Abstract
Escherichia coli RecBCD helicase unwinds blunt-end duplex DNA to repair damaged DNA molecules in the homologous recombination pathway. Previous single-molecule experiments showed that RecBCD recognizes an 8 nt DNA sequence, χ, and lowers its unwinding rate afterward under saturating ATP condition. We have developed a single-molecule force-tethered particle motion (FTPM) method, which is modified from the conventional TPM method, and applied it to study RecBCD motion in detail. In the FTPM experiment, a stretching force is applied to the DNA-bead complex that suppresses the bead's Brownian motion, resulting in an improved spatial resolution at long DNA substrates. Based on the equipartition theorem, the mean-square displacement of the bead's Brownian motion measured by FTPM correlates linearly to DNA extension length with a predicted slope, circumventing the difficulties of conventional TPM experiments, such as nonlinearity and low resolution of long DNA substrates. The FTPM method offers the best resolution in the presence of only a small stretching force (0.20 pN). We used the FTPM method to investigate RecBCD helicase motion along 4.1 kb long χ-containing duplex DNA molecules, and observed that the translocation rate of RecBCD changes after the χ sequence under limited ATP concentrations. This suggests that χ recognition by RecBCD does not require saturating ATP conditions, contrary to what was previously reported.
Introduction
Escherichia coli RecBCD helicase is responsible for initiating double-strand-break DNA repair through the homologous recombination pathway. RecBCD recognizes nearly blunt end duplex DNA, and binds and unwinds the DNA molecules to generate single-stranded DNA substrate for RecA binding (1). There are two helicase subunits in the RecBCD complex: RecB and RecD (2,3). In addition to its helicase activity, RecBCD also functions as a nuclease that is responsible for degrading foreign DNA substrates. The delicate interplay between the helicase and nuclease activities, and the regulation between RecB and RecD helicases are modulated by an 8 nt DNA sequence, χ (5′-GCTGGTGG-3′) (1). Several single-molecule experiments have been used to study RecBCD motion along χ-containing DNA substrates. For example, translocation of individual RecBCD helicases along χ-containing duplex DNA was monitored by means of fluorescence dyes intercalated within duplex DNA. Using this strategy, it was found that RecBCD pauses and lowers its translocation velocity after recognizing the χ sequence, under saturating ATP concentrations (1 mM, KMATP ∼150 μM) (4,5). However, in another single-molecule experiment, no translocation rate change was observed upon χ at low ATP concentrations (<50 μM) (6). We developed a force-tethered particle motion (FTPM) experiment, modified from conventional TPM experiments, to directly monitor RecBCD movement along 4.1 Kb DNA containing the χ sequence, with improved resolution to test whether χ recognition requires saturating ATP conditions, as previously suggested (4,5). The FTPM experiments directly detect individual RecBCD molecules lowered their translocation rates once they encountered the χ sequence, even under limited ATP concentrations.
TPM is a simple and powerful method for monitoring DNA length changes at the single-molecule level. This method has been used to investigate the interaction of DNA with different DNA enzymes, including RNA polymerases, DNA recombinases, DNA helicases, and lac repressor, as well as kinetics at the Holliday junction (6–14). In a TPM experiment the surface-anchored DNA molecules are bound by a submicron-sized microsphere bead at its distal end (11,14). In solution these tethered DNA-bead molecules experience Brownian motion, and the amplitude of the bead's Brownian motion is thought to be limited by its DNA tether length. Several recent studies suggested that the relationship between the bead's Brownian motion and the DNA tether length is not as linear as previously thought (14–16). Therefore, it is necessary to establish an empirical relationship between DNA length and bead Brownian motion. Because of this nonlinearity, conventional TPM experiments are limited to using DNA lengths shorter than 2 kb, where the deviation from linearity is minimal.
To circumvent these difficulties, we modified the existing TPM method by introducing a stretching force to the tethering beads through a continuous flow. This small stretching force suppresses the bead's Brownian motion and thus increases the precision of the determination of bead positions. This same strategy was previously employed for magnetic tweezers (17) and flow-stretched experiments (18). In addition, based on the equipartition theorem, the mean-square displacement (MSD) of the bead's Brownian motion is linearly correlated with the DNA extension length (17,18). Therefore, there is a predicted linear relationship between MSD values and DNA length, and an empirical calibration curve of the bead's Brownian motion at various DNA lengths is not required. In addition, the linear relationship ensures that much longer DNA substrates can be used in FTPM experiments. The best resolution for FTPM is a compromise between the precision and sensitivity of the FTPM measurements, which occurs at an intermediate force (∼0.20 ± 0.01 pN for the 220 nm beads used here). Under this condition, the FTPM experiment is able to resolve an ∼50 bp difference in 433 bp long DNA at a time resolution of 6 s. Because of its multiplex nature, FTPM offers efficient data acquisition and improved resolution with the application of a very small force.
Materials and Methods
DNA and protein preparation
Double-stranded DNA substrates (433, 836, 1551, 1893, and 4190 bp) were prepared by means of polymerase chain reaction (PCR) using pBR322 as the template, with one 5′ digoxigenin-labeled primer and one 5′ biotinylated-labeled primer to achieve the specified DNA lengths. χ-Containing DNA substrates were prepared using a 5′ digoxigenin-labeled primer and a non-labeled primer in the 3χH3χF template (6) to generate a blunt-end entry for RecBCD helicase. χ-Containing DNA was constructed in an orientation such that the χ sequence would be correctly recognized by RecBCD from the 3′ entry end. The positions of the χ site were at 1145 bp from the surface-immobilized end for the 1967 bp long DNA, and at 1404 bp for the 4141 bp long DNA. Overexpression and purification of biotinylated E. coli RecBCD enzymes were carried out as described previously (6), except that additional purification steps of ammonium sulfate precipitation and a Q-sepharose column were used before a home-made monomeric avidin column.
Slide preparation and single-molecule TPM measurements
Round coverglasses (40 mm and 30 mm round; Warner Instruments, Hamden, CT) were cleaned by sonication in 2 M KOH solution for 1 min; rinsed with water, ethanol, and water; and then dried with N2 gas. These two coverglasses were assembled into a flow cell with a chamber volume of ∼50 μL (RC-30WA; Warner Instruments). The chamber was first washed with RecBCD buffer (25 mM Tri-HCl, 1 mM Mg(OAc)2, 1 mM DTT, 80 mM NaCl, pH = 7.5). Then 20 μg/mL anti-digoxigenin (Roche, Indianapolis, IN) was introduced into the chamber and incubated for 30 min at room temperature. To prevent nonspecific interactions of DNA or protein to the glass surface, the reaction chamber was washed and incubated with RecBCD buffer containing casein (BioFx, Owings Mills, MD) for 30 min at room temperature. Subsequently, 1 nM dsDNA (digoxigenin-biotin-labeled or digoxigenin-labeled) was specifically attached to the anti-digoxigenin-decorated surface for 30 min at room temperature. Finally, 300 pM streptavidin-coated beads were flowed into the cell for measurement and tracking. For the RecBCD unwinding experiment, biotinylated RecBCD molecules were preincubated with 300 pM streptavidin beads offline for 30 min at room temperature, and then introduced to the DNA-bound surface in the presence of specified ATP concentration and ATP regeneration system. Before images were acquired, excess beads were washed out with RecBCD reaction buffer.
A stretching force was introduced by a continuous laminar flow using a syringe pump (PHD 2000; Harvard Apparatus, Holliston, MA) with the constant flow rate of 0–500 mL/h. The forces associated with these flow rates correspond to 0–16 pN in our experimental geometry. The forces have been found to correlate linearly with flow rates up to 100 mL/h. All experiments were carried out at 22°C ± 1°C.
Streptavidin-coated beads
Streptavidin-coated beads were prepared from 0.22 μm diameter carboxylated modified polystyrene beads (Bangs Laboratories, Fishers, IN) that were covalently linked with biotin and later mixed with excess streptavidin (19). Unbound, excess streptavidin molecules were removed by spinning the beads down at 14 krpm for 6 min, followed by washing in PBS buffer containing 0.1% tween-20. The spin/wash processes were repeated four to five times before use. These washed beads could be used for a period of 1 week.
Imaging acquisition and analysis
Brownian motion of tethered beads was observed by means of an inverted optical microscope (IX-71; Olympus, Center Valley, PA), using the differential interference contrast (DIC) method. The images were acquired with a CCD camera (Cascade 512B; Roper Scientific, Tucson, AZ) using custom software written in Labview with an exposure time of 10 ms and a frame interval of 63 ms. The centroid positions of the chosen tethered beads in individual frames were determined to nanometer precision by fitting the bead intensity profile with a two-dimensional Gaussian curve. The standard deviation (SD) in determining the bead centroid position is 4.2 nm for beads preabsorbed to the glass surface. The stage drift was corrected by subtracting the average movement of the two best-correlated beads that were preabsorbed onto the surface. Drift-corrected centroid positions of the tethering beads were used to calculate the MSD for the consecutive 40 image frames in the RecBCD unwinding experiments. The MSD value did not change when it was calculated from more than 40 consecutive frames, suggesting that Brownian fluctuations were fully sampled in the phase space, even for the long substrates used. For the resolution comparison, the MSD values were calculated from consecutive 200 frames. This process was repeated for ∼80–100 different beads at each DNA length to establish a histogram of the MSD values (see Fig. 3). This histogram was fitted with a Gaussian distribution with a central MSD value (mean) and width. The mean MSD values of several different known DNA lengths were compiled to establish the MSD-DNA tether length plot at various flow rates (see Fig. 4). The width (SD) of the Gaussian fits from the MSD histogram (see Fig. 3) dictated the precision of the MSD measurement in our experiment.
Figure 3.
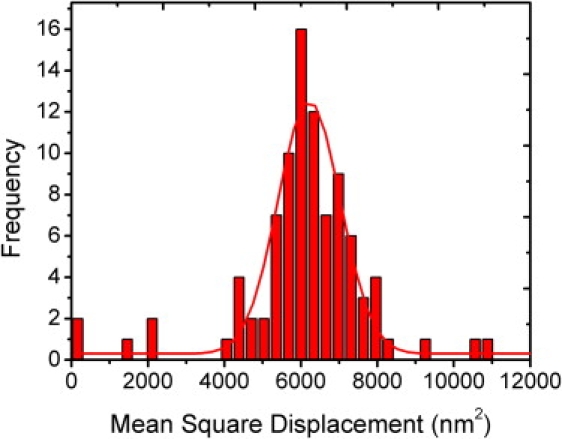
The histogram (N = 88) of MSD values of 220 nm beads tethered to 1551 bp dsDNA molecules under a stretching force of 0.20 pN. The histogram was fitted with a Gaussian distribution with a mean MSD value of 6215 nm2 (± 52 nm2) and an SD of 1631 nm2 (± 110 nm2).
Figure 4.
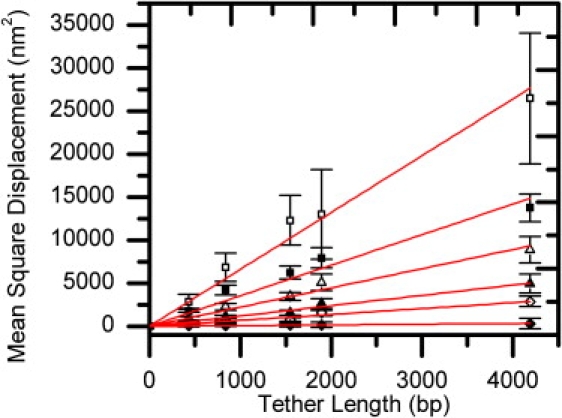
The MSD values of 220 nm bead Brownian motion and DNA tether lengths are linearly correlated at various forces. Note that the slope is inversely proportional to the force. Each data point comes from a Gaussian-fitted histogram curve of ∼80–100 tethered beads from Fig. 3. □ = 0.06 ± 0.01 pN (4 mL/h); ■ = 0.20 ± 0.01 pN (15 mL/h); Δ; = 0.41 ± 0.01 pN (30 mL/h); ▴ = 0.86 ± 0.03 pN (60 mL/h); ⋄ = 1.62 ± 0.09 pN (100 mL/h); ♦ = 16.25 ± 0.50 pN (500 mL/h).
The anchor point where the DNA molecules were attached to the surface was determined by the mean position of the same tethering beads when the flow was introduced in two opposing directions under high flow rate. With the known anchor position, the DNA extension length is the difference between the anchor point and the bead centroid position under specific flow rates. The DNA extension length changes at different stretching forces (i.e., different flow rates). To determine the force, a plot of MSD values at different DNA extension lengths under a specific flow rate is fitted with a least-squares linear line (see Results and Discussion).
RecBCD translocation experiments
For RecBCD unwinding experiments, the 1 nM duplex DNA that was labeled with digoxigenin at only one end was attached to the surface so that the RecBCD enzyme could enter through the distal end of the DNA. Biotinyated RecBCD molecules were preincubated with streptavidin-coated beads in reaction buffer containing 10 mM DTT at 37°C water bath for 10 min before introduction into the chamber. The final volume of 60 μL containing the RecBCD/bead mixture (300 pM), ATP (35 μM), and ATP regeneration system (20 U/mL pyruvate kinase and 1.5 mM phosphoenolpyruvate; Sigma, St. Louis, MO) was flowed into the chamber to initiate unwinding at room temperature (22°C ± 1°C). The unwinding experiment was conducted under the optimal flow rate of 15 mL/h. The force experienced by the RecBCD-DNA-bead complex at this flow rate corresponds to ∼0.20 pN. Enzyme positions along the DNA in the unwinding time trace were obtained by calculating the MSD values of 40 frame duration (∼2 s) and converting the MSD values into DNA tether lengths (see Fig. 4). The unwinding rates were determined by the least-squares linear fits.
Results and Discussions
FTPM method
The FTPM experiment is based on the equipartition theorem. In this modified TPM experiment, we apply a small stretching force to the tethered beads by introducing a continuous flow into the multiplex TPM measurement. The stretching force can also be introduced through other means, such as using optical tweezers for latex beads or a magnetic field for magnetic beads. In the FTPM experiment, the bead's Brownian motion in the direction perpendicular to the force is suppressed by the stretching force. The surface-tethered DNA bead, which is pulled by this stretching force (F) to the extension length of l, is equivalent to a damped pendulum (17,20). At thermal equilibrium, the mean potential energy of the tethered bead in the direction perpendicular to the stretching force, , is equal to the thermal energy (), according to the equipartition theorem (where is the Boltzmann constant, and T is the temperature in Kelvin). Thus, the MSD (〈Δx2〉) of the tethered bead in the x-direction is directly proportional to the DNA extension length (l): (17). The linear relationship between the MSD of the bead's Brownian motion and DNA extension length is predetermined (with a slope of ). In contrast to the conventional TPM experiment, there is no need to conduct empirical calibration experiments for various DNA lengths in the FTPM experiment, as long as the force is fixed and determined. This predetermined slope also ensures a true linear relationship between the DNA extension length and the MSD of the bead's Brownian motion, suggesting that a much longer DNA substrate can be used.
In our FTPM experiment, the DNA-tethered beads were extended by a stretching force that was introduced by a continuous flow through a syringe pump. The anchor point where the DNA molecules were attached to the surface was determined by the mean position when the flow was introduced (under the same high flow rate) in opposing directions. The DNA extension length (l) was determined by the distance between the DNA anchor point and the bead centroid position under flow. Due to errors associated with the determination in the centroid positions of the beads under flow, there were errors in determining the DNA anchor position, which led to the error in determining the DNA extension length. The pulling geometry in Fig. 1 suggests that geometrical correction is required to account for the difference between the DNA extension length and the measured projected extension, which is affected by several factors such as the bead radius and the vertical position of the bead. Our experimental error in determining the anchor position is comparable to the error associated with the correction of the requisite angle of the DNA to the surface anchor point (both ∼8% relative error). Therefore, these errors do not allow us to make a quantitative, accurate correction to account for the deviation from the pulling geometry. The correlation coefficients (R2 value) of the linear least-squares fit in the MSD values/extension plot are the same (0.9913 for 4 mL/h flow rate) whether or not it passes through zero, within our measurement uncertainty. Previous studies with the similar flow-stretched geometry also considered this correction error to be negligible (21).
Figure 1.
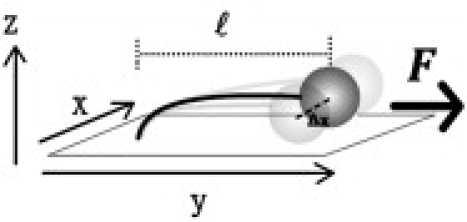
Schematic representation of the FTPM method. A stretching force (F) is applied to tethered beads along the y axis through a continuous flow. The bead Brownian motion along the x direction (Δx) is suppressed by a stretching force. Its MSD values are proportional to the DNA extension length (l) based on the equipartition theorem.
The FTPM measurement allows us to determine the bead centroid position as a function of time (x, y, t). Once we know the anchor position of the DNA molecules, we are able to directly determine both the DNA extension length (l, from y) and MSD values (〈Δx2〉, from x) at the same time. The magnitude of the stretching force () is determined from the MSD values of the beads (〈Δx2〉) and the DNA extension length (l) at a specific flow rate. By varying the magnitude of the flow rates, we can stretch the same duplex DNA molecules at different forces. The stretching force depends only on the flow rate and the size of the tethering bead. In our experimental geometry of the chamber volume ∼50 μL and bead size of ∼0.22 μm, the flow rate of 0–500 mL/h corresponds to the applied stretching force of 0–16 pN.
To verify the reliability to generate stretching forces (0–16 pN) accurately in our setup, we conduct the force (F) versus extension (l) curve on 4190 bp long duplex DNA molecules (see Fig. 2). The data can be fitted with a worm-like chain model, returning a persistence length of 44.13 ± 5.85 nm, and a contour length of 1314.23 ± 4.97 nm (1424.60 nm expected for B-form DNA). The fitted persistence length is consistent with previously reported values (20,22). The difference between the fitted and predicted contour lengths can be attributed to the errors associated with the DNA extension length measurement and the actual pulling geometry, as discussed above.
Figure 2.
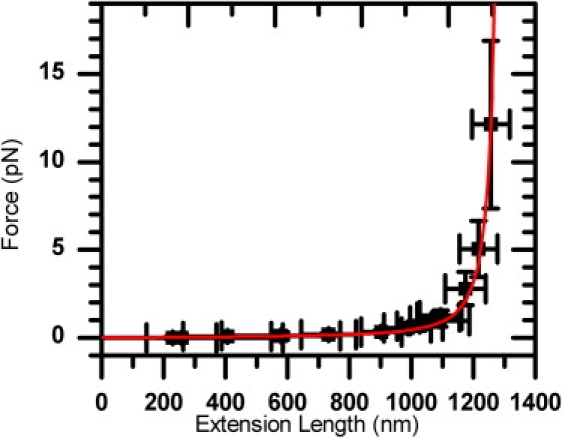
Force-extension curve of 4190 bp long dsDNA molecules tethered with 220 nm beads under various flow rates. This curve is fitted with a worm-like chain model to a persistence length of 44.13 (± 5.85) nm and contour length of 1314.23 (± 4.97) nm.
To determine the MSD values of the bead's Brownian motion, 200 frames were recorded and analyzed by means of image processing. This process was repeated for ∼80–100 beads at each DNA length and different flow rates. A histogram of these compiled MSD values was fitted with a Gaussian distribution with a peak MSD value and an SD. For example, 88 measurements were used in the analysis of 1551 bp long DNA at the flow rate of 15 mL/h (a stretching force of 0.20 pN), with a peak MSD value of 6215 nm2 and SD of 1631 nm2, as shown in Fig. 3. The SD of this Gaussian fit offers a measure of the precision of our FTPM measurement, i.e., how accurate the FTPM experiments are in determining the DNA length.
Because DNA extension length is a function of the applied force, we constructed an MSD-DNA extension length (l, in nm) plot (not shown) to validate its linear relationship at various forces. As predicted from the equipartition theorem, the MSD-DNA extension length relationship was fitted linearly at all stretching forces. For example, the fitted slope at 0.06 pN (4 mL/h) was 66.54 ± 4.40 nm, consistent with the prediction (kBT/F) of 67.83 nm. To facilitate comparisons with other methods, a plot of MSD-DNA tether length (in bp) is shown in Fig. 4. At fixed force, the DNA tether length and its extension length are described by the worm-like chain model at the force range used in FTPM, so the slope of the curve can still be predicted from the equipartition theorem. Consistent with the fact that the stretching force suppresses the bead's Brownian motion, the MSD values of bead's Brownian motion at a specific DNA length, as well as error bars of the MSD values, decrease as the forces increase from 0.06 pN to 16 pN. At forces larger than 0.50 pN, the error bars are rather small and insignificant, and the fitted slopes become very small.
Sensitivity, precision, and resolution of FTPM
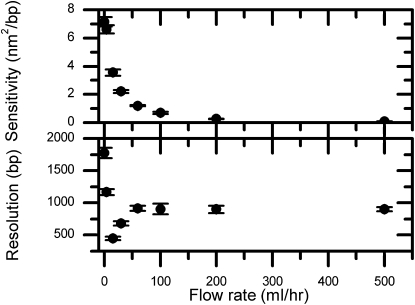
The ability to refer to DNA length accurately indicates the sensitivity of FTPM experiments. However, we are more interested in knowing which DNA length the measured MSD value corresponds to—500 bp or 550 bp (i.e., the resolution). Apparently, this resolution depends not only on the precision of the MSD measurement, but also on the stretching force used (i.e., the sensitivity). The larger the stretching force, the more the Brownian motion is suppressed, and thus the better is the precision. However, the larger the stretching force, the smaller the slope of MSD-DNA tether length plot (resulting in less sensitivity, as shown in Fig. 4). The small slope of the MSD-DNA tether length plot suggests little differences for the MSD values at various DNA lengths, and thus demolishes the discrimination power of DNA length. Therefore, the best resolution of the FTPM experiments would result from the better precision of the MSD measurement (i.e., the smaller the error bars), and the larger the difference of the bead's Brownian MSD values among DNA lengths (i.e., the larger slope). We previously defined the precision as the error bar of MSD values in the MSD-DNA length plot, which is determined by the SD in the Gaussian fitting of Fig. 3. The sensitivity of FTPM experiments is defined by the slopes of the MSD-DNA tether length plot (see Fig. 4), as the slope describing how the magnitude of MSD value changes with DNA length. The sensitivity of FTPM (the slope, kBT/F) is a function of the stretching force and temperature, and decreases with the increasing force, as shown in Fig. 5 (top). However, the bead's Brownian motion is much noisier at smaller forces (as seen in the larger error bar, thus precision is less). Therefore, the best resolution of an FTPM experiment is a compromise between the sensitivity and the precision of the bead's Brownian motion. We used the ratio of precision (error bar in Fig. 4) and sensitivity (slope of Fig. 4) to determine the condition for the best resolution of the FTPM experiments (Fig. 5, bottom). As shown in Fig. 5, the best resolution is achieved at an intermediate stretching force (15 mL/h, ∼0.20 pN). Fig. 5 was constructed using 4190 bp long DNA, and choices of other DNA lengths do not change the optimal condition that achieves the best resolution in our FTPM experiments.
Figure 5.
(Top) The slope of the MSD-DNA tether length plot (Fig. 4) defines the sensitivity (nm2/bp) of FTPM experiments at specific force. The higher the flow rate (the larger stretching force), the smaller the sensitivity. (Bottom) The resolution of FTPM experiments is defined as the ratio of sensitivity and the precision of each measurement. The precision is the width of the Gaussian fit in Fig. 3, or the error bars in Fig. 4. Resolution is a compromise between the sensitivity and precision of the bead's Brownian motion. This plot shows the resolution of various flow rates at 4190 bp long DNA. The best resolution is achieved at the flow rate of 15 mL/h, which corresponds to 0.20 pN in our geometry.
At this optimal flow condition (15 mL/h), our FTPM experiment has the resolution of 422 bp at 4190 bp long DNA, whereas in the conventional TPM experiment (at almost zero force), the resolution is fourfold worse (1771 bp). We used this empirically determined flow condition to conduct further FTPM experiments to achieve the best resolution. This optimal flow condition depends only on the bead size used, and therefore only needs to be determined once for the specific bead size.
Comparison with other stretching methods
There are several existing single-molecule methods that use stretching forces to monitor DNA length change during enzyme-catalyzed reactions (2,5,18,21,23,24). These methods monitor the change in the extension length of DNA molecules (l) either through the fluorescence dyes intercalated in duplex DNA (2,23,24) or through the position of the bead attached to the distal end of DNA (5,18,21). Since FTPM experiments can track the centroid positions of tethering beads, we can determine the DNA extension lengths and the MSD values of the same DNA tethers simultaneously for comparison purposes.
We compared the resolution of DNA length determined from the MSD measurement (FTPM) and from the extension measurements at various stretching forces (see Fig. 6 and Table 1). At the same low stretching force of 0.20 pN, the resolution of FTPM measurement (422 bp) is twofold better than that of the extension measurement (1035 bp) at a long DNA length of 4190 bp, and fivefold better at a short DNA length of 433 bp (56 bp vs. 247 bp). To accurately determine the DNA extension length, it is necessary to apply high stretching forces. Since the FTPM experiment and extension length measurement have different optimal forces, it might be useful to compare the resolutions at their own optimal conditions: high stretching force (16 pN) for the extension measurement, and low force (0.20 pN) for the FTPM experiment. At the long DNA length of 4190 bp, the resolutions of the two methods are almost identical (416 vs. 422 bp). However, at the shorter DNA length of 433 bp, the resolution for the FTPM method (56 bp) is more accurate than that of the extension method (128 bp). Therefore, FTPM can provide improved resolution at short DNA lengths compared to the stretching methods, and FTPM requires only minimal stretching forces of 0.20 pN. Since many enzyme-catalyzed reactions are force-dependent, the high forces (16 pN) that are required to achieve the best resolution in the extension measurement may present additional energy barriers for these enzymatic processes.
Figure 6.
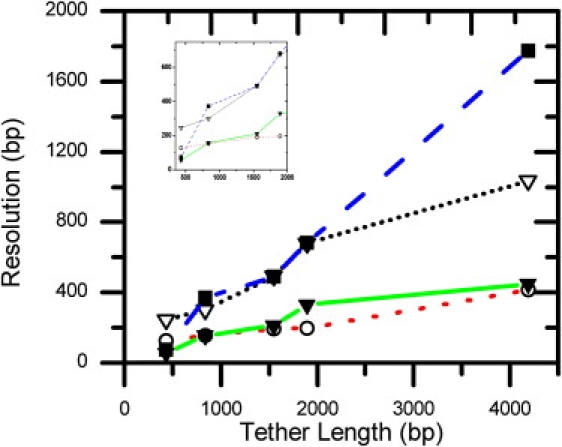
Comparison of the resolution of DNA length determination by DNA extension length, FTPM, and zero-force TPM experiments. ■ = resolution for MSD determination at zero force (resembling conventional TPM); ▾ = FTPM determination at its optimal force (0.20 pN); ∇ = extension length determination at low force (0.20 pN); and ○ = extension length determination at high force (16 pN).
Table 1.
Resolution (in bp) of DNA tether length determination
| DNA length (bp) | Extension |
MSD |
||
|---|---|---|---|---|
| F = 16 pN F = 0.20 pN | F = 0.20 pN F = 0 pN | |||
| 433 | 128 | 247 | 56 | 74 |
| 836 | 154 | 300 | 157 | 372 |
| 1551 | 194 | 494 | 211 | 491 |
| 1893 | 197 | 679 | 330 | 681 |
| 4190 | 416 | 1035 | 422 | 1771 |
Conventional TPM experiments are carried out at almost zero force range (<0.05 pN, no stretching force). We also compared the resolution of conventional TPM at zero force and FTPM at 0.20 pN force. Instead of analyzing the SD of bead centroid positions for the conventional TPM method, we analyzed the MSD values of the bead Brownian motion, as in our FTPM experiments. The resolutions at short DNA lengths were similar for 0.20 pN (FTPM) and 0 pN (conventional TPM) forces (56 vs. 74 bp resolution for 433 bp long DNA). However, the resolution at zero force became progressively worse as the DNA tether length increased, as previously reported. At the longest DNA length used (4190 bp), the resolution of FTPM was fourfold better than that of the conventional TPM (422 bp vs. 1771 bp). Therefore, FTPM provides improved resolution compared to the conventional TPM methods at long DNA lengths.
Overall improvement from the conventional TPM method
Regular TPM methods take advantage of the Brownian motion of submicron-sized beads, and correlate their Brownian motion to the DNA tethering length at the almost zero force region. The longer the DNA, the larger the Brownian motion is presumed to be. The dependence of Brownian motion magnitude and DNA tether length has been shown to be approximately linear in empirical observations (14), whereas other experiments have shown nonlinearity (15,16). A few image-processing methods are currently used to quantitatively describe the bead Brownian motion in conventional TPM experiments, such as the width of averaged bead motion (14), the mean excursion length of bead motion (16), and the SD of bead centroid positions (15); however, all of these methods require a series of measurements of known DNA lengths and bead Brownian motion at a specific bead size to establish the empirical calibration curve. Recently, Nelson et al. (16) developed a mathematical model based on a worm-like chain model to describe tethered DNA conformations in conventional TPM experiments. Using Monte Carlo simulation, they quantitatively described the bead motion at various DNA lengths. Their model is consistent with the reported data within experimental error, but their model also predicts a nonlinear relationship of bead mean excursion and DNA tether length, especially in the longer DNA length. This nonlinear dependence of DNA tether length places constraints on single-molecule TPM experiments, such as a required empirical calibration curve of bead Brownian motion and DNA length, and a DNA length limitation. Most conventional TPM measurements are carried out at short DNA length (<2 kb), where the nonlinearity is minimized.
In our simple modification to the existing TPM method, FTPM applies a small stretching force (∼0.20 pN) to suppress the bead's Brownian motion. Based on the equi-partition theorem, FTPM experiments measure the MSD values of the bead Brownian motion in the direction perpendicular to the stretching force, and predict a linear relationship between DNA length and MSD values. Therefore, FTPM offers several improvements over the conventional TPM method: 1), the theorem-based linear dependence of DNA tether length and the MSD values of bead Brownian motion; 2), the improved resolution of DNA tether length determination at long DNA lengths; and 3), the ability to use an extended range (>2 Kb) of DNA length substrates. These improvements can be achieved by simply applying a small stretching force without the need for complex instrumentation.
RecBCD unwinding experiment
We used the improved resolution of FTPM to revisit the motion of individual RecBCD helicases along duplex DNA. The E. coli RecBCD helicase/nuclease complex preferentially binds to blunt-end duplex DNA molecules, unwinds them, and generates ssDNA substrates for RecA binding in the homologous recombinational repair pathway. Among other DNA helicases, the RecBCD complex is unique in its high processivity and high unwinding rates. RecBCD contains two helicase subunits: RecB is a 3′–5′ helicase and RecD is a 5′–3′ helicase. The properties of the RecBCD complex can be regulated by an 8 nt DNA sequence, χ. The bipolar nature of the RecBCD helicase and its χ regulation have been shown to play a functional role in repairing double-strand DNA breaks in E. coli (1,2).
Bulk biochemical experiments showed that the χ recognition by RecBCD is dependent on the ratio of magnesium ions to ATP. In the presence of free magnesium ions (Mg2+ >> ATP), χ-specific DNA fragments are observed (25,26). Earlier single-molecule RecBCD experiments carried out under a saturating ATP condition (1 mM ATP and 2 mM Mg2+) showed that once RecBCD encountered, recognized, and responded to the χ sequence during its unwinding process, it paused a few seconds and lowered its unwinding rate (4,5). It was proposed (4) that either the whole helicase complex undergoes conformational changes at the χ site, or one of the helicase domains is turned off at the χ sequence. Despite the differences between the two models, it is thought that the pause and unwinding rate change at the χ site should occur under all ATP conditions. However, previous single-molecule TPM experiments carried out under the limiting-ATP condition (1 mM ATP, 1 mM ADP, and 1 mM Mg2+) did not detect that RecBCD paused or changed rates at the χ site, even though the enzyme should have recognized the χ sequence with the statistical efficiency measured from the bulk ensemble measurements (6). Spies et al. (4) reconciled these observations by suggesting that the pause duration may be too short to detect, or the unwinding rate change may be insignificant at the low ATP conditions in (6). Handa et al. (5) also proposed that χ recognition by RecBCD requires high ATP conditions. With the improved resolution of the FTPM method, we reinvestigated the RecBCD motion on χ-containing DNA under ATP-limited conditions in the presence of excess magnesium ions, and tested whether χ recognition by RecBCD required saturating ATP concentrations.
Our experiment is similar to the previous single-molecule TPM experiment studying RecBCD motion (6) except that the 1.5 kb DNA is replaced by a 4.1 Kb DNA containing a triple χ repeat sequence. In this experiment, the linear 4.1 kb long duplex DNA is anchored at the surface, with its distal end free of labels so that it can be bound by a RecBCD/bead complex. The χ sequence is located ∼1.4 Kb from the surface with the correct orientation to ensure recognition by RecBCD (see Fig. 7 A). The unwinding reaction is initiated by introducing a continuous flow (15 mL/h, ∼0.20 pN) with buffer containing 35 μM ATP and 1 mM Mg2+. As a result of the flow, any unbound RecBCD/beads are washed out from the chamber. Some RecBCD/bead complexes bound to the free end of the anchored dsDNA molecules will begin to translocate and unwind along dsDNA substrates. As an active RecBCD molecule binds to and unwinds a single dsDNA molecule, the tethered bead's Brownian motion will start to decrease. The decreasing Brownian motion directly measures the unwinding and translocating process catalyzed by RecBCD against the flow direction. The centroid positions of the tethering beads are determined to nanometer precision using image processing, and the MSD values are calculated. The time course of the MSD values is then converted into the DNA tether length by the slope of Fig. 4 to calculate the RecBCD unwinding rate (as shown in Fig. 7). Since the interval between frames is around 63 ms and the MSD calculation is based on 40 frames, the current temporal resolution in our FTPM experiment is around 2 s. The spatial resolution of the FTPM experiments increases from 422 bp at long DNA (4.1 Kb) to 56 bp at short DNA length (433 bp), as listed in Table 1.
Figure 7.
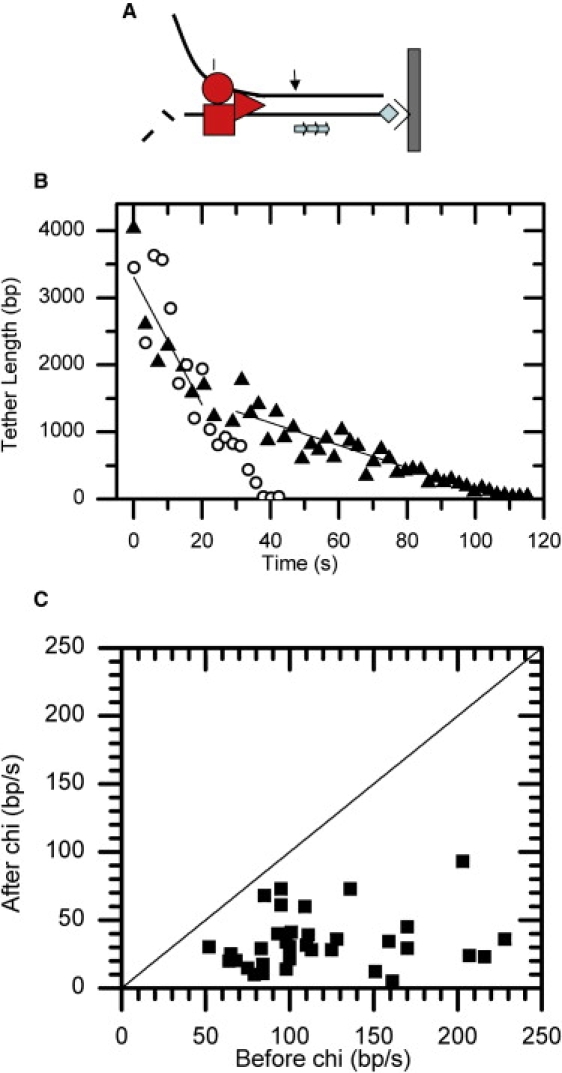
(A) Schematic drawing of a RecBCD helicase translocate along an immobilized DNA molecule. (B) Unwinding time courses of RecBCD helicase moving along a 4.1 kb long duplex χ-containing DNA (▴) and χ-zero DNA (○) were determined by FTPM. The χ sequence is located at 1404 bp from the surface. The arrow indicates the first χ position where RecBCD would encounter the χ sequence. The solid line represents the linear fittings of unwinding rates before (95 bp/s) and after (17 bp/s) the χ position for χ-containing DNA. The unwinding rate for a χ-zero DNA does not change (93 bp/s). This experiment was carried out at 35 μM ATP and 1 mM magnesium, below the KM of ATP for RecBCD. (C) Of 55 RecBCD unwinding processes observed, 34 lowered the translocation rate at the χ sequence. The plot includes data from both 1967 bp and 4141 bp DNA substrates containing the χ sequence. All of the rate changes at χ position lead to a slower rate, as they all fall into the bottom-right section.
As shown in Fig. 7 B, under the limited-ATP condition (35 μM), individual RecBCD molecules translocated along χ-zero duplex DNA substrates at a uniform rate (open circles, unwinding rate of 93 bp/s). This translocation rate is similar to what was reported in previous single-molecule experiments at similar ATP and magnesium concentrations without any external force, within the experimental uncertainty (6). Since earlier optical-tweezers experiments indicated that the RecBCD translocation rate is constant up to 6 pN (27), it is unlikely that a small 0.20 pN force could have an additional effect on the translocation rate. However, when RecBCD translocates along a χ-containing DNA at the same limited-ATP condition (Fig. 7 B, solid triangle), the unwinding curve cannot be fitted with a single unwinding rate. The unwinding curve is best fitted with two unwinding rates (95 bp/s from 0 to 20 s, and 17 bp/s from 30 to 110 s), with the unwinding rate changes around the χ position within our experimental resolution (∼200 bp for 1.4 Kb long DNA). This translocation rate change upon the χ site is similar to what was observed before under the saturating ATP condition (4,5), suggesting that the RecBCD molecules had recognized the χ sequence and had been modified by that sequence. We also used a χ-containing DNA of 1.9 kb length, with χ located 1.1 kb from the surface, and observed a similar translocation rate change at the χ sequence (data not shown). Of the 55 RecBCD molecules we studied at these two different χ DNA lengths, 34 (∼60%) exhibited the translocation rate change upon χ recognition at 35 μM ATP, and 40% of them translocated along χ DNA with a uniform rate without change. Earlier single-molecule in vitro experiments using similar triple-χ repeat DNA substrates observed that ∼50% of RecBCD changed its translocation speed upon χ sequences (4,5), consistent with our χ-recognition efficiency reported here. When we compared the RecBCD translocation rate before and after encountering the χ sequence, we found that all 34 RecBCD molecules lowered their rates upon recognizing χ, as shown in Fig. 7 C. The fact that RecBCD changes its unwinding rate at the χ position under the limited-ATP (35 μM) condition directly proves that a saturating ATP concentration is not required for χ recognition by RecBCD, contrary to what was proposed previously (4,5).
Previous single-molecule RecBCD experiments performed under the saturating ATP condition also observed a pause with a mean duration ∼5 s at the χ site (4). Unfortunately, the resolution of our current FTPM experiments (∼200 bp at 1.4 Kb long DNA) prevents us from unambiguously identifying the pause (and thus the pause duration) of RecBCD molecules upon χ recognition. This is likely due to the slow translocation rates in the low ATP condition used here. Experiments done under saturating ATP conditions have higher translocation rates (>500 bp/s) and thus can distinguish the run state and pause state clearly. The previous TPM experiment (6) likely did not have sufficient resolution to detect the rate change of RecBCD unwinding χ DNA under reaction conditions similar to that used here. The FTPM experiments presented here used longer DNA with improved resolution, allowing more accurate determination of pre-χ unwinding rates. Together with the improved resolution at the χ position (see Fig. 6), the FTPM experiments allow us to determine the rate change clearly.
In addition to its helicase activity, RecBCD is also responsible for degrading invading DNA sequences by means of its nuclease activity. Recognition of χ allows RecBCD to switch from a destructive mode to a constructive mode in repairing E. coli genomic DNA. The crystal structure suggests that the 3′-entry single strand unwound by RecB is threaded through the tunnel in RecC for the scanning DNA sequence to allow χ recognition (28). Consistent with this view, our data imply that any active DNA translocation by RecBCD helicase will allow χ recognition, regardless of the ATP concentrations used.
Conclusions
The FTPM technique is based on the equipartition theorem and provides improved resolution to study many protein-DNA interactions. The equipartition theorem dictates that the MSD of tethered bead Brownian motion is linearly proportional to its DNA tether length, with the known slope at specific bead size and stretching force. The best resolution in FTPM experiments occurs at an intermediate stretching force (∼0.20 pN for 220 nm beads), obviating the need for high stretching forces to accurately determine DNA extension lengths, as required in other stretching methods. The multiplex nature of FTPM, as opposed to other optical optical-tweezers methods, allows easier and faster data acquisition for single-molecule studies. With the improved resolution at long DNA lengths, we were able to accurately measure the translocation rate of RecBCD helicases along 4.1 kb DNA molecules containing the χ sequence. We determined that RecBCD lowered its translocation rate at the χ sequence, even at limited ATP concentrations. This result directly shows that χ recognition by RecBCD does not require saturating ATP concentrations. Other means of applying stretching forces (such as magnetic forces) may be able to provide better instrument stability and further improve the FTPM measurements.
Acknowledgments
The authors thank Jeff Gelles for providing Labview programs for tracking, and Wen-Han Kao for preparing Fig. 1.
The earlier work was supported by grants from the Canadian Foundation for Innovation, McGill University Start-up Fund, National Sciences and Engineering Research Council of Canada (to H.-W.sL.), and later from National Taiwan University and National Science Council of Taiwan (to H.-W.L.).
References
- 1.Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 2.Dillingham M.S., Spies M., Kowalczykowski S.C. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 3.Taylor A.F., Smith G.R. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 4.Spies M., Bianco P.R., Dillingham M.S., Handa N., Baskin R.J. A molecular throttle: the recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 5.Handa N., Bianco P.R., Baskin R.J., Kowalczykowski S.C. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after χ recognition. Mol. Cell. 2005;17:745–750. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Dohoney K.M., Gelles J. χ-Sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 7.Guerra R.F., Imperadori L., Mantovani R., Dunlap D.D., Finzi L. DNA compaction by the nuclear factor-Y. Biophys. J. 2007;93:176–182. doi: 10.1529/biophysj.106.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mumm J.P., Landy A., Gelles J. Viewing single λ site-specific recombination events from start to finish. EMBO J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pouget N., Turlan C., Destainville N., Salome L., Chandler M. IS911 transpososome assembly as analysed by tethered particle motion. Nucleic Acids Res. 2006;34:4313–4323. doi: 10.1093/nar/gkl420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian H. A mathematical analysis for the Brownian dynamics of a DNA tether. J. Math. Biol. 2000;41:331–340. doi: 10.1007/s002850000055. [DOI] [PubMed] [Google Scholar]
- 11.Schafer D.A., Gelles J., Sheetz M.P., Landick R. Transcription by single molecules of RNA-polymerase observed by light-microscopy. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 12.Tolic-Norrelykke S.F., Engh A.M., Landick R., Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 13.Vanzi F., Broggio C., Sacconi L., Pavone F.S. Lac repressor hinge flexibility and DNA looping: single molecule kinetics by tethered particle motion. Nucleic Acids Res. 2006;34:3409–3420. doi: 10.1093/nar/gkl393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H., Landick R., Gelles J. Tethered particel motion method for studying transcript elongation by a single RNA-polymerase molecule. Biophys. J. 1994;67:2468–2478. doi: 10.1016/S0006-3495(94)80735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pouget N., Dennis C., Turlan C., Grigoriev M., Chandler M. Single-particle tracking for DNA tether length monitoring. Nucleic Acids Res. 2004;32:e73. doi: 10.1093/nar/gnh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson P.C., Zurla C., Brogioli D., Beausang J.F., Finzi L. Tethered particle motion as a diagnostic of DNA tether length. J. Phys. Chem. B. 2006;110:17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 17.Strick T.R., Allemand J.F., Bensimon D., Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Oijen A.M., Blainey P.C., Crampton D.J., Richardson C.C., Ellenberger T. Single-molecule kinetics of λ exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 19.Young E.C., Berliner E., Mahtani H.K., Perezramirez B., Gelles J. Subunit interactions in dimeric kinesin heavy-chain derivatives that lack the kinesin rod. J. Biol. Chem. 1995;270:3926–3931. doi: 10.1074/jbc.270.8.3926. [DOI] [PubMed] [Google Scholar]
- 20.Smith S.B., Finzi L., Bustamante C. Direct mechanical measurements of the elasticity of single DNA-molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 21.Kim S.J., Blainey P.C., Schroeder C.M., Xie X.S. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat. Methods. 2007;4:397–399. doi: 10.1038/nmeth1037. [DOI] [PubMed] [Google Scholar]
- 22.Bustamante C., Marko J.F., Siggia E.D., Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 23.Prasad T.K., Yeykal C.C., Greene E.C. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J. Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 24.Bianco P.R., Brewer L.R., Corzett M., Balhorn R., Yeh Y. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 25.Anderson D.G., Churchill J.J., Kowalczykowski S.C. χ-Activated RecBCD enzyme possesses 5′->3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by χ is not simply ejection of the RecD subunit. Genes Cells. 1997;2:117–128. doi: 10.1046/j.1365-2443.1997.1130311.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor A.F., Smith G.R. Strand specificity of nicking of DNA at χ-sites by RecBCD enzyme—modulation by ATP and magnesium levels. J. Biol. Chem. 1995;270:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- 27.Perkins T.T., Li H.W., Dalal R.V., Gelles J., Block S.M. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]