Abstract
A growing number of proteins have been shown to adopt knotted folds. Yet the biological roles and biophysical properties of these knots remain poorly understood. We used protein engineering and atomic force microscopy to explore the single-molecule mechanics of the figure-eight knot in the chromophore-binding domain of the red/far-red photoreceptor, phytochrome. Under load, apo phytochrome unfolds at forces of ∼47 pN, whereas phytochrome carrying its covalently bound tetrapyrrole chromophore unfolds at ∼73 pN. These forces are not unusual in mechanical protein unfolding, and thus the presence of the knot does not automatically indicate a superstable protein. Our experiments reveal a stable intermediate along the mechanical unfolding pathway, reflecting the sequential unfolding of two distinct subdomains in phytochrome, potentially the GAF and PAS domains. For the first time (to the best of our knowledge), our experiments allow a direct determination of knot size under load. In the unfolded chain, the tightened knot is reduced to 17 amino acids, resulting in apparent shortening of the polypeptide chain by 6.2 nm. Steered molecular-dynamics simulations corroborate this number. Finally, we find that covalent phytochrome dimers created for these experiments retain characteristic photoreversibility, unexpectedly arguing against a dramatic rearrangement of the native GAF dimer interface upon photoconversion.
Introduction
For decades, it was assumed that proteins do not adopt knotted folds, which is to say that pulling on the C-terminus and N-terminus of a protein would yield an uncomplicated linear polypeptide. However, recent structural biology studies (1–3), including the rapid accumulation of three-dimensional coordinates via structural genomics efforts and new computational tools to mine the Protein Data Bank for previously unrecognized knots (4–6), have upended this assumption and made clear that some proteins are indeed knotted in their native state. There are now approximately a dozen independent protein folds with recognized knots, not including shallow knots in which only a few residues pass under a loop (7,8).
Phytochrome is an evolutionarily conserved, photoreversible red/far-red light photoreceptor with a covalently attached linear tetrapyrrole chromophore. Plant phytochromes mediate germination, flowering, shade-avoidance, and senescence, among other light responses, by trafficking from the cytoplasm to the nucleus upon red (∼660-nm) light irradiation (9). Cyanobacterial, eubacterial, and fungal phytochromes were more recently described (10,11). Phytochromes' roles in microbes are less well-understood than in plants. Deinococcus radiodurans provided the first phytochrome (DrBphP) for which an x-ray crystallographic structure was determined (1). This structure of the red (r) light-absorbing Pr form of the chromophore-binding domain (CBD) of DrBphP comprises an N-terminal extension harboring the covalent attachment site (Cys-24) for the biliverdin IXα (BV) chromophore, a PAS domain, and a GAF domain that surrounds the largely buried tetrapyrrole. The native protein contains two additional domains not represented in the CBD, a PHY domain that stabilizes the far-red (fr) light-absorbing Pfr form of phytochromes, and a histidine kinase domain proposed to mediate phosphorylation events triggered by red-light absorption (9). The DrBphP CBD (DrCBD) structure revealed an unanticipated knotted topology for which there was no previous evidence (1). An ∼29-residue insertion between secondary-structure elements of a canonical GAF domain forms a lasso for the N-terminal 34 amino acids to create a figure-eight polypeptide knot (Fig. 1, A and B) (7,12)
Figure 1.

Design and characterization of a DrCBD polymer. (A) Cysteines (yellow spheres) were engineered within DrCBD (one monomer displayed based on Protein Data Bank code 2o9c, with loops smoothed for clarity, colored by reverse rainbow from blue N-terminus to red C-terminus, with BV as gray sticks) at L314 in the GAF domain dimer interface and at E18 near a symmetry contact in the P21212 crystal form (Protein Data Bank code 1ztu). (B) DrCBD figure-eight knot has four crossovers (numbered sequentially in this simplification; orientation as in A). (C) Depiction of four DrCBD-E18C/L314C subunits in our designed disulfide-bridged polymer (color scheme and orientation of right-hand monomer as in A and B). (D) DrCBD-L314C/E18C oligomerizes in a redox-dependent manner. Lane 1, holoprotein; lane 2, holoprotein + dithiothreitol (DTT); lane 3, apoprotein; lane 4, apoprotein + DTT; lane 5, molecular weight markers; lane 6, pseudocrystallized CBD polymers. The mass of the CBD monomer is 37 kDa; the strong dimer band represents the natural GAF domain interaction, represented by the two colored subunits in C.
We investigated the mechanical characteristics of the knot in phytochrome. The forced unfolding of proteins by atomic force microscopy (AFM) yields insights into forces required to unfold a protein, maps of folding subdomains within complicated protein structures, and the free-energy landscapes of folding (13–16). AFM is well-suited to the study of mechanical unfolding of knotted proteins, because it provides a way to control the open ends of a protein knot (17,18). Experimentally, researchers have used single-molecule techniques such as optical traps to construct and manipulate knots for polymeric structures such as DNA helices or actin filaments (19,20). Wang and Ikai (21) and Wang et al. (22) reported on mechanical unfolding experiments with carbonic anhydrase, containing a shallow knot in its structure. However, those authors found that the mechanical stability of this knotted protein was too high to unfold the native state in AFM experiments (21,22). We describe what to our knowledge is the first successful example of single-molecule unfolding of an intrinsically knotted protein. Applying single-molecule force spectroscopy to the phytochrome figure-eight knot, we determined the unfolding force of DrCBD and the size of the tightly pulled knot in the peptide chain, and compared the results with molecular-dynamics (MD) simulations.
Materials and Methods
Creating variant phytochromes
Disulfide bridge locations were engineered, based on a visual inspection of crystal packing (23) within two published forms of DrCBD (Protein Data Bank codes 1ztu and 2o9c) and verification using Disulfide by Design (24).
Beginning with a pET-based overexpression vector (Novagen, Gibbstown, NJ) encoding the 321 amino-acid DrBphP CBD with an N-terminal T7-tag and C-terminal hexahistidine tag (1), rolling-circle mutagenesis was performed in several steps, to achieve the plasmid encoding CBD-E18C/L314C. First, CBD-L314C was created using forward primer 5′- CCG CTT GCT GAG CTG TCA AGT TCA GGT CAA GG-3′ and reverse primer 5′- CCT TGA CCT GAA CTT GAC AGC TCA GCA AGC GG-3′. The same strategy was used to create this change in the pET vector that encodes the full-length DrBphP (1). Because the site-directed mutation (E18C) was unsuccessful several times, we created an intermediate vector with several silent mutations to remove a predicted 8-basepair hairpin surrounding the E18 site (forward primer, 5′-GGT GGC CCG GAA ATT ACG ACG GAG AAC TGC GAG CGC-3′; reverse primer, 5′-GCG CTC GCA GTT CTC CGT CGT AAT TTC CGG GCC ACC-3′), and then created the desired final gene within this framework (forward primer, 5′- GCG TTT ACC TTG GTG GCC CGT GTA TTA CGA CGG AGA ACT GC-3′; reverse primer, 5′-GCA GTT CTC CGT CGT AAT ACA CGG GCC ACC AAG GTAAAG CG-3′). All sequences were confirmed by automated DNA sequencing (University of Wisconsin Biotechnology Center, Madison, WI).
The expression and purification of full-length and CBD, wild-type sequence, and variant phytochromes were performed under green safe lights via Nickel affinity and hydrophobic interaction chromatography, as previously described (12), with 1 mM Tris(2-carboxyethyl) phosphine added during the in vitro chromophore ligation step. This small change led to a significantly higher fraction of phytochrome containing covalently attached chromophore. For force measurements, protein buffer was exchanged by dialysis to 30 mM Tris, pH 8.0, and 50 mM NaCl without added reductant, and was allowed to oxidize for several weeks at 4°C. In some cases, protein-crystallization droplets were set up and, although crystals were never observed, this more completely polymerized protein (Fig. 1 B) was used for AFM measurements.
Ultraviolet/visible spectroscopy
Spectroscopy measurements were performed on a PerkinElmer Life Sciences (Waltham, MA) Lambda 650 spectrophotometer (PerkinElmer Life Sciences. The Pr spectra were measured on proteins that had never been illuminated, whereas Pfr spectra were measured after saturating irradiation with 690-nm light from a 10-nm half-bandwidth interference filter (Andover Corp., Salem, NH).
Force measurements
Single-molecule unfolding experiments were performed on a custom-built atomic force microscope at room temperature. For all measurements, we used gold-coated cantilevers (Type B Bio-Lever, Olympus, Tokyo, Japan) with spring constants of 6 pN/nm. Before commencing measurements, ∼20 μL of protein solution (0.5 mg/mL in phosphate-buffered saline) were placed on a clean glass surface and incubated for 5 min. For all force-extension curves, the lever was retracted at a constant pulling speed of 1 μm/s. Force-extension traces were recorded at 20,000 points/s.
Contour-length calculation
The expected contour-length gain upon mechanically unfolding proteins can be calculated as:
| (1) |
where daa is the contour length of a single amino acid, i and j are the anchoring amino-acid positions (18 and 314 in this case), and di,j is the distance between the anchoring points i and j in the folded structure (25). For a polypeptide persistence length of p = 0.5 nm, daa= 0.365 ± 0.002 nm was determined to very high precision (14,15,25,26), and di,j can be measured in the x-ray crystal structure as d18,314 = 3.9 nm.
Molecular dynamics
We performed standard MD simulations using the software AMBER9.0 with the ff03 force field (27) at fixed pressure (1 bar) and temperature (300 K), involving ∼8000 atoms in a periodically repeated and anisotropic simulation box with Ewald electrostatics. Polypeptides were generated using the AMBER tleap tool. Figure-eight knots were tied into them, using interactive MD in visual MD (28). Thereafter, the system was equilibrated for ∼5 ns with Langevin dynamics, solvated with TIP3P water and neutralizing Na+ ions, and further equilibrated by a ∼5-ns MD simulation. For peptide pulling, we used the AMBER-steered MD tool with a constant velocity of 0.01 nm/ns to drive the first and last atoms of the backbone in opposite directions. Knot lengths and their errors were estimated from the calculated force-extension F(l) curves and their fluctuations, respectively. The average force F was found to increase monotonically with extension l. At the experimentally relevant force of ∼70 pN, we measured an amino-acid contour length of 0.365 ± 0.001, in accordance with published experiments.
Results
Design and characterization of polymeric phytochromes
Successful protein unfolding by AFM has relied on natural polymers or on engineered protein interfaces to create covalent homopolymeric proteins. This allows the AFM unfolding event for each domain to be observed multiple times within a single unfolding experiment. We designed phytochrome polymers by substituting cysteine for two side chains that closely approach their symmetry mates in the crystal lattices of DrCBD (1,12,23,29). The first nonnative cysteine was introduced at residue 314 (Fig. 1, A and B), and was predicted to form a disulfide bond across the substantial CBD dimer interface (Fig. 1 C). The second bridge was engineered at the solvent-accessible residue 18 (Fig. 1, A and B), which is in close proximity to its counterpart in a neighboring subunit in one crystal lattice. These nonnative polymers are expected to consist of alternating head-to-head and tail-to-tail disulfide bridges (Fig. 1 C). The double-cysteine-substituted variant is a soluble protein that forms covalent dimers and higher-order multimers when oxidized (Fig. 1 D).
The existence of a strong CBD dimer mediated by GAF helices involves a relatively new understanding of phytochrome dimerization (12), which was long suspected to occur predominantly via the histidine kinase domain (12,30–32). We thus also created the L314C variant in the full-length DrBphP containing PHY and histidine kinase domains. DrBphP L314C became a tool to study the large-scale conformational changes that might occur during photoconversion. This protein readily forms covalent dimers in the absence of a reducing agent (Fig. 2), and these covalent dimers include the wild-type ground-state Pr spectrum. Moreover, covalent dimers undergo a spectrally normal photocycle in both directions (Fig. 2). This result implies that the orientation of phytochrome's CBD dimer interface remains relatively constant during photoconversion of the full-length protein, both for the red light-driven forward reaction from Pr → Pfr and the dark reversion from Pfr → Pr.
Figure 2.

Covalently dimerized full-length DrBphP L314C undergoes normal photoconversion upon illumination with 690-nm light (gray dashed line, DrBphP; black solid line, DrBphP L314C). (Inset) SDS-PAGE gel of illuminated proteins. Lane 1, DrBphP + DTT; lane 2, DrBphP; lane 3, DrBphP L314C + DTT; lane 4, DrBphP L314C (d and m, 84-kDa monomer and corresponding dimer, respectively). Preilluminated and dark-reverted proteins have same gel profile (not shown).
Determination of knot size and DrCBD mechanical stability
Force-extension curves of the CBD polymer with bound BV were measured. In some cases, multiple unfolding peaks were visible, indicating that the polymer contained several CBD units (Fig. 3 A). Worm-like chain (WLC) fits were used to measure the length gain of the polypeptide chain upon unfolding to nanometer-scale precision (15,26,33). To achieve reproducible results, we held the persistence length p = 0.5 nm constant, and only fit the contour length (). The distribution of 47 measured contour-length increases had an average value of 98.1 ± 1.0 nm (Fig. 3 B). This value is significantly shorter than the expected contour-length gain for the transition from a fully folded to a fully extended state of DrCBD. The expected contour-length gain can be calculated using formula 1 from above (25). The predicted value is = 104.1 ± 0.6 nm for the total unfolding of CBD, a value larger by 6.2 nm than our experimentally measured one. Hence the stretched polypeptide of DrBphP CBD appears shortened by 17 ± 3 amino acids. A straightforward explanation for this observation is that the tightly pulled knot contains 17 amino acids that make the stretched chain appear shorter than an unknotted chain.
Figure 3.

AFM unfolding of DrBphP CBD. (A) Force-extension trace (red) of a single CBD polyprotein with bound chromophore. Theoretical fits are according to WLC model (33) (blue). (B) Histogram of contour-length increases caused by total unfolding of CBD units with bound chromophore. Vertical black line at 104 nm represents calculated contour-length increase for an unknotted CBD. (C) Force-extension trace of a single apo-DrCBD polyprotein. (D) Histogram of contour-length increases for total unfolding of the apo-form.
Because the folding pathway in vivo for the knotted phytochrome is completely unexplored, it is a formal possibility that the knotting of the protein requires the previous attachment of the tetrapyrrole chromophore. To explore this issue, we performed the same measurements on apo-CBD polymers that had been expressed and purified without the addition of BV (Fig. 3, C and D). The measured contour-length increase was 97.5 ± 1.8 nm, identical within error to the holoprotein case.
Although the contour length assigned to the knot is the same for apoprotein versus holoprotein, there is a striking and significant difference in the unfolding forces measured in the two experiments, i.e., ∼73 pN versus ∼47 pN, respectively, for holo-CBD and apo-CBD (cf. Fig. 3 A versus 3 C or Fig. 4 C versus 4 G). The higher forces needed in the presence of chromophore clearly show that BV, covalently attached at residue 24, is buried within the protein structure until the protein reaches the major unfolding peak. Based on this observation, we can exclude the possibility that amino acids after Cys-18 have unfolded prematurely at low forces before the experimental measurements, because this would have led to BV being pulled out of the binding pocket. This scenario would have increased the distance between residues 18 and 314 within the monomer, and thus would also lead to an apparent shortening of the unfolded polypeptide, which we attribute to the tightened knot.
Figure 4.

Unfolding pathway of DrBphP CBD. (A) Zoom into single holo-CBD unfolding event (framed in Fig. 3A). Rising force connects to a stable intermediate state, with WLC fit shown as green line. (B) Histogram of contour-length increases from totally folded to intermediate state . (C) Histogram of forces needed to unfold first part of structure (A, red circles). Average unfolding forces are = 73.3 ± 6.6 pN. (D) Histogram of forces needed to unfold intermediate state to fully extended protein (A, gray circles). The average unfolding forces are = 32.8 ± 4 pN. (E) Zoom into a single apo-CBD unfolding event (framed in Fig. 3C). as fit by WLC is shown as green line. (F) Histogram of . (G) Histogram of forces needed to unfold first part of structure (E, red circles). Average unfolding force is = 47.1 ± 3.2 pN. (H) Histogram of forces needed to unfold intermediate state (E, gray circles), with average value of = 27.7 ± 2.8 pN.
Unfolding pathway of phytochrome
Between the major unfolding peaks of both the holo and apo forms of DrCBD, the force-extension traces persistently exhibit a further unfolding event at ∼30 pN (Fig. 3, A and C). These events indicate an intermediate on the mechanical unfolding pathway. The fact that this intermediate is more apparent in apo than in holo traces can be readily explained by the higher unfolding forces of the major peaks for holo-CBD that mask the subsequent low force peaks. We used WLC fits to measure the length of the polypeptide chain unraveled in going from the native to the intermediate state, (Fig. 4, A and E). We obtained an average contour-length increase of = 68.0 ± 2.2 nm for CBD with bound BV (Fig. 4 B), and = 65.3 ± 2.6 nm for the apoprotein (Fig. 4 F). These results are identical within error, and lead to an estimate that ∼180 amino acids unfold in going from the native to the intermediate state. The 186-amino-acid GAF-domain is hence the most likely candidate for the domain that unravels first, leaving the remaining 90-amino-acid PAS domain to form the unfolding intermediate. This interpretation is corroborated by the sensitivity of the unfolding forces of the major unfolding peak to the presence of chromophore (Fig. 4, C and G), whereas the unfolding force distribution for the intermediate peak that marks subsequent unfolding of the PAS domain is independent of chromophore (Fig. 4, D and H). Because the tetrapyrrole contacts the GAF domain exclusively in the folded protein, the major unfolding peak must be associated with this domain. Moreover, the PAS domain in the bacterial phytochrome shows only few electrostatic and hydrophobic interactions with the GAF domain (12). It is therefore very likely that the subdomains unfold sequentially, with the GAF domain the first to unfold. The observation that unfolding intermediates follow domain boundaries is reminiscent of the concept of mechanical unfoldons in maltose-binding protein, in which quasistable intermediates along the mechanical unfolding pathway of a full-length protein were linked by the sequential, independent unfolding of structural building blocks (so-called unfoldons) (14).
Validation by molecular-dynamics calculations
To corroborate our experimental result of 17 ± 3 amino-acid residues comprising a taut figure-eight knot, we performed steered MD simulations of short 41-knotted polypeptide chains with atomistic resolution of solutes and water. Because the final experimental knot location is unknown, we randomly selected two (30 amino acids [aa] long) stretches of phytochrome, i.e., I), EPIHIPGSIQ PHGALLTADG HSGEVLQMSL (aa 41–70), and II), KFAPDATGEV IAEARREGLH AFLGHRFPAS (aa 191–220). Force-extension curves were calculated during peptide pulling, and the contour-length differences between the knotted peptide and the unknotted analog were evaluated at relevant pulling forces. At a force of ∼70 pN, we find 7.11 ± 0.4 nm and 7.25 ± 0.4 nm equating to 19 ± 1 aa and 20 ± 1 aa involved in the tight figure-eight knot of peptides I and II, respectively (Fig. 5). This result is within the error of our experimental measurements. The lack of sequence specificity suggests that packing effects of the backbone dominate the tightly pulled knot structure, an idea to be investigated further elsewhere. Interestingly, we observed only a weak force dependence of knot-tightening for pulling forces larger than ∼70 pN. The increase to a strong force of ∼1 nN causes knot lengths to shrink only one amino-acid residue further for both peptides, indicating extremely tight peptide packing and steric hindrance at experimentally and physiologically relevant unfolding forces. These tightly pulled knots are very long-lived; they do not dissolve (unknot) in a ∼150-ns free MD simulation.
Figure 5.
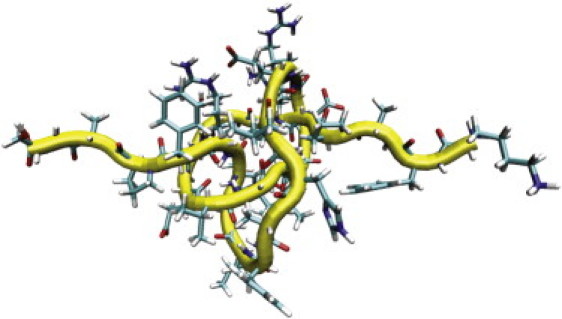
MD snapshot of tightly pulled peptide II (DrBphP residues 191–220).
The sequence independence of the structure of the tightly pulled knot is not equivalent to a lack of sequence requirement for forming the knot initially. Recent computational work suggests that threading in knotted proteins may be an early event in folding, and may potentially depend on interactions that are not part of the final folded structure (34).
Discussion
Very little is known about the mechanical properties of protein knots, the evolutionary advantages they confer (if any), or the mechanisms by which they fold. It was suggested that protein stability and/or function are influenced in a beneficial way by the presence of knots (35,36).
The overall mechanical stability of apo-DrCBD (∼47 pN) is similar to or lower than that of other proteins whose unfolding was studied by single-molecule methods (ranging from ∼20–50 pN for maltose-binding protein, through ∼130–300 for titin Ig domains, to ∼100–600 pN for green fluorescent binding protein) (13,14,26,37). Thus, the mechanical stability of the folded protein alone has not driven the evolution of a knotted phytochrome.
The functional role of the DrBphP CBD knot has not been determined. However, a PAS-less and therefore unknotted phytochrome from a thermophilic cyanobacterial species was characterized (38), and its red and far-red photoreversibility is comparable to that of longer and more well-characterized plant and bacterial proteins. Similarly, there exists a widespread class of phytochrome-like proteins that is missing the 29-residue lasso loop (11), and thus cannot contain the same knot. These are chromophore-bound, blue-green photoreversible photoreceptors (39,40). Thus, photoreversibility is also not the sole role of the knot.
We favor another functional role for the knot in phytochrome, i.e., it limits the motions the protein domains undergo relative to one another when light energy is absorbed by the chromophore and used to do the work of conformational change on the protein (1). Free vibrations would be lost as heat, and unrestricted motions would be inefficient for signal transduction via the multiple domains of phytochrome and its interacting cellular partners. The constraints imposed by the knot between the PAS and GAF domains may limit motion along the proper trajectory for the exposure of appropriate side chains of the GAF domain, e.g., to interact productively with the PHY domain and signaling partners downstream of the phytochrome itself. The construction of permuted phytochromes lacking the knot should be possible, and would constitute one way to test this hypothesis.
With respect to the folding mechanisms of knotted proteins, it is estimated, using a random distribution, that more proteins should be knotted than are observed, and it remains largely unclear why nature steers some proteins to form a complicated knotted structure, while discouraging most (41,42). Moreover, how the energy landscape of folding must be tuned to allow the formation of such a knot in finite time remains a puzzle (34,43). Mallam et al. (43) and Mallam and Jackson (44) rigorously showed that the thermodynamic and kinetic unfolding and refolding pathways of the knotted, dimeric methyl transferase YibK are governed by the same parameters known for unknotted proteins. The rate-limiting steps in YibK folding are proline isomerization and dimerization, comparable to the rate-limiting steps in well-studied unknotted proteins. The fate of the knot itself during folding and unfolding is difficult to address in these studies. One likely possibility is that formation of the knot is an early and nonrate-limiting step in protein folding (42). Our report paves the way for single-molecule unfolding studies of other classes of knots in other knotted proteins, and moreover sets the stage for refolding experiments (45), in which it should be possible to answer the unresolved question of whether the knot forms as a first step in protein folding or as a later step. As the number of known protein knots continues to grow, this work becomes generally applicable, and can reveal fundamental properties about the statistical mechanical behavior of polypeptides.
Acknowledgments
Computing time on the HLRBII was provided by Leibniz Rechenzentrum Munich. Dr. J. R. Wagner created the DrCBD L314C-encoding pET plasmid. K.T.F. acknowledges the Max Planck Institut für Plasma Physik for hosting her as a visiting scientist during the time this project was initiated.
This work was supported by the National Science Foundation (grant MCB-0424062 to K.T.F. and Richard Vierstra) and the Deutsche Forschungsgemeinschaft (grant RI 99013-1 to M.R., with support from the Emmy-Noether-Program to J.D.).
Contributor Information
Matthias Rief, Email: mrief@ph.tum.de.
Katrina T. Forest, Email: forest@bact.wisc.edu.
References
- 1.Wagner J.R., Brunzelle J.S., Forest K.T., Vierstra R.D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 2.Misaghi S., Galardy P.J., Meester W.J., Ovaa H., Ploegh H.L. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 3.Shi D., Gallegos R., DePonte J., 3rd, Morizono H., Yu X. Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 Å resolution. J. Mol. Biol. 2002;320:899–908. doi: 10.1016/S0022-2836(02)00539-9. [DOI] [PubMed] [Google Scholar]
- 4.Khatib F., Weirauch M.T., Rohl C.A. Rapid knot detection and application to protein structure prediction. Bioinformatics. 2006;22:e252–e259. doi: 10.1093/bioinformatics/btl236. [DOI] [PubMed] [Google Scholar]
- 5.Lai Y.L., Yen S.C., Yu S.H., Hwang J.K. pKNOT: the protein KNOT web server. Nucleic Acids Res. 2007;35:W420–W424. doi: 10.1093/nar/gkm304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolesov G., Virnau P., Kardar M., Mirny L.A. Protein knot server: detection of knots in protein structures. Nucleic Acids Res. 2007;35:W425–W428. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virnau P., Mirny L.A., Kardar M. Intricate knots in proteins: function and evolution. PLoS Comput Biol. 2006;2:e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeates T.O., Norcross T.S., King N.P. Knotted and topologically complex proteins as models for studying folding and stability. Curr. Opin. Chem. Biol. 2007;11:595–603. doi: 10.1016/j.cbpa.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockwell N.C., Su Y.S., Lagarias J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes J., Lamparter T., Mittmann F., Hartmann E., Gartner W. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 11.Karniol B., Wagner J.R., Walker J.M., Vierstra R.D. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner J.R., Zhang J., Brunzelle J.S., Vierstra R.D., Forest K.T. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 13.Oberhauser A.F., Carrion-Vazquez M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertz M., Rief M. Mechanical unfoldons as building blocks of maltose-binding protein. J. Mol. Biol. 2008;378:447–458. doi: 10.1016/j.jmb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 15.Mickler M., Dima R.I., Dietz H., Hyeon C., Thirumalai D. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proc. Natl. Acad. Sci. USA. 2007;104:20268–20273. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman J.R., Clarke J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr. Opin. Struct. Biol. 2007;17:58–66. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Taylor W.R., Lin K. Protein knots: a tangled problem. Nature. 2003;421:25. doi: 10.1038/421025a. [DOI] [PubMed] [Google Scholar]
- 18.Sulkowska J.I., Sulkowski P., Szymczak P., Cieplak M. Tightening of knots in proteins. Phys. Rev. Lett. 2008;100:058106. doi: 10.1103/PhysRevLett.100.058106. [DOI] [PubMed] [Google Scholar]
- 19.Bao X.R., Lee H.J., Quake S.R. Behavior of complex knots in single DNA molecules. Phys. Rev. Lett. 2003;91:265506. doi: 10.1103/PhysRevLett.91.265506. [DOI] [PubMed] [Google Scholar]
- 20.Arai Y., Yasuda R., Akashi K., Harada Y., Miyata H. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 21.Wang T., Ikai A. Protein stretching III: force-extension curves of tethered bovine carbonic anhydrase b to the silicon substrate under native, intermediate and denaturing conditions. Jpn. J. Appl. Phys. 1999;38:3912–3917. [Google Scholar]
- 22.Wang T., Arakawa H., Ikai A. Reversible stretching of a monomeric unit in a dimeric bovine carbonic anhydrase B with the atomic force microscope. Ultramicroscopy. 2002;91:253–259. doi: 10.1016/s0304-3991(02)00106-7. [DOI] [PubMed] [Google Scholar]
- 23.Yang G., Cecconi C., Baase W.A., Vetter I.R., Breyer W.A. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. USA. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dombkowski A.A. Disulfide by design: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics. 2003;19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 25.Dietz H., Rief M. Protein structure by mechanical triangulation. Proc. Natl. Acad. Sci. USA. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietz H., Berkemeier F., Bertz M., Rief M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case D.A. University of California; San Francisco: 2006. AMBER9.0. [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Dietz H., Bertz M., Schlierf M., Berkemeier F., Bornschlogl T. Cysteine engineering of polyproteins for single-molecule force spectroscopy. Nat. Protocols. 2006;1:80–84. doi: 10.1038/nprot.2006.12. [DOI] [PubMed] [Google Scholar]
- 30.Noack S., Michael N., Rosen R., Lamparter T. Protein conformational changes of Agrobacterium phytochrome Agp1 during chromophore assembly and photoconversion. Biochemistry. 2007;46:4164–4176. doi: 10.1021/bi602419x. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto K.T., Tokutomi S. Formation of aggregates of tryptic fragments derived from the carboxyl-terminal half of pea phytochrome and localization of the site of contact between the fragments by amino-terminal amino acid sequence analysis. Photochem. Photobiol. 1989;50:113–120. doi: 10.1111/j.1751-1097.1989.tb04136.x. [DOI] [PubMed] [Google Scholar]
- 32.Jones A.M., Erickson H.P. Domain structure of phytochrome from Avena sativa visualized by electron microscopy. Photochem. Photobiol. 1989;49:479–483. doi: 10.1111/j.1751-1097.1989.tb09198.x. [DOI] [PubMed] [Google Scholar]
- 33.Bustamante C., Marko J.F., Siggia E.D., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 34.Wallin S., Zeldovich K.B., Shakhnovich E.I. The folding mechanics of a knotted protein. J. Mol. Biol. 2007;368:884–893. doi: 10.1016/j.jmb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lua R.C., Grosberg A.Y. Statistics of knots, geometry of conformations, and evolution of proteins. PLoS Comput Biol. 2006;2:e45. doi: 10.1371/journal.pcbi.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor W.R. Protein knots and fold complexity: some new twists. Comput. Biol. Chem. 2007;31:151–162. doi: 10.1016/j.compbiolchem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Linke W.A., Grutzner A. Pulling single molecules of titin by AFM–recent advances and physiological implications. Pflugers Arch. 2008;456:101–115. doi: 10.1007/s00424-007-0389-x. [DOI] [PubMed] [Google Scholar]
- 38.Ulijasz A.T., Cornilescu G., von Stetten D., Kaminski S., Mroginski M.A. Characterization of two thermostable cyanobacterial phytochromes reveals global movements in the chromophore-binding domain during photoconversion. J. Biol. Chem. 2008;283:21251–21266. doi: 10.1074/jbc.M801592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshihara S., Katayama M., Geng X., Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 40.Rockwell N.C., Njuguna S.L., Roberts L., Castillo E., Parson V.L. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry. 2008;47:7304–7316. doi: 10.1021/bi800088t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virnau P., Kantor Y., Kardar M. Knots in globule and coil phases of a model polyethylene. J. Am. Chem. Soc. 2005;127:15102–15106. doi: 10.1021/ja052438a. [DOI] [PubMed] [Google Scholar]
- 42.Mallam A.L., Onuoha S.C., Grossmann J.G., Jackson S.E. Knotted fusion proteins reveal unexpected possibilities in protein folding. Mol. Cell. 2008;30:642–648. doi: 10.1016/j.molcel.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Mallam A.L., Jackson S.E. Probing nature's knots: the folding pathway of a knotted homodimeric protein. J. Mol. Biol. 2006;359:1420–1436. doi: 10.1016/j.jmb.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 44.Mallam A.L., Jackson S.E. Folding studies on a knotted protein. J. Mol. Biol. 2005;346:1409–1421. doi: 10.1016/j.jmb.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 45.Rief M., Gautel M., Oesterhelt F., Fernandez J.M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]


