Abstract
Many of the physiological functions of von Willebrand Factor (VWF), including its binding interaction with blood platelets, are regulated by the magnitude of applied fluid/hydrodynamic stress. We applied two complementary strategies to study the effect of fluid forces on the solution structure of VWF. First, small-angle neutron scattering was used to measure protein conformation changes in response to laminar shear rates (G) up to 3000/s. Here, purified VWF was sheared in a quartz Couette cell and protein conformation was measured in real time over length scales from 2–140 nm. Second, changes in VWF structure up to 9600/s were quantified by measuring the binding of a fluorescent probe 1,1′-bis(anilino)-4-,4′-bis(naphtalene)-8,8′-disulfonate (bis-ANS) to hydrophobic pockets exposed in the sheared protein. Small angle neutron scattering studies, coupled with quantitative modeling, showed that VWF undergoes structural changes at G < 3000/s. These changes were most prominent at length scales <10 nm (scattering vector (q) range >0.6/nm). A mathematical model attributes these changes to the rearrangement of domain level features within the globular section of the protein. Studies with bis-ANS demonstrated marked increase in bis-ANS binding at G > 2300/s. Together, the data suggest that local rearrangements at the domain level may precede changes at larger-length scales that accompany exposure of protein hydrophobic pockets. Changes in VWF conformation reported here likely regulate protein function in response to fluid shear.
Introduction
Von Willebrand Factor (VWF) is a large multimeric protein found in normal human blood at concentrations of 10–20 μg/mL (1). In solution, this protein appears as a loosely packed ellipsoidal molecule with intramolecular interactions within the protein that likely stabilizes the protein solution structure (2). Besides being a soluble protein in circulation, VWF is also expressed in the secretory granules of vascular endothelial cells and platelets. By acting as a bridge/adaptor molecule that aids the binding of platelets to sites of endothelial denudation or vascular injury, VWF plays a critical role in regulating the progress of atherothrombosis. The regulation of VWF structure and function is also relevant in the context of vascular diseases that are associated with elevated VWF activity. Examples of such pathological conditions include acute coronary syndromes, von Willebrand disease type 2B; thrombotic thrombocytopenic purpura; hemolysis, elevated liver enzymes, low platelets; and antiphospholipid syndrome (3,4).
Several lines of evidence suggest that VWF structure and function may be altered by hydrodynamic stresses in circulation. In this regard, fluid forces enhance VWF binding to the platelet receptor GpIbα (5), its susceptibility to proteolysis by the metalloprotease ADAMTS13 (6–8), and its contribution to thrombus growth (9,10). Exposure of VWF to physiological fluid shear down to 2000/s for 12 s in a capillary tube has been shown to augment VWF A2-domain proteolysis by ADAMTS13 (6). Fluid shear above 2300–6000/s promotes the aggregation or self-association of purified VWF (11). Such shear conditions >2000–5000/s also allow novel disulfide bond formation in the protein (12), promoting the self-assembly of VWF into a network of fibers on a collagen matrix (13). In recent studies, protein conformation changes in solution have been thought to precede VWF immobilization on collagen substrates (14).
In this study, we examine the role of hydrodynamic or fluid forces in regulating the solution conformation of VWF. In this regard, whereas the biochemical evidence cited above suggests that VWF may undergo conformation changes in solution in response to fluid shear, only one previous investigation has examined changes in VWF solution structure in response to hydrodynamic shear (14). Using a novel microfluidic device and fluorescently labeled VWF, that investigation showed that VWF undergoes a large-scale transition from a folded to a stretched conformation at shear rates above 2000–5000/s. The resolution of that study was limited by the fluorescence microscope used (∼1–2 μm). Because structural features in VWF span a wide length scale from individual domains with 3–5 nm diameter to the entire multidomain, multimeric protein with a radius of gyration of 100–150 nm (2), we suggest that the use of higher-resolution small angle neutron scattering (SANS) spectroscopy to probe VWF conformation changes is appropriate. Two major advantages of SANS over the previous study are: a), SANS studies protein conformation changes in real time, in the absence of protein labeling and at high resolution down to 2 nm; and b), SANS is ideally suited to measure changes in protein conformation over a wide size range, from domain-level features (∼2 nm) to overall protein shape (∼100–150 nm). To complement the SANS experiments and to study VWF conformation changes at shear rates up to 9600/s, we also introduced the use of environment-sensitive fluorescent probe 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (bis-ANS) to study VWF conformation changes. The fluorescence of this dye depends on its local environment, and it is markedly augmented when bound to hydrophobic/nonpolar pockets within proteins (15).
The SANS studies suggest that VWF may undergo subtle changes in molecular structure when exposed to physiological shear rates. These changes may involve rearrangement of protein domains located within the globular section of VWF. At shear rate >2300–6000/s, more dramatic changes are observed that result in increased exposure of protein hydrophobic domains and bis-ANS binding. Overall, our data support a model where local, domain-level rearrangements at physiological shear rates precede changes at larger length scales that accompany exposure of protein hydrophobic pockets.
Materials and Methods
SANS
Both human VWF purified from plasma cryoprecipitate available from the Community Blood Bank (Erie, PA) and bovine serum albumin (BSA) (Sigma Chemicals, St. Louis, MO) were subjected to fluid shear in the 0.5 mm gap Oak Ridge National Laboratory Couette shear cell (16). The shear cell consisted of a rotating outer cylinder (inner radius = 25.00 mM) and a stationary inner cylinder (outer radius = 24.50 mM). It was aligned in the path of the 30-m NG-3 SANS spectrometer neutron beam at the National Institute of Standards and Technology (NIST) Center for Neutron Research (Gaithersburg, MD). VWF was dialyzed extensively against HEPES buffer (30 mM HEPES, 110 mM NaCl, 10 mM KCl, 1 mM MgCl2, pH 7.3) containing 99.9% D2O before experimentation (2). Seven mL VWF at ∼200 μg/mL, pooled from 3–4 cryoprecipitate units, was used in each run. Western blots confirmed that the multimer distribution of VWF used in the runs was similar. Although higher than physiological levels, this concentration of VWF was necessary to collect scattering data efficiently in reasonable experimental time. Five mg/mL BSA dissolved in phosphate buffer made with 99.9% D20 was used in control runs.
SANS experiments produce data that quantify scattering intensity, I(q), as a function of the scattering vector, q. In general, q is a measure of instrument magnification (2), and it is defined as 4 πSin(θ/2)/λ, with θ representing the scattering angle and λ the neutron wavelength. The I(q) versus q plot contains information on protein solution structure. Structural features in the size-range from 2–140 nm were examined in this study, corresponding to q varying from 0.045–3.169/nm (length scale studied, ∼2 π/q). Change in intensity profile upon shear application in a certain q range is indicative of structural changes in that particular size-range within the protein. Additional material in the Supporting Material section provides details on SANS experimentation and data reduction protocols.
Protein concentration was determined at various time points using the Coomassie/Bradford protein assay kit (Pierce Biotechnology, Rockford, IL). Efforts were made to minimize the time between sample collection and concentration analysis. In many cases, protein measurements were performed within 1–2 min of sample withdrawal by directly pipetting protein from the Couette cell into the 96-well plate used for concentration assays. Independent controls were performed to confirm that stoppage of shear for protein sampling does not affect the conclusions of this work. Finally, studies that compared the results of bradford analysis with amino-acid analysis were undertaken to confirm that the Coomassie dye in the Bradford assay binds similarly to both sheared and unsheared VWF ((2), data not shown). Using the protein measurements above, in some instances, the scattering data were normalized with respect to the solution concentration of VWF in the Couette cell to obtain I/c versus q plot.
Molecular model of VWF response to fluid shear
A computer model was simulated to relate changes observed in SANS experiments with potential biomolecular changes in VWF conformation (see the Supporting Material for details). A VWF protomer for this model was constructed using geometry information available from published experimental data (Fig. 1 A). This dimeric protomer consists of a pair of 34 × 2 nm rods, each linked to five nonoverlapping spheres (Fig. 1 B). Here, the rods represent the B1-B2-B3-C1-C2 domains of VWF, whereas the spheres are the D′D3-A1-A2-A3-D4 domains. Based on the number of amino acids in each domain, three 5 nm and two 4 nm diameter spheres were placed at the end of each rod of the protomer. One of the 5 nm spheres was fixed at the end of the rod and it was assumed to represent the protein's D4 domain. Three different scenarios were simulated using this model. Case A simulated VWF elongation by changing the angle ϕ between the monomer units of the dimeric protomer.2ϕ, ranging from 24–68° in this computer model. Case B simulated the effect of domain rearrangement within the globular section of the protomer. Here, the distance between two of the 5 nm spheres and the fixed D4 domains was increased from 3.5 nm and 10 nm “before shear” to 5.5 nm and 13.5 nm “after shear”, respectively. In addition, a small 1nm sphere was introduced to represent scattering due to interdomain regions that may be separated from the fixed spheres as a result of domain rearrangement. Case C simulated the effect of multimerization. Here, two protomer units were attached at their N-terminus to simulate multimeric VWF. Scattering from the multimer was compared to that from the protomer. Computer-predicted I(q) versus q data in all cases were compared with experimental data.
Figure 1.
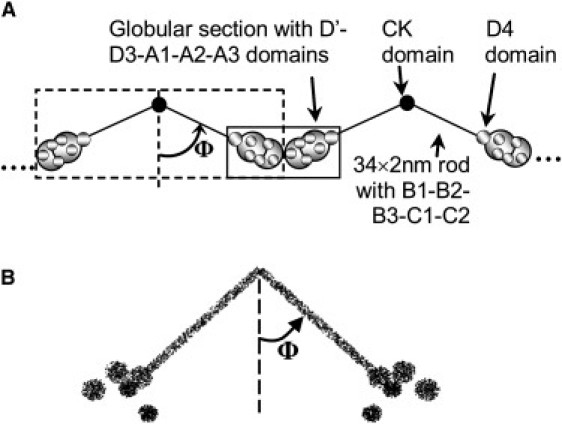
VWF structure. (A) Schematic of multimeric protein based on previous electron microscopy studies (32,33) (see the Supporting Material for details). (B) VWF protomer generated by computer. This figure is composed of 20,000 individual dots, each dot representing a unique grid-point in the computer-generated VWF.
Bis-ANS experiments
The fluorescent probe bis-ANS (Molecular Probes, Eugene, OR) was dissolved (0.1% w/v) in phosphate buffer (8 mM Na2HPO4, 2 mM KH2PO4, 140 mM NaCl; pH 7.2) and equilibrated for three days before use in experiments. 210 μL of protein/buffer was sheared at varying shear rates (0/s–9600/s) for 300 s, using a cone-and-plate viscometer (Haake VT 550, Thermo Scientific, Waltham, MA; 0.5° cone angle, 5 cm diameter). One hundred forty μL of this sheared sample was withdrawn at indicated time points and incubated with 15 μL of bis-ANS (at a final concentration of 14 μM) for precisely 60 s in a quartz cell before obtaining fluorescence measurements using a QuantaMaster Fluorometer (PTI, Birmingham, NJ). Bis-ANS spectra were recorded by exciting the sample at 375 nm and collecting emission from 420–620 nm. Studies were performed with both ∼60 μg/mL VWF and ∼50 μg/mL BSA dissolved in HEPES buffer at room temperature. Independent experiments were performed to confirm that the bis-ANS signal under these conditions varies linearly with both probe and protein concentration (E. Themistou, I. Singh, S. V. Balu-Iyer, P. Alexandridis, and S. Neelamegham, unpublished).
Absolute fluorescence (in counts per second) was measured to detect the binding of bis-ANS to protein. At a given wavelength λ, this absolute fluorescence for protein subjected to a shear rate of G is denoted by F(G)(λ). Subtraction of residual fluorescence due to HEPES buffer and normalization based on protein concentration (c) resulted in a new parameter, Fc(G)(λ) = [F(G)(VWF)(λ) − F(G) (HEPES) (λ) ]/c. Comparison of the effect of shear was further performed by normalizing sheared sample fluorescence with that of unsheared control (G = 0/s) at the wavelength where bis-ANS fluorescence was maximum (max): Fc(G)max/Fc(G = 0/s)max.
Student's t-test was used for all statistical comparisons. p < 0.05 was considered significant.
Results
SANS studies reveal changes in protein structure under physiological fluid shear
Six independent experiments were undertaken over a range of laminar fluid shear protocols to assess conformation changes in purified VWF. Changes in neutron scattering were measured in real time while the protein was being sheared in a Couette shear cell (Fig. 2 A). Functional assays performed earlier showed that the purified protein responds to fluid shear in a manner similar to the response of native protein in blood plasma (2,11). VWF molecular mass ranges from 500 kDa (dimer/protomer-VWF) to >10,000 kDa (multimer-VWF) in these assays.
Figure 2.
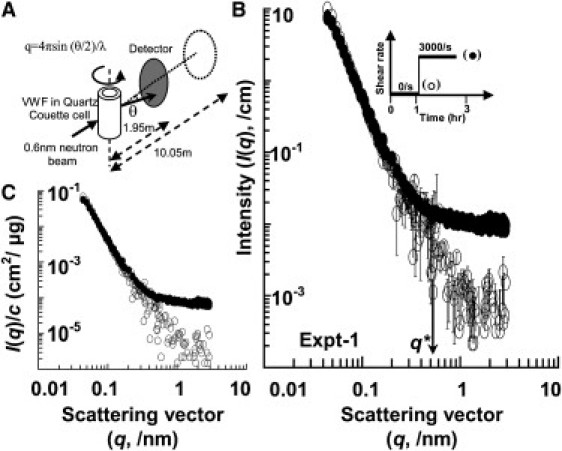
SANS analysis of VWF subjected to fluid shear. (A) Setup for shear experiments employing SANS. 0.6 nm neutrons are scattered by protein sample placed in a Couette cell. Area detectors placed at either 1.95 or 10.05 m detect scattered neutrons. (B) I(q) versus q (q = 4 πsin(θ/2)/λ) plot for experiment 1. Scattering intensity before shear is shown by empty circles and during shear (G = 3000/s) by solid circle. The applied shear profile is in the inset. Changes in scattering intensity are observed at both low-q (<0.08, large length scales) and high-q (>0.6, small length scales). q∗ denotes the highest scattering vector value beyond which intensity increases upon shear application. (C) Normalization of scattering intensity (I(q)) measured in panel B by concentration (c) measured during the experiment (i.e., I/c versus q plot). Change in scattering intensity at high-q (q > 0.6/nm) suggest alterations in structural features at length scales <10 nm. Error bars in panel B are standard deviations. These error bars are very small and are thus not visible in the case of most data points.
In one run (Fig. 2 B), shear application is shown to result in a drop in scattering intensity at low-q (q < 0.08/nm) and an increase at high-q (q > 0.6/nm; length scale < 10 nm). This change in scattering was accompanied by an ∼7% drop in VWF solution concentration. Upon normalizing with respect to the protein concentration (i.e., plotting I/c versus q; Fig. 2 C), our data present evidence of structural changes taking place at small length scales <10 nm (high-q, q > 0.6/nm). The normalized data do not reveal significant changes in scattering at large length scales (low-q, q < 0.08/nm), which would be expected had the protein unraveled.
Similar observations were made in five other runs (Fig. S1 in the Supporting Material). In each run, a decrease in the scattering intensity at low-q (q < 0.08/nm) and an increase in intensity at high-q (q > 0.6/nm) was observed. Further, normalizing intensity data for 7–30% protein concentration drop in individual runs (i.e., plotting I/c versus q as in Fig. 2 C) revealed that structure changes were more prominent at small length scales (high-q). In all cases, changes at large length scales (i.e., low-q) could not be clearly discerned. In the context of these experiments we note that: a), placing VWF in the shear cell under static conditions alone does not result in protein adhesion to the Couette cell wall over extended time periods (>5 h). Further, neither changing the Couette shear cell described in this manuscript (Oak Ridge National Laboratory shear cell) to an alternative quartz shear cell (termed NIST-Boulder shear cell), nor our attempts to passivate the surface reduced the extent of protein loss. b), Negligible scattering was also observed when the entire 7 mL protein sample was withdrawn from the Couette cell after a given run, and scattering due to residual-surface immobilized VWF was measured. Thus, scattering reported here primarily arises from the protein in solution. c), VWF loss under shear is not unique to our experiments, and this aspect is also discussed by other investigators (6,12). A summary of changes in the high-q region in all six experiments (Fig. 2 and Fig. S1) is shown in Table 1. This table reports a), the q-value beyond which shear data do not match static data (q∗ = 0.61 + 0.07 (data presented as root mean-square + standard deviation)) and b), the change in average slope of I(q) versus q in this region (−2.41 + 0.493 before shear to −0.80 + 0.16 after shear application).
Table 1.
Summary of shear studies
| Experiment No. | q∗ (/nm) | Slope (before shear) | Slope (after shear) |
|---|---|---|---|
| 1 | 0.55 | −2.74 | −0.87 |
| 2 | 0.63 | −2.29 | −0.9 |
| 3 | 0.60 | −2.94 | −0.7 |
| 4 | 0.66 | −2.81 | −0.96 |
| 5 | 0.50 | −1.93 | −0.53 |
| 6 | 0.70 | −1.76 | −0.81 |
Fluid shear was applied on VWF in six independent runs, experiment 1 in Fig. 2 and experiments 2–6 in Fig. S1. The q value, above which scattering intensity increased (q∗), was measured. Change in scattering intensity slope at q = q∗ was also quantified before (0/s) and after shear application. Postshear results are presented at G = 3000/s in all experiments except for experiment 3, in which the maximum applied G was 1500/s.
A variety of fluid shear protocols were applied in our study (Fig. S1, insets). Whereas some of these runs involved a slow stepwise increase in shear from 0/s to 3000/s, shear was abruptly increased to 3000/s in others. Stepwise increase in shear caused a progressive increase in neutron scattering (Fig. S1, panels B and D). Stopping shear did not result in the return of scattering intensity to levels before shear application, and this suggests that structural changes observed were not reversible in the time course of our study. Further, although the NG-3 is among the brightest neutron sources available today for SANS studies, the flux rate is still low and it takes ∼30 min to acquire high-quality data at a single magnification. In experiments performed where SANS data at the high-q range were collected in 10-min time slices, we consistently observed that a majority of the changes in neutron scattering occurred in the first 10 min.
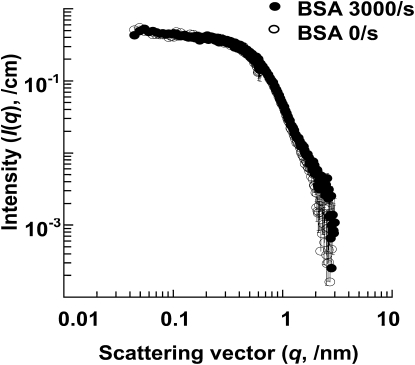
In control runs, changes in scattering were not observed upon shearing either the protein BSA (Fig. 3) or HEPES buffer in the absence of protein (data not shown). Additional shear studies were also attempted with a large human blood protein called fibrinogen. These studies, however, could not be completed because of the instability of fibrinogen in the deuterated media/solvent when solution temperature was dropped from 37°C to 25°C in 12.9 mM sodium citrate buffer (pH 7.3).
Figure 3.
SANS analysis with BSA 5mg/mL under shear at 3000/s (solid circles) and without shear (empty circles) show similar intensity profiles. Error bars represent standard deviations. These error bars are very small in most cases and are thus not visible.
Rearrangement of domains within VWF explain SANS observations
Computer modeling was performed with the goal of suggesting potential mechanisms that can account for the observed changes in VWF scattering at high-q upon fluid shear application. Both the protomer/dimer VWF (Fig. 4, A–F) and the multimeric VWF (Fig. 4, G–I) were simulated. The latter computer modeling quantified scattering by the tetrameric form of VWF.
Figure 4.
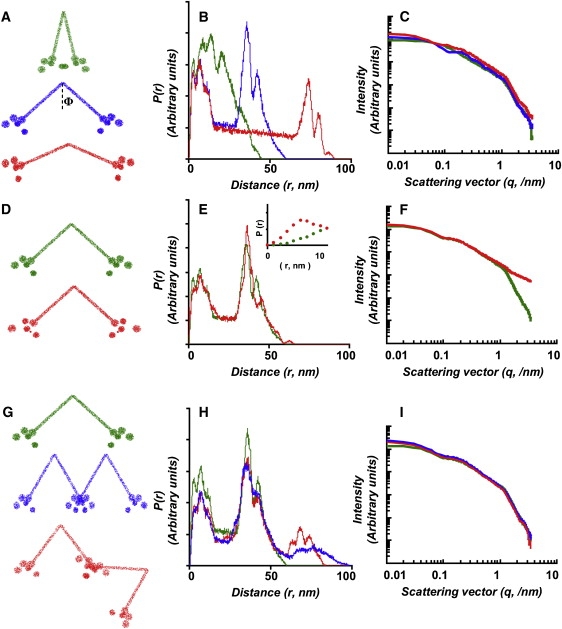
Computational modeling of VWF response to shear. Scattering profile due to protomer and multimer VWF was simulated. Results from three types of hypothetical changes are presented. In panels A–C, the effect of VWF elongation is examined by changing the angle ϕ linking two monomers as shown in panel A from ϕ =12° to 36° and 84°. In panels D–F, the effect of changing the arrangement of domains in the globular section of VWF was examined. Here, the distance between the two 5 nm spheres from the fixed D4 domain was increased from 3.5 to 5.5 nm and from 10 to 13.5 nm, as illustrated. In addition, a 1 nm sphere was introduced 6 nm away from the D4 domain. In G–I, the effect of VWF multimerization was simulated. These last computations compare the scattering of dimer VWF with two different tetramer-VWF configurations. As seen, increase in the molecular dimensions of VWF (panel A) cause an expansion in the length scale of the distance distribution function P(r) plot (panel B). Changes in I(q) are observed at both the low- and high-q range (panel C). Alterations in domain-level features (panel D) result in a decrease in scattering intensity slope in the VWF protomer at high-q (>0.6/nm, green to red in panel F). Multimerization of VWF (panel G) results in an expanded range of the distance distribution function (panel H). This results in an increase in scattering intensity at large length scales (low-q) but not in the small length scales (high-q). Changes in scattering intensity in panel F, but not in C or I, more closely resemble experimental data in Figs. 2 and S1.
Three different cases were simulated. In Fig. 4, A–C, the effect of VWF elongation was examined by changing the angle 2ϕ linking two monomers (panel A). In Fig. 4, D–F, the effect of changing the arrangement of domains in the globular section of VWF was examined. In Fig. 4, G–I, the effect of VWF multimerization was studied. Figures are color coded (green, blue, and red) so that specific conformation changes simulated in Fig. 4, A, D, and G can be related to changes in the distance distribution function (Fig. 4, B, E, and H) and in the scattering intensity profile (Fig. 4, C, F, and I). Schematics in panels 4, A, D, and G are to scale, because individual points are from actual computer modeling. As expected, an increase in the molecular dimensions of VWF (Fig. 4 A) resulted in an expansion in the length scale of P(r) (Fig. 4 B). In the I(q) versus q plot, scattering intensity is observed to change at both the high- and low- q values, as would be expected if the protein elongated. Increase in interdomain distances and appearance of a smaller domain (Fig. 4 D, green to red) resulted in a modest change in P(r) (Fig. 4 E) and a localized change in scattering intensity slope only at high-q (>0.6/nm, green to red in Fig. 4 F). In this computer modeling, a rearrangement of domains (5 and 4 nm spheres) in the protein's globular section alone was sufficient to cause changes in scattering intensity at q > 0.6/nm. Introduction of the feature at small length scale (1 nm sphere) was necessary to fit I(q) data at q ∼1.8–3/nm. Such small features may appear as a result of changes in interdomain distances at small length scales. Multimerization of VWF dimer to tetramer resulted in intensity increase at low-q (<0.04/nm) only while structural features in the high-q range remain identical in both dimer and tetramer VWF. Because changes in scattering intensity in Fig. 4 F, but not in Fig. 4, C or I, more closely resemble experimental data in Fig. 2 and Fig. S1, our computer modeling suggests that shear application below 3000/s may cause domain-level alterations in VWF structure without altering the overall size of the protein.
While considering the implications of the above computer modeling, it is important to note that these computations do not suggest that the unique change illustrated in Fig. 4 D are responsible for our experimental observations. They are simply meant to illustrate the fact that changes at small length scale that do not alter the overall shape of the protein likely occur at low shear rates <3000/s. Such changes may be due to changes in the relative position of domains within the globular section of the protein. Also, these calculations show that simulation of multimeric protein (Fig. 4, G–I) is not necessary for explaining experimental changes observed at small length scale.
Hydrophobic domains within VWF are exposed at high shear rates
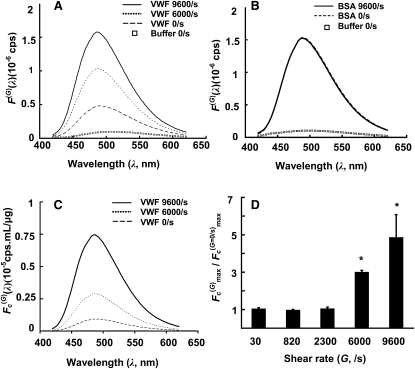
We determined if the binding of bis-ANS to VWF was altered upon shear application. Cone-and-plate viscometry was used to shear protein and bis-ANS was added just after shear. Offline fluorescence measurements were made 1 min after dye addition. A representative plot (Fig. 5 A) shows that bis-ANS exhibits low fluorescence in a HEPES buffer. Addition of protein in the absence of fluid shear increases the magnitude of the fluorescence signal (F(G)(λ)), and it results in peak emission at a lower wavelength, 489 nm. Application of fluid shear at 6000/s and 9600/s resulted in an additional increase in measured absolute fluorescence. This was accompanied by a blue-shift in the peak emission wavelength to 485 nm. In control runs, the application of fluid shear to ∼50 μg/mL BSA dissolved in either HEPES buffer or PBS did not result in changes in bis-ANS fluorescence (Fig. 5 B).
Figure 5.
Shear induced conformation changes in VWF detected using bis-ANS. (A) Representative spectra for an experiment performed with 60 μg/mL VWF. Bis-ANS binding to VWF increases absolute fluorescence (F(G)(λ), counts per second) by 5–10-fold over the HEPES buffer signal (squares). Application of shear for 5 min at 6000 or 9600/s further increased the absolute bis-ANS signal by 110–220%, compared to results obtained with no-shear control (G = 0/s). (B) Shearing 50 μg/mL BSA does not change signal due to bis-ANS BSA interaction. (C) Concentration normalized spectra (Fc(G)(λ)) for VWF. Raw data are from panel A. (D) Comparison of Fc(G)max/Fc(G = 0/s)max for VWF under different shear conditions. Data in panels A–C are representative of eight independent experiments. Results in panel D are presented as mean ± SE (N ≥ 3 at each shear rate). ∗p < 0.05 with respect to shear rate = 30/s.
The increased probe binding to VWF upon shear application is likely caused by an increase in the exposure of nonpolar/hydrophobic domains within the protein (15). Concentration normalized data are shown in Fig. 5 C, and these suggest that bis-ANS fluorescence increased by ∼200% upon application of shear at 6000/s and it was augmented by ∼700% at 9600/s. The observation that absolute fluorescence was higher in sheared protein compared to fluorescence with VWF placed in no-flow/static conditions was consistently observed in eight independent experiments performed with different donor samples. In these runs, fluorescence of sheared protein was consistently higher than fluorescence of unsheared VWF when comparing: a), measured absolute fluorescence values (F(G)(λ)) regardless of protein concentration loss to viscometer surface, b), concentration normalized fluorescence (Fc(G)(λ)), and c), when runs were performed in which unsheared VWF concentration was adjusted to the same value as that of the sheared protein, before fluorescence measurements. Root mean-square Fc(G)max/Fc(G = 0/s)max data are presented in Fig. 5 D. These results show that a critical shear rate between 2300–6000/s is necessary to enhance bis-ANS VWF binding interactions.
In a previous publication that quantified the magnitude of fluid shear sufficient to induce shear-induced platelet activation (11), we noted that a threshold shear rate of ∼6000/s is necessary for efficient platelet activation after VWF-platelet GpIb binding. Bis-ANS signal is observed to increase at a similar shear range in the current work. Taken together, the data suggest a correlation between the increased bis-ANS signal caused by VWF conformation change and the ability of this protein to activate blood platelets in functional studies.
Discussion
Shear stress applied on VWF because of fluid motion regulates the size and activity of the protein. Such regulation then impacts the role of VWF during thrombosis in the vasculature. We applied small angle scattering to measure protein solution structure in real time at shear rates up to 3000/s. A fluorescent spectroscopy assay employing bis-ANS was also developed to probe nonpolar/hydrophobic VWF domains that are exposed upon shear application at shear rates up to 9600/s. These experimental modalities are distinct from those of previous studies that assay VWF conformation in solution (14,17) and on substrates (18,19), because our studies probe the volume-averaged properties of VWF. This is unlike previous studies that employed atomic force microscopy, microfluidics, and fluorescence microscopy to study individual protein dynamics. The length scales we probe are also significantly smaller (2–140 nm), compared to those probed in previous investigations. Finally, our experimental methods studied the native protein without fluorescence labeling or immobilization. Our findings on the length scale, shear dependence, and reversibility of VWF conformation change are discussed below.
Length scale
In our study, protein conformation changes were observed over the range of physiological and pathological shear rates. We report, based on SANS and associated modeling, that VWF may undergo subtle domain-level changes at length scales below 10 nm (q > 0.6 nm) when exposed to physiological shear rates below 3000/s. Similar changes in scattering intensity at q > 0.6 nm were noted previously when VWF was treated with 1.25 M Guanidine HCl (2). Biochemical studies showed that this concentration of denaturant Guanidine HCl is sufficient to alter VWF solution conformation such that the protein was now susceptible to proteolysis by ADAMTS13 (2,20). Further, VWF proteolysis by ADAMTS13 is reported in capillaries at wall shear rates down to 2000/s (volume-averaged shear rate of 1333/s) (6). Taken together, these data support the possibility that physiological levels of fluid shear and Guanidine HCl may cause similar structural changes in VWF solution structure.
That subtle changes at the domain level may have important functional consequences is supported by recent studies that have demonstrated functional and specific interactions between protein domains located within the globular-head section of VWF. In this regard, Martin et al. (21) showed that the VWF A2 domain interacts with the A1 domain, and that it may block VWF-GpIbα interactions in the native protein. Nishio et al. (22) also suggested that the A1 domain hinders the action of the metalloprotease ADAMTS13 on the VWF A2-domain. Binding interactions via the A1 domain enhance the rate of proteolysis mediated by ADAMTS13. Ulrichts et al. (23) presented evidence for the shielding of the VWF A1 domain by the D′D3 domain of VWF. Finally, Factor VIII binding to VWF is shown to accelerate proteolytic cleavage of VWF by ADAMTS13 under vortexing-shear conditions (24). Our SANS data now suggest that fluid shear forces may be able to support structural alterations at the domain level of VWF. This may then augment both A1-domain binding function and A2-domain cleavage rates.
Shear dependence
The extent of VWF conformation change increased with shear in a stepwise manner. This was observed in the SANS experiments, in which the amount of neutron scattering increased with shear rate. In the bis-ANS studies, also, fluorescence signal increased with shear rate. Whereas only low levels of change were observed in both the SANS and bis-ANS studies below 2300/s, application of higher shear rates led to exposure of hydrophobic domains within the protein, which enhanced the bis-ANS signal. A recent computer simulation study (25) also predicted the transition of VWF binding to platelets occurring in a similar range of applied share rates. Others, using fluorescently tagged VWF, have reported that the protein undergoes a sharp transition from a compact to an elongated structure above a critical shear rate that lies between 2000–5000/s. Taken together, the diverse experiments suggest that, whereas it is possible that large length scale changes that exaggerate protein structure and expose hydrophobic domains within the protein may occur at high shear rates (>2300/s), smaller changes due to the rearrangement of domain level features manifest themselves at lower, physiological shear rates (<3000/s). These subtle rearrangements may be a prerequisite that precedes larger changes in protein structure. Studies that assess the conformation stability and unfolding of the VWF A domain (26) suggest that the VWF A2 domain may be most susceptible to unfolding.
The proposition that low physiological levels of shear can induce changes in protein structure is reasonable and consistent with biophysical theory and experimentation. Other studies have shown that DNA molecules with size similar to VWF undergo elongations under shear conditions that are equivalent to 30/s (27). Whereas one study suggests that extraordinarily high shear rates (107/s) are required for protein unfolding under shear (28), these observations are likely limited to small, single-domain proteins (such as BSA in our control experiments). Large molecules with multiple domains such as VWF are susceptible to conformation change under simple shear, as demonstrated here and elsewhere (14). Detailed hydrodynamic calculations also show that directed maximum normal forces due to convective flow in the order of 0.1–0.5 pN are applied on large multimeric proteins like VWF in solution at a shear rate of 300–600/s, with the magnitude of applied forces scaling linearly with the shear rate (29). Although these forces are small compared to the force applied to break typical bio-molecular bonds (>10 pN), they are directed, cyclical forces (as opposed to random Brownian forces) that can affect protein structure.
Time dependence and reversibility
In SANS studies, we observed changes in neutron scattering intensity at the earliest time point where data were collected, i.e., 10 min after fluid shear initiation. In addition, using Annexin-V as a marker of platelet activation, we have previously reported that even 10 s of high shear application (G = 9600/s) is sufficient to mediate significant platelet activation via GpIb-VWF interactions (11). Exposure to shear for larger times and higher shear rates increased the extent of platelet activation in this study. Other studies have also made similar observations (30). Taken together, the data suggest that VWF may undergo shear dependent conformation changes that occur at small timescales and that these structural changes may be amplified with time of exposure and magnitude of fluid shear.
With respect to VWF relaxation, SANS studies did not reveal reversibility in protein conformation change at length scale <10 nm. In studies performed at higher shear rates using bis-ANS as a probe of protein conformation, some reversibility in VWF conformation was observed after stoppage of shear (E. Themistou, I. Singh, S. V. Balu-Iyer, P. Alexandridis, and S. Neelamegham, unpublished). These observations are consistent with the findings of other researchers (17), who stretched VWF into a thin fiber using atomic force microscopy and then observed its relaxation over time using microscopy. These authors suggested that VWF exhibits two characteristic relaxation times. The protein partially returns to a compressed state in the first 2 s, and this is followed by a slower relaxation process over a period of minutes. The protein in this study (17) did not recover its native configuration over the duration of the study. Taken together, the data suggest that VWF may undergo partial relaxation after stretch/stress application. It may be expected that, whereas larger length scale features may revert to their native state, domain-level relaxation either takes place relatively slowly or does not take place at all.
In summary, this report demonstrates the use of SANS and fluorescence spectroscopy to study protein/VWF dynamics in response to shear flow. Changes in solution structure reported here may act in synergy with similar modifications occurring in VWF that is immobilized/bound to collagen (31), endothelial cell (7), and platelet (8) surfaces. Together, these conformation changes regulate platelet adhesive function in human circulation.
Acknowledgments
We thank Drs. Sathy V. Balu-Iyer and Paschalis Alexandridis (SUNY-Buffalo) for discussions on the use of Bis-ANS and SANS to probe protein conformation change.
This work was supported by National Institutes of Health grants HL77258 and HL77261. The NIST neutron research facility used in this work is supported in part by the National Science Foundation under Agreement No. DMR-0454672.
Supporting Material
References
- 1.Ruggeri Z.M., Mendolicchio G.L. Adhesion mechanisms in platelet function. Circ. Res. 2007;100:1673–1685. doi: 10.1161/01.RES.0000267878.97021.ab. [DOI] [PubMed] [Google Scholar]
- 2.Singh I., Shankaran H., Beauharnois M.E., Xiao Z., Alexandridis P. Solution structure of human von Willebrand factor studied using small angle neutron scattering. J. Biol. Chem. 2006;281:38266–38275. doi: 10.1074/jbc.M607123200. [DOI] [PubMed] [Google Scholar]
- 3.Groot E., de Groot P.G., Fijnheer R., Lenting P.J. The presence of active von Willebrand factor under various pathological conditions. Curr. Opin. Hematol. 2007;14:284–289. doi: 10.1097/MOH.0b013e3280dce531. [DOI] [PubMed] [Google Scholar]
- 4.Spiel A.O., Gilbert J.C., Jilma B. von Willebrand factor in cardiovascular disease: focus on acute coronary syndromes. Circulation. 2008;117:1449–1459. doi: 10.1161/CIRCULATIONAHA.107.722827. [DOI] [PubMed] [Google Scholar]
- 5.Goto S., Salomon D.R., Ikeda Y., Ruggeri Z.M. Characterization of the unique mechanism mediating the shear-dependent binding of soluble von Willebrand factor to platelets. J. Biol. Chem. 1995;270:23352–23361. doi: 10.1074/jbc.270.40.23352. [DOI] [PubMed] [Google Scholar]
- 6.Tsai H.M., Sussman I.I., Nagel R.L. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83:2171–2179. [PubMed] [Google Scholar]
- 7.Dong J.F., Moake J.L., Nolasco L., Bernardo A., Arceneaux W. ADAMTS13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 8.Shim K., Anderson P.J., Tuley E.A., Wiswall E., Sadler J.E. Platelet-VWF complexes are preferred substrates of ADAMTS13 under fluid shear stress. Blood. 2008;111:651–657. doi: 10.1182/blood-2007-05-093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donadelli R., Orje J.N., Capoferri C., Remuzzi G., Ruggeri Z.M. Size regulation of von Willebrand factor-mediated platelet thrombi by ADAMTS13 in flowing blood. Blood. 2006;107:1943–1950. doi: 10.1182/blood-2005-07-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shida Y., Nishio K., Sugimoto M., Mizuno T., Hamada M. Functional imaging of shear-dependent activity of ADAMTS13 in regulating mural thrombus growth under whole blood flow conditions. Blood. 2008;111:1295–1298. doi: 10.1182/blood-2007-09-110700. [DOI] [PubMed] [Google Scholar]
- 11.Shankaran H., Alexandridis P., Neelamegham S. Aspects of hydrodynamic shear regulating shear-induced platelet activation and self-association of von Willebrand factor in suspension. Blood. 2003;101:2637–2645. doi: 10.1182/blood-2002-05-1550. [DOI] [PubMed] [Google Scholar]
- 12.Choi H., Aboulfatova K., Pownall H.J., Cook R., Dong J.F. Shear-induced disulfide bond formation regulates adhesion activity of von Willebrand factor. J. Biol. Chem. 2007;282:35604–35611. doi: 10.1074/jbc.M704047200. [DOI] [PubMed] [Google Scholar]
- 13.Barg A., Ossig R., Goerge T., Schneider M.F., Schillers H. Soluble plasma-derived von Willebrand factor assembles to a haemostatically active filamentous network. Thromb. Haemost. 2007;97:514–526. [PubMed] [Google Scholar]
- 14.Schneider S.W., Nuschele S., Wixforth A., Gorzelanny C., Alexander-Katz A. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc. Natl. Acad. Sci. USA. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosinski-Collins M.S., King J. In vitro unfolding, refolding, and polymerization of human ΓD crystallin, a protein involved in cataract formation. Protein Sci. 2003;12:480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porcar L., Hamilton W.A., Butler P.D. A vapor barrier Couette shear cell for small angle neutron scattering measurements. Rev. Sci. Instrum. 2002;73:2345–2354. [Google Scholar]
- 17.Steppich D.M., Angerer J.I., Sritharan K., Schneider S.W., Thalhammer S. Relaxation of ultralarge VWF bundles in a microfluidic-AFM hybrid reactor. Biochem. Biophys. Res. Commun. 2008;369:507–512. doi: 10.1016/j.bbrc.2008.02.062. [DOI] [PubMed] [Google Scholar]
- 18.Siedlecki C.A., Lestini B.J., Kottke-Marchant K.K., Eppell S.J., Wilson D.L. Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood. 1996;88:2939–2950. [PubMed] [Google Scholar]
- 19.Novak L., Deckmyn H., Damjanovich S., Harsfalvi J. Shear-dependent morphology of von Willebrand factor bound to immobilized collagen. Blood. 2002;99:2070–2076. doi: 10.1182/blood.v99.6.2070. [DOI] [PubMed] [Google Scholar]
- 20.Tsai H.M. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 21.Martin C., Morales L.D., Cruz M.A. Purified A2 domain of von Willebrand factor binds to the active conformation of von Willebrand factor and blocks the interaction with platelet glycoprotein Ibalpha. J. Thromb. Haemost. 2007;5:1363–1370. doi: 10.1111/j.1538-7836.2007.02536.x. [DOI] [PubMed] [Google Scholar]
- 22.Nishio K., Anderson P.J., Zheng X.L., Sadler J.E. Binding of platelet glycoprotein Ibalpha to von Willebrand factor domain A1 stimulates the cleavage of the adjacent domain A2 by ADAMTS13. Proc. Natl. Acad. Sci. USA. 2004;101:10578–10583. doi: 10.1073/pnas.0402041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrichts H., Udvardy M., Lenting P.J., Pareyn I., Vandeputte N. Shielding of the A1 domain by the D′D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J. Biol. Chem. 2006;281:4699–4707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- 24.Cao W., Krishnaswamy S., Camire R.M., Lenting P.J., Zheng X.L. Factor VIII accelerates proteolytic cleavage of von Willebrand factor by ADAMTS13. Proc. Natl. Acad. Sci. USA. 2008;105:7416–7421. doi: 10.1073/pnas.0801735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mody N.A., King M.R. Platelet adhesive dynamics. Part II: high shear-induced transient aggregation via GPIbα-vWF-GPIbα bridging. Biophys. J. 2008;95:2556–2574. doi: 10.1529/biophysj.107.128520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auton M., Cruz M.A., Moake J. Conformational stability and domain unfolding of the Von Willebrand factor A domains. J. Mol. Biol. 2007;366:986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 27.Smith D.E., Babcock H.P., Chu S. Single-polymer dynamics in steady shear flow. Science. 1999;283:1724–1727. doi: 10.1126/science.283.5408.1724. [DOI] [PubMed] [Google Scholar]
- 28.Jaspe J., Hagen S.J. Do protein molecules unfold in a simple shear flow? Biophys. J. 2006;91:3415–3424. doi: 10.1529/biophysj.106.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankaran H., Neelamegham S. Hydrodynamic forces applied on intercellular bonds, soluble molecules, and cell-surface receptors. Biophys. J. 2004;86:576–588. doi: 10.1016/S0006-3495(04)74136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J.N., Bergeron A.L., Yu Q., Sun C., McIntire L.V. Platelet aggregation and activation under complex patterns of shear stress. Thromb. Haemost. 2002;88:817–821. [PubMed] [Google Scholar]
- 31.Bonnefoy A., Romijn R.A., Vandervoort P.A., Van Rompaey I., Vermylen J. von Willebrand factor A1 domain can adequately substitute for A3 domain in recruitment of flowing platelets to collagen. J. Thromb. Haemost. 2006;4:2151–2161. doi: 10.1111/j.1538-7836.2006.02111.x. [DOI] [PubMed] [Google Scholar]
- 32.Fowler W.E., Fretto L.J., Hamilton K.K., Erickson H.P., McKee P.A. Substructure of human von Willebrand factor. J. Clin. Invest. 1986;76:1491–1500. doi: 10.1172/JCI112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fretto L.J., Fowler W.E., McCaslin D.R., Erickson H.P., McKee P.A. Substructure of human von Willebrand factor. Proteolysis by V8 and characterization of two functional domains. J. Biol. Chem. 1986;261:15679–15689. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.