set determines the penetration depth to within ± 1.1 Å, as shown in Fig. 5 A by a rigorous evaluation of parameter confidence limits with a Monte Carlo resampling technique (Supporting Material). The α-HL stem is thus located ∼15 Å away from the Au interface. Control experiments with α-HL in a tBLM prepared with a longer tether (i.e., 60 Å) indicated that the proximity of the Au surface to the WC14-based tBLM does not alter the association of α-HL with the membrane (Supporting Material).
Figure 4.
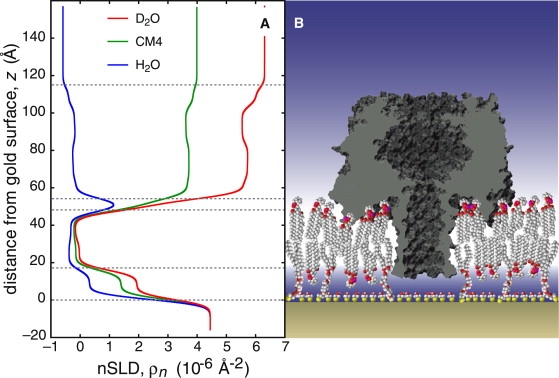
Supramolecular model of the tBLM reconstituted with α-HL. Simultaneous fitting of a molecular model to five NR data sets (two for the tBLM and three for the protein-reconstituted tBLM) leads to five nSLD profiles that are all consistent with one supramolecular structure. (A) The three profiles for the tBLM reconstituted with α-HL. These profiles contain the calculated contribution of the α-HL x-ray crystal structure (21) at a lateral density and insertion depth derived from the model fit. (B) A scaled cartoon of the system.
Figure 5.
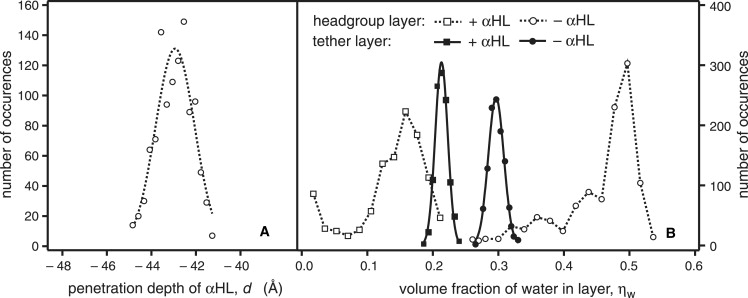
Parameter estimation and confidence limits for the supramolecular structure of α-HL reconstituted into a tBLM, determined with NR and quantified by Monte Carlo resampling (Supporting Material). Five individual data sets were linked in a composition-space modeling approach (23) that took advantage of the known α-HL crystal structure (21). Histograms of parameters describing the resampled data indicate that the protein can be localized along the surface normal with a precision of ± 1.1 Å within the fluid bilayer (A). (B) Both the lipid headgroups of the outer membrane leaflet (open symbols; dashed lines) and the content of the submembrane layer (solid symbols), including the membrane anchor and lipid headgroups of the inner leaflet, are significantly dehydrated upon reconstitution of α-HL. Parameter distributions for the submembrane layer hydration and protein insertion depth were approximately Gaussian (fitted lines). In contrast, the parameter distributions for the hydration of the outer lipid headgroup layer show extended tails and were evaluated by their mean values and standard deviations (for the full parameter set, see Table S3 in the Supporting Material).
A quantitative evaluation of the nSLD profile shows that the α-HL channel reconstitutes into the tBLM at ∼33% of the maximum hexagonal packing density, as set by the dimensions of the cap domain (25). The incorporation of such a large amount of protein without bilayer collapse is another indicator of the tBLM's robustness.
In H2O, a region of high nSLD around z = 52 Å (Fig. 4 A) provides the location of the outer membrane headgroup layer. Incorporation of α-HL leads to a significant nSLD increase in this region, indicating a strong dehydration of the headgroups. The loss of hydration expected from the compression of the lipid bilayer by the α-HL stem would be on the order of 3%, based on the observed lateral density of the cap domain near the membrane and the protein geometry. NR shows that the α-HL channel removes most of the water from the lipid headgroups (Fig. 5 B). This suggests that the protein cap interacts strongly with lipid headgroups and significantly perturbs their conformations, consistent with predictions based on the crystal structure (25). The overhang between the membrane-penetrating stem segment and the cap domain is lined with hydrophilic residues that may form a suitable environment for zwitterionic phospholipid headgroups. Moreover, the edge of the cap contains hydrophobic residues that may interact with the acyl chains of the lipid bilayer. These results should aid Poisson-Nernst-Planck modeling of these systems because dehydration of the cleft between the cap domain and membrane will influence the dielectric properties of the channel-membrane system and could affect the channel conductance (32).
Conclusions
Although crystallography remains the technique of choice for determining protein structures with full atomic detail, a great deal of complementary information can be elucidated from disordered structures with other techniques, such as neutron reflectometry. Inspired by earlier work (7,18), we developed what to our knowledge is a new methodology for structural measurements on transmembrane proteins, verification of their functionality, and probing of their interactions with disordered membranes. The α-HL protein ion channel was chosen as a model system for proof-of-concept because its structure is known to high resolution (25). The technology we describe here will permit a broad range of biomedical investigations where the interaction of proteins with membranes is of immediate interest, such as studies into toxicology (33,34), Alzheimer's disease (35), cell signaling involving lipids (36), or laminopathies (37).
Although the data interpretation described here utilized a known crystal structure, the techniques developed in this work can be readily extended to the use of other structural motifs as a basis. For instance, NR-derived structures of proteins with established functionality in a tBLM could be used in conjunction with computer models to discriminate between disparate solutions in the modeling. We anticipate that this technology will complement existing methods for both structure and functional measurements of membrane proteins.
Acknowledgments
Support by the National Institute of Standards and Technology (U.S. Department of Commerce) in providing the neutron research facilities used in this work is gratefully acknowledged. We thank Dr. Hirsh Nanda for helpful discussions, Rima Budvytyte for assistance with EIS experiments, and Dr. Paul A. Kienzle for support in the NR data analysis.
This work was supported by the National Science Foundation (CBET-0555201 and 0457148), the National Institutes of Health (1 RO1 RR14182 and 1 P01 AG032131-01), the American Health Assistance Foundation (A2008-307), the Lithuanian State Science and Studies Foundation (T-31/07), a National Institute of Standards and Technology-NRC Research Associateship to J.W.F.R., the National Institute of Standards and Technology Single Molecule Manipulation and Measurement Program, and the National Institute of Standards and Technology Office of Law Enforcement Standards.
Certain commercial materials, instruments, and equipment are identified in this manuscript to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials, instruments, or equipment identified is necessarily the best available for the purpose.
Footnotes
Duncan J. McGillivray and Gintaras Valincius contributed equally to this work.
Duncan J. McGillivray's present address is Dept. of Chemistry, The University of Auckland, Auckland, New Zealand.
Wilma Febo-Ayala's present address is Dept. of Chemistry, University of Pennsylvania, Philadelphia, PA.
Supporting Material
References
- 1.Henderson R., Unwin P.N.T. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–31. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 2.Deisenhofer J., Epp O., Miki K., Huber R., Michel H. Structure of the protein subunits in the photosynthetic reaction center of Rhodopseudomonas viridis at 3 Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 3.White S.H. The progress of membrane protein structure determination. Protein Sci. 2004;13:1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis B.A., Harbison G.S., Herzfeld J., Griffin R.G. NMR structural analysis of a membrane protein – Bacteriorhodopsin peptide backbone orientation and motion. Biochemistry. 1985;24:4671–4679. doi: 10.1021/bi00338a029. [DOI] [PubMed] [Google Scholar]
- 5.Liang B., Tamm L.K. Structure of outer membrane protein G by solution NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raguse B., Braach-Maksvytis V.L.B., Cornell B.A., King L.B., Osman P.D.J. Tethered lipid bilayer membranes: Formation and ionic reservoir characterization. Langmuir. 1998;14:648–659. [Google Scholar]
- 7.Tanaka M., Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 8.McGillivray D.J., Valincius G., Vanderah D.J., Febo-Ayala W., Woodward J.T. Molecular-scale structural and functional characterization of sparsely tethered bilayer lipid membranes. Biointerphases. 2007;2:21–33. doi: 10.1116/1.2709308. [DOI] [PubMed] [Google Scholar]
- 9.Schiller S.M., Naumann R., Lovejoy K., Kunz H., Knoll W. Archaea analogue thiolipids for tethered bilayer lipid membranes on ultrasmooth gold surfaces. Angew. Chem. Int. Ed. Engl. 2003;42:208–211. doi: 10.1002/anie.200390080. [DOI] [PubMed] [Google Scholar]
- 10.Vockenroth I.K., Atanasova P.P., Jenkins A.T.A., Köper I. Incorporation of α-hemolysin in different tethered bilayer lipid membrane architectures. Langmuir. 2008;24:496–502. doi: 10.1021/la7030279. [DOI] [PubMed] [Google Scholar]
- 11.Vockenroth I.K., Ohm C., Robertson J.W.F., McGillivray D.J., Lösche M. Stable insulating tethered bilayer membranes. Biointerphases. 2008;3:FA68–FA73. doi: 10.1116/1.2912097. [DOI] [PubMed] [Google Scholar]
- 12.Dura J.A., Pierce D., Majkrzak C.F., Maliszewskyj N., McGillivray D.J. AND/R: a neutron diffractometer/reflectometer for investigation of thin films and multilayers for the life sciences. Rev. Sci. Instrum. 2006;77:074301. doi: 10.1063/1.2219744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menestrina G. Ionic channels formed by Staphylococcus aureus α-toxin. Voltage-dependent inhibition by divalent and trivalent cations. J. Membr. Biol. 1986;90:177–190. doi: 10.1007/BF01869935. [DOI] [PubMed] [Google Scholar]
- 14.Glazier S.A., Vanderah D.J., Plant A.L., Bayley H., Valincius G. Reconstitution of the pore-forming toxin a-hemolysin in phospholipid/18-octadecyl-1-thiahexa(ethylene oxide) and phospholipid/n-octadecanethiol supported bilayer membranes. Langmuir. 2000;16:10428–10435. [Google Scholar]
- 15.Sackmann E. Supported membranes - scientific and practical applications. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 16.Montal M., Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bezrukov S.M., Kasianowicz J.J. Current noise reveals protonation kinetics and number of ionizable sites in an open protein ion channel. Phys. Rev. Lett. 1993;70:2352–2355. doi: 10.1103/PhysRevLett.70.2352. [DOI] [PubMed] [Google Scholar]
- 18.Cornell B.A., Braach-Maksvytis V.L.B., King L.B., Osman P.D.J., Raguse B. A biosensor that uses ion-channel switches. Nature. 1997;387:580–583. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 19.Valincius G., McGillivray D.J., Febo-Ayala W., Vanderah D.J., Kasianowicz J.J. Enzyme activity to augment the characterization of tethered bilayer membranes. J. Phys. Chem. B. 2006;110:10213–10216. doi: 10.1021/jp0616516. [DOI] [PubMed] [Google Scholar]
- 20.Als-Nielsen J., Kjaer K. X-ray reflectivity and diffraction studies of liquid surfaces and surfactant monolayers. In: Riste T., Sherrington D., editors. Phase Transitions in Soft Condensed Matter. Plenum Press; New York: 1989. pp. 113–138. [Google Scholar]
- 21.Kienzle, P. A., M. Doucet, D. J. McGillivray, K. V. O'Donovan, N. F. Berk, et al. 2000–2009. ga_refl. http://www.ncnr.nist.gov/reflpak/garefl.html.
- 22.Parratt L.G. Surface studies of solids by total reflection of x-rays. Phys. Rev. 1954;95:359–369. [Google Scholar]
- 23.Vaknin D., Kjaer K., Als-Nielsen J., Lösche M. Structural properties of phosphatidylcholine in a monolayer at the air/water interface. Neutron reflection study and reexamination of x-ray reflection experiments. Biophys. J. 1991;59:1325–1332. doi: 10.1016/S0006-3495(91)82347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiener M.C., White S.H. Fluid bilayer structure determination by the combined use of x-ray and neutron diffraction. II. “Composition-space” refinement method. Biophys. J. 1991;59:174–185. doi: 10.1016/S0006-3495(91)82209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L., Hobaugh M.R., Shustak C., Cheley S., Bayley H. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 26.Connolly M.L. Analytical molecular-surface calculation. Appl. Crystallogr. 1983;16:548–558. [Google Scholar]
- 27.Raistrick I.D., Franceschetti D.R., Macdonald J.R. Theory. In: Barsoukov E., Macdonald J.R., editors. Impedance Spectroscopy: Theory, Experiment, and Applications. 2nd ed. Wiley; New York: 2005. pp. 27–117. [Google Scholar]
- 28.Redwood W.R., Pfeiffer F.R., Weisbach J.A., Thompson T.E. Physical properties of bilayer membranes formed from a synthetic saturated phospholipid in decane. Biochim. Biophys. Acta. 1971;233:1–6. doi: 10.1016/0005-2736(71)90351-8. [DOI] [PubMed] [Google Scholar]
- 29.Dilger J.P., McLaughlin S.G.A., McIntosh T.J., Simon S.A. Dielectric constant of phospholipid bilayers and the permeability of membranes to ions. Science. 1979;206:1196–1198. doi: 10.1126/science.228394. [DOI] [PubMed] [Google Scholar]
- 30.Bezrukov S.M., Vodyanoy I., Brutyan R.A., Kasianowicz J.J. Dynamics and free energy of polymers partitioning into a nanoscale pore. Macromolecules. 1996;29:8517–8522. [Google Scholar]
- 31.Majkrzak C.F., Berk N.F., Krueger S., Dura J.A., Tarek M. First-principles determination of hybrid bilayer membrane structure by phase-sensitive neutron reflectometry. Biophys. J. 2000;79:3330–3340. doi: 10.1016/S0006-3495(00)76564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coalson R.D., Kurnikova M.G. Poisson-Nernst-Planck theory approach to the calculation of current through biological ion channels. IEEE Trans. Nanobioscience. 2005;4:81–93. doi: 10.1109/tnb.2004.842495. [DOI] [PubMed] [Google Scholar]
- 33.Halverson K., Panchal R.G., Nguyen T.L., Gussio R., Little S.F. Anthrax biosensor, protective antigen ion channel asymmetric blockade. J. Biol. Chem. 2005;280:30056–30062. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 34.Kent M.S., Yim H., Murton J.K., Satija S., Majewski J. Oligomerization of membrane-bound diphtheria toxin (CRM197) facilitates a transition to the open form and deep insertion. Biophys. J. 2008;94:2115–2127. doi: 10.1529/biophysj.107.113498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valincius G., Heinrich F., Budvytyte R., Vanderah D.J., Sokolov Y. Soluble amyloid oligomers affect dielectric membrane properties by bilayer insertion and domain formation: Implications for cell toxicity. Biophys. J. 2008;95:4845–4861. doi: 10.1529/biophysj.108.130997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wymann M.P., Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 37.Liu B., Zhou Z. Lamin A/C, laminopathies and premature ageing. Histol. Histopathol. 2008;23:747–763. doi: 10.14670/HH-23.747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


