Abstract
P2X receptors are non-selective cation channels operated by extracellular ATP. Currently, little is known concerning the functions of these receptors during development. Previous work from our lab has shown that zebrafish have two paralogs of the mammalian P2X3 receptor subunit. One paralog, p2rx3.1, is expressed in subpopulations of neural and ectodermal cells in the embryonic head. To investigate the role of this subunit in early cranial development, we utilized morpholino oligonucleotides to disrupt its translation. Loss of this subunit resulted in craniofacial defects that included malformation of the pharyngeal skeleton. During formation of these structures, there was a marked increase in cell death within the branchial arches. In addition, the epibranchial (facial, glossopharyngeal, and vagal) cranial sensory ganglia and their circuits were perturbed. These data suggest that p2rx3.1 function in ectodermal cells is involved in purinergic signaling essential for proper craniofacial development and sensory circuit formation in the embryonic and larval zebrafish.
Keywords: P2X, Zebrafish, Morpholino, Chondrogenesis, Sensory ganglia
Introduction
Purinergic receptors are expressed in nearly every tissue in adult animals [1], and the physiological roles that these receptors play in ATP signaling are an area of major interest. Extracellular ATP signaling occurs via P2 receptors, which fall into two classes: the metabotropic P2Y and the ionotropic P2X receptors [2]. Most investigations into purinergic signaling have utilized tissues derived from post-natal animals, resulting in little insight into the role(s) that ATP plays during development. Recent reports show that purinergic signaling mediated via P2Y receptors plays a role in the development of the eye [3] and regulates the proliferation of neural progenitors in the retina [4] and cortex [5]. Surprisingly, there have only been a few reports describing the expression of P2X receptors in embryonic animals, and still fewer that have investigated any functional aspects of their presence (for recent overviews, see [1, 6–9]). However, the expression of these receptors in embryonic tissue suggests the hypothesis that P2X receptors are mediating extracellular signaling by ATP during embryogenesis and can thus expand the potential targets through which this purine may regulate important developmental processes.
Activation of P2X receptors can result in the depolarization of cells [10, 11] and, for some members of the gene family, directly cause rises in intracellular calcium concentrations due to their relatively high permeability to this ion [12]. These properties make P2X receptors exquisitely poised to induce or facilitate release of bioactive molecules involved in cell–cell signaling. Such intercellular signaling is a critical component of craniofacial development [13–16]. These signals, which take place between cells present in one or more of the germ layers, are mediated via direct cellular contact or by intercellular signals that can act locally and/or distally from their source. The molecules that mediate such signaling are varied in nature, ranging from large glycoproteins to small lipid or amino acid derivatives [17–19]. A common feature of these molecules is that they must be released into the extracellular space through the action of secretagogues. The presence of P2X receptors in these tissues indicates that extracellular ATP could perform such a function.
We have previously reported the expression of P2X receptor subunit genes in the embryonic zebrafish [20]. Based on those findings, we have begun to investigate the roles that these receptors play during embryonic development using zebrafish as a model organism. In a recent report, we showed that a P2X receptor subunit gene, p2rx3.1, is expressed by non-neuronal cells in the head of zebrafish embryos [21]. In this study, we determined that this non-neural gene expression is in cranial ectodermal cells and that reduction of purinergic signaling in this population via knockdown of the P2X3.1 subunit leads to defects in both skeletogenesis and neurogenesis in the head.
Materials and methods
Fish husbandry Zebrafish strains used in this study included AB, Tg[p2rx3.2:eGFP]sl1 [21] and Tg[fli1:eGFP]y1 [22]. Embryos were produced by pairwise matings and were raised and cared for as described by Westerfield [23] and staged according to hours post-fertilization (hpf) [24]. Embryos used for in situ hybridization, immunocytochemistry and microscopy were treated with 0.003% phenylthiourea to reduce pigmentation. For our experiments, special care was taken to stage the embryos using both the eye-to-otic-vesicle distance as well as the angle formed by the eye and ear (per [24]) so that any gross developmental delay due to injection of MO and/or RNA could be accounted for.
In situ hybridization Embryos/larvae were anesthetized in 0.4% tricaine (3-amino-benzoic acid ethyl ester), fixed in 4% paraformaldehyde for 24 h and then stored in 100% methanol at −20°C. For in situ hybridization, the embryos were processed as previously described [21]. Plasmids were linearized with the appropriate restriction enzyme and cRNA preparation was performed using the AmpliScribe Transcription Kit (EpiCentre, Madison, WI). cRNAs were synthesized in the presence of digoxigenin-UTP (Roche) or modified by covalent modification using either d-nitrophenol (DNP) or fluorescein with the LabelIt Kit (Mirus Bio Corporation, Madison, WI). cRNAs were incubated in the presence of whole embryos at 65°C overnight in hybridization buffer (50% formamide, 5X SSC, 50 μg/ml heparin, 500 μg/ml tRNA, 9.2 mM citric acid and 0.1% Tween). Embryos were washed and incubated with alkaline-phosphatase-conjugated anti-DNP (Vector Laboratories, Burlingame, CA), anti-digoxigenin, or anti-fluorescein antibody (Roche). For cryosectioning, embryos were embedded in 1.5% agar/30% sucrose and frozen in 2-methyl-butane chilled by liquid nitrogen. Ten-micrometer-thick transverse, serial sections were cut using a cryotome, collected on glass slides and mounted in 75% glycerol for microscopy and storage.
Immunocytochemistry Embryos/larvae were anesthetized as described above and fixed in 4% paraformaldehyde for 3 h at room temperature and washed 3 × 5 min in phosphate-buffered saline/0.2% Triton X-100 (PBS-Triton). After incubation in a blocking solution (10% normal goat serum in PBS-Triton) for 1 h at room temperature, embryos were incubated in primary antibody (in 2% goat serum/PBS-Triton) overnight at 4°C. Anti-HuC antibody (16A11, Molecular Probes, Eugene, OR), zn-12, zn-8 antibodies (obtained from the Developmental Studies Hybridoma Bank at U. Iowa) or anti-GFP antibody (Molecular Probes) were used at dilutions of 1:1,000, 1:200, 1:200, and 1:500, respectively. Embryos were washed extensively in PBS-Triton and antibody:antigen complexes visualized using a secondary antibody (1:500) conjugated with either horseradish peroxidase (Sigma-Aldrich, St. Louis, MO), Alexa 488 or Alexa 594 (Molecular Probes). For brightfield imaging, embryos were mounted in 3% methylcellulose and images were captured with a Nikon TE200 inverted microscope equipped with a Nikon DN100 digital camera. For confocal microscopy, embryos/larvae were mounted in 2% agarose and covered with fish water. Confocal images were obtained on an Olympus FV500 or FV1000 microscope using long-working-distance water immersion lenses. Images collected in the z-dimension were collapsed into one maximal intensity projection or were rendered into three dimensions (MetaMorph, Universal Imaging Corp). Final brightness and/or contrast values of images were manipulated in Adobe Photoshop 6.0.
Morpholino injections Morpholino oligonucleotides (MOs; GeneTools, Philomath, OR, or Open Biosystems, Huntsville, AL) were designed against three regions of the p2rx3.1 receptor subunit mRNA. The first anti-sense MO (MO-start: t gaa gcc cag cac tct ggg agc cat) was designed against the first 25 base pairs of the open-reading frame including the translation start site; the corresponding mismatch oligonucleotide was t gaa Ccc Gag caG tcA ggg agc Gat, with capital letters indicating substitutions. The second and third MOs were designed against 25 base-pair regions of the gene that includes the splice donor (I9E10: a caa tca tct tac caa tcc cac tga) and acceptor (E10I10: tta aac ttg cct gcc tgc aaa tag a) sites of exon 10. MOs were dissolved in 0.1 M KCl, 20 mM HEPES (pH 7.4) and 0.01% phenol red and injected into single-cell embryos. Two to five nl were injected using a Picospritzer III (General Valve Corporation, Fairfield, NJ) attached to a broken capillary. Post injection, embryos were allowed to develop in fish water at 28.5°C.
RNA synthesis for microinjection A plasmid containing the cDNA encoding rp2rx3 [25] was linearized with EcoR1, capped RNA transcribed in vitro using the T7 Message Machine kit and poly-adenylated using a Poly(A) tailing kit (Ambion Inc., Austin, TX). RNA was dissolved in diethylpyrocarbonate-treated distilled H2O and mixed 1:1 with the previously described injection buffer. Approximately 20–50 pg of RNA was injected into each single-cell embryo. For MO/RNA co-injections, embryos were first injected with the MO and then re-injected with the RNA.
Single embryo RNA isolation/cDNA production RNA was isolated from single zebrafish embryos utilizing the Chomczynski single-step RNA isolation method [26]. cDNA was made from total RNA utilizing the PowerScript Reverse Transcriptase Kit (Clontech Inc, Mountain View, CA).
Cartilage staining Five days post-fertilization (dpf) control or MO-injected larvae were anesthetized as described above and fixed in 4% paraformaldehyde for 2 days. They were re-hydrated in 50% ethanol for 24 h and 100% ethanol for 24 h. They were placed in 0.03% Alcian Blue (Fisher Scientific) in 70% ethanol/30%acetic acid for 24 h at room temperature and subsequently digested in 0.5 mg/ml trypsin at 37°C for 1 h and bleached in a solution of 6% hydrogen peroxide/1% KOH for 1 h. They were washed twice with PBS (pH7.5) and fixed in 4% paraformaldehyde overnight. Embryos were mounted in 2% methylcellulose and photos taken using a Nikon DN-100 digital camera attached to a zoom stereomicroscope.
Results
Cranial ectodermal cells express p2rx3.1
Previous work from our lab has shown that the P2X receptor subunit gene p2rx3.1 is expressed in the head of 12 hpf embryos in two distinct populations of cells [21]. The first consists of the neurons that contribute to the trigeminal ganglia (gV) and the second is a more diffuse non-neuronal group.
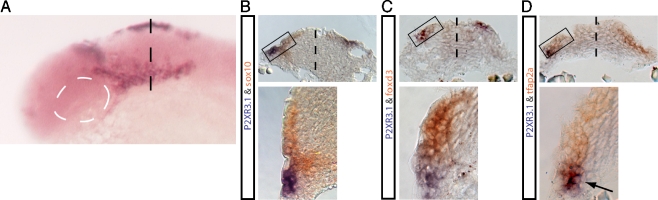
The non-neuronal p2rx3.1+ cells were located lateral to the neural plate and extended from the nascent eye field to the region of the developing trigeminal ganglia (Fig. 1a). This expression pattern is similar to that of the cranial neural crest, but could also be accounted for by tissues such as the ectoderm or mesoderm. To investigate which of these cell types was responsible for the non-neural expression of p2rx3.1, we first examined the co-expression of this gene with that of two transcription factors, sox10 and foxd3, that are selectively expressed by neural crest cells early in development [27, 28]. At 12 hpf, p2rx3.1+ cells (purple) were ventral to and not co-labeled with either sox10 or foxd3 (brown; Fig. 1b and c). These findings rule out the possibility that p2rx3.1 is expressed by the cranial neural crest, whereas the presence of p2rx3.1 mRNAs in the lateral margin of the head suggested that ectodermal cells might be the source of expression. To test this possibility, we next investigated whether p2rx3.1 mRNA was co-localized with that of tfap2a, a transcription factor expressed by both cranial ectoderm and neural crest [29]. At 12 hpf, two distinct classes of cells expressing these markers were observed: those that only expressed tfap2a (brown) and were seen in the same location as the sox10- and foxd3-expressing neural crest cells, and a ventrolateral population that expressed both p2rx3.1 and tfap2a (Fig. 1d). From these data, we conclude that p2rx3.1 is expressed by lateral cranial ectodermal cells.
Fig. 1.
The p2rx3.1 gene is expressed in cranial ectoderm at 12 hpf. Whole-mount in situ hybridization shows p2rx3.1 expression (purple) in the head at 12 hpf (a). The dotted white line shows position of eye field, the dotted black line is approximate level of transverse sections shown in panels (b–d). In b–d, the top images are of complete transverse section, with the dotted line indicating the midline and the box outlining magnified region shown below. Brown labeling is sox10 in b, foxd3 in c, and tfap2a in d. Purple labeling in all three panels is p2rx3.1, and arrow in d points to double-labeled cells
P2X3.1 function is required for normal craniofacial development
The expression of a P2X receptor subunit by the lateral ectoderm suggests that purinergic signaling could be occurring in or around this site beginning about 12 hpf. Many studies have shown that the ectoderm plays an instructive role in craniofacial development, as well as providing progenitor cells that give rise to many of the peripheral sensory neurons of the head [30–33]. Therefore, we set out to test the hypothesis that perturbing p2rx3.1 function would disrupt craniofacial development and sensory neurogenesis.
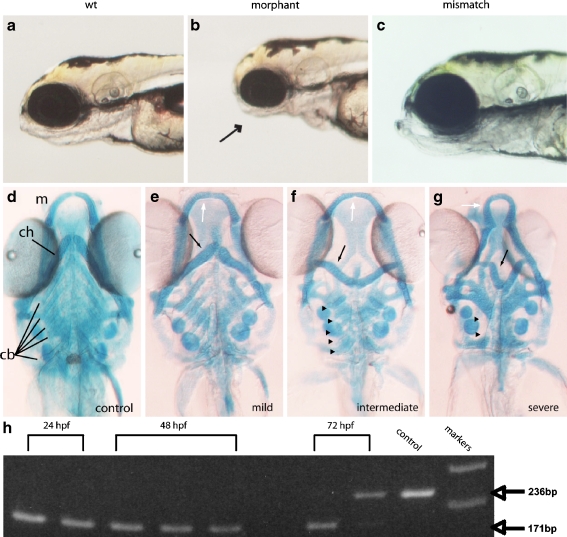
The p2rx3.1 gene is composed of 13 exons and exists as a single copy in the zebrafish genome (data not shown) making it amenable to the use of morpholino-oligonucleotide (MO)-mediated knockdown. We therefore designed and tested the effects of MOs that were directed against the p2rx3.1 gene. The first MO, MO-start, was designed against the first 25 base pairs of the open-reading frame of the p2rx3.1 gene so as to prevent translation of the P2X3.1 protein. In MO-start-injected embryos, approximately two-thirds (n = 454/698 injected) exhibited a distinct morphological phenotype that was characterized by a smaller head. This phenotype was first discernible between 36 and 48 hpf and fully apparent by 72 hpf and by 5 dpf, brightfield visual inspection demonstrated that the lower jaw and branchial arch regions were malformed in comparison to WT siblings (Fig. 2a and b). Upon inspection of other structures including the eyes, otocysts, pectoral fins, and notochord, we determined that the gross defects were limited to the head/pharyngeal region of the larvae. This phenotype was not observed when a control (five mismatched bases) MO [34] (Fig. 2c) or buffer alone (data not shown) was injected.
Fig. 2.
Loss of p2rx3.1 signaling results in defective pharyngeal skeleton formation by 5 dpf. Panel a shows a brightfield micrograph of a wild-type embryo, panel b is a morphant larva injected with MO-start, and panel c shows a larva injected with a control (mismatch) MO. The arrow points to the underdeveloped pharyngeal region in the morphant. Panel d shows a 5-dpf Alcian-Blue-stained control animal. Panels e–g show 5-dpf morphant larvae with differing degrees of cartilage deformations; e is a morphant larva with mild defects seen in the mandible (white arrow) and ceratohyal (black arrow) but with normal ceratobranchials, whereas f shows a morphant with more severe defects in the mandibular (white arrow) and ceratohyal (black arrow) cartilages but little disruption of ceratobranchials (arrowheads). However, in g, a morphant larvae shows severe affects on all pharyngeal cartilages- mandible (white arrow), ceratohyal (black arrow) as well as the near-total loss of ceratobranchial cartilages (arrowheads; m mandible, ch ceratohyal, cb ceratobrachials). PCR-mediated assessment of splice-morpholino efficacy is shown in panel h. cDNA was prepared from individual embryos at either 24, 48, or 72 hpf. Bands are PCR products generated from reactions containing either wild-type or MO-e10i10 morphant embryo-derived cDNAs using oligonucleotide primers that target sequences in exons 9 and 11. The distance spanning the primers in a wild-type cDNA containing exon10 is 236 bp, whereas a cDNA lacking exon 10 is 171 bp. By 72 hpf, the MO effects are being lost, most probably due to intracellular dilution brought about through multiple cell divisions, and properly spliced mRNA can be detected (upper band in second 72-hpf lane). Markers are 100-bp ladders corresponding to 200 and 300 bp
Knockdown of P2X3.1 results in defective pharyngeal skeleton development
One explanation for the reduced head size in p2rx3.1 morphants could be defective development of the cartilages that form the lower jaw and arches. To test this hypothesis, MO-injected larvae were fixed at 5 dpf and stained using Alcian Blue to visualize the cartilages of the developing upper and lower jaws. When compared to control larvae (Fig. 2d), staining revealed that morphant embryos had morphological alterations in most of the jaw and branchial arch cartilages (MO-start, n = 59/87; Fig. 2e–g). In all morphants (n = 59), defects were most apparent in the cartilages derived from the first and second branchial arches, whereas in a smaller percentage of morphants (n = 11/59) morphological changes in cartilages derived from branchial arches 3–7 were observed as well (Fig. 2g).
Examination of the first-arch derivatives (Meckel’s and palatoquadrate cartilages) in morphant embryos revealed that both of these components did not extend as far anteriorly as in controls (Fig. 2d–g). In addition, Meckel’s cartilage was often fused at the midline and exhibited a ‘flattened’ morphology. The ceratohyal cartilages (derived from the second arch) showed more dramatic effects (Fig. 2e–g). In some morphants (n = 48/59), they did not project rostrally but instead projected transversely (Fig. 2e and f). In more severely affected morphants, they were inverted and projected caudally (n = 11/59; Fig. 2g). Additionally, in these larvae, the ceratobranchial cartilages, derived from branchial arches 3–7, displayed phenotypes that ranged from a slight shortening (n = 34/59; Fig. 2f) to a complete loss (n = 11/59; Fig. 2g). Defects in cartilage formation were specific to the jaw and arches, as the neurocranium and pectoral fin cartilages appeared normal (data not shown).
To verify the role of this receptor subunit in craniofacial development, we examined the ability of additional MOs to generate the same phenotype and tested whether the morphant phenotype could be rescued by injecting RNA encoding a rat (r) P2X3 receptor subunit. To determine if we could phenocopy the morphological defects observed with MO-start, we designed MOs that disrupted protein function by interfering with the appropriate splicing of the p2rx3.1 pre-mRNA. Previous work from our lab has shown that exon 10 of p2rx genes encodes a major portion of the second transmembrane domain that is critical for forming the ion-permeation pathway of the receptor [35]. This same domain is also essential for subunit assembly into a functional receptor complex [36]. Thus, mRNAs lacking this exon encode proteins that are unable to assemble into functional receptors. To generate such mRNAs, we designed two splice MOs that were complementary to the splice donor (M0-i9e10) and acceptor (MO-e10i10) sites of exon 10. Injection of these MOs into embryos should result in exclusion of exon 10 from mature mRNAs during splicing of the p2rx3.1 hnRNAs. RT-PCR analysis of cDNA from individual MO-injected embryos verified that each splice MO successfully prevented inclusion of exon 10 into mRNA (results for MO-i9e10 shown in Fig. 2h; MO-e10i10 data not shown), and injection of either MO into embryos yielded morphant phenotypes indistinguishable from that of MO-start (e.g., MO-i9e10 = 54/87). Our next step was to carry out rescue experiments using RNA encoding the rP2X3 receptor subunit. This subunit has functional properties very similar to those of P2X3.1 [25, 37, 38], suggesting that it could act as a substitute in cells lacking the native subunit and does not contain target sequence for any of the MOs used in this study. In larvae co-injected with rP2X3 RNA and any of the individual p2rx3.1 MOs, a partial rescue of the morphant phenotypes, including cartilage development, was observed (Table 1). We did not observe any phenotype when rP2X3 RNA was injected alone (n = 89/89). Taken together, these findings demonstrate that loss of p2rx3.1 function specifically perturbs craniofacial development.
Table 1.
MO rescue by rat P2X3 mRNA
| MO-start | Splice MO | rP2X3 RNA | n | % affected |
|---|---|---|---|---|
| + | − | − | 98/179 | 55 |
| + | − | + | 42/179 | 24* |
| − | + | − | 58/150 | 39 |
| − | + | + | 23/139 | 17* |
*p < 0.0001 compared to MO alone
Cartilages from 5 dpf embryos were stained with Alcian Blue and examined using brightfield microscopy. Embryos were scored as disrupted if any branchial cartilages were not present, reduced in size or did not form correct orientations. Between 4–8 ng of MO was injected into each embryo; for co-injection experiments RNA encoding the rP2X3 was injected at 20–50 pg/embryo. Statistical analysis was done using Fisher’s exact chi-square test
Loss of p2rx3.1 signaling results in aberrant neural crest behaviors in the branchial arches
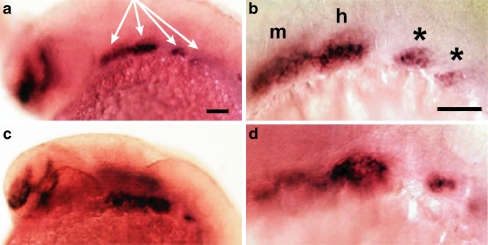
Defects in jaw cartilage development often arise from perturbations in migration or differentiation of cranial neural crest cells [39]. Cells destined to become chondrocytes (pre-chondrogenic neural crest cells) are located in the branchial arches and express the transcription factor dlx2a [40, 41]. These cells typically migrate laterally and then ventrally from the dorsal neural plate region into the branchial arches, where they condense to form mesenchymal branchial masses. They then proliferate, and begin the processes of chondrogenesis and morphogenesis [39, 42]. In 24 hpf MO-start injected embryos, dlx2a expression patterns revealed that the migration of pre-chondrogenic crest cells was indistinguishable from wild-type controls (Fig. 3). However, there were subtle differences in the appearances of the mandibular and hyoid branchial masses in the MO-injected embryos compared to controls (disrupted: MO-start; n = 35/69, MO-e10i10; n = 18/33, control; n = 0/27; Fig. 3), which suggested a defect in crest cell condensation.
Fig. 3.
Migration of pre-chondrogenic neural crest is unaffected in p2rx3.1 morphants. In situ hybridization was used to examine expression of the pre-chondrogenic neural crest marker dlx2a. In all panels, purple marks the presence of dlx2a mRNA. Panels a and b: 24 hpf control embryos, c and d: 24 hpf embryos injected with MO-start. b and d are higher-magnification images taken from a and c. Anterior is to the left, dorsal at top, m mandibular condensation, h hyoid condensation, asterisks are the beginnings of gill arch condensations
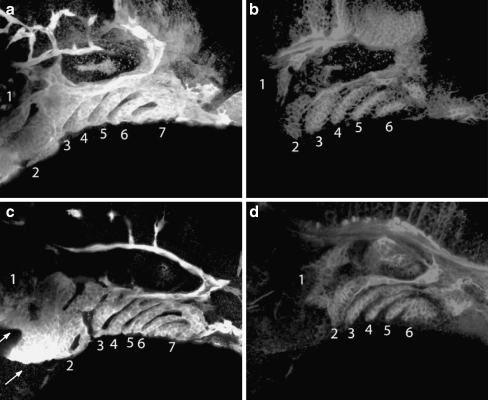
To investigate to what extent the mandibular and hyoid branchial masses were perturbed in p2rx3.1 morphant embryos, we utilized the transgenic fish line TG(fli1::eGFP)y1 in which eGFP is expressed in neural crest cells, and their derivatives, which contribute to the cartilages of the pharyngeal skeleton and vascular tissue [22]. By 52 hpf in control embryos, eGFP+ cells were present in all seven branchial arches (Fig. 4a). The non-fluorescent stripes between the masses of eGFP+ cells in the arches contain endoderm, which is labeled by an antibody specific to zn8 (Fig. 4b). At 52 hpf in MO-injected embryos, the shape and size of the first and second arches were altered compared to controls (Fig. 4c). Additionally, fluorescent cells in arches 3–7 were present but the levels of fluorescence appeared to be less robust than those in the control animals (Fig. 4c). These findings are consistent with the Alcian Blue results described above. The loss of p2rx3.1 function appeared to selectively affect the crest cells as the surrounding endoderm was unaffected (Fig. 4d) with the exception of its shape, which can be explained by the morphological changes in the arches themselves.
Fig. 4.
Branchial arch defects in neural crest condensation, but not endoderm formation, are observed in morphant embryos. Fifty-four hours post-fertilization fli1:eGFPy1 embryos were co-immunostained with anti-GFP (a,c) and zn-8 (b,d) antibodies and examined using confocal microscopy. Panels a and b are a control embryo, panels c and d are a MO-injected embryo. a,c branchial arches are numbered 1–6; b,d pharyngeal pouches are numbered 1–6. Arrows in c point to defective first and second arches
The in situ hybridization results for dlx2a expression in the arches, in combination with the data obtained using the Tg[fli1:eGFP]y1 embryos, demonstrated that the condensations of neural crest cells in the first and second arches were abnormal in p2rx3.1 MO-injected embryos. Abnormal neural crest condensations can arise from changes in cell–cell signaling that leads to improper packing of the crest cells, loss of cells due to death, inability of the cells differentiate, or a combination of these. To test for changes in cell survival in p2rx3.1 morphants, we injected acridine orange (AO) to detect cell death [43]. At the 12 somite stage, both control and MO-injected embryos displayed AO+ cells (Fig. 5a and d) scattered throughout the embryos, which is consistent with other published reports [e.g., 44, 45]. However, there were noticeably more AO+ cells in the head and branchial arch region of MO-injected embryos when compared to control embryos (Fig. 5d; embryos with increased numbers of labeled cells: MO-start; n = 99/161, controls; n = 2/95). By 24 hpf, the number of AO+ cells in control embryos decreased to nearly undetectable levels (Fig. 5b) whereas MO-injected embryos maintained high levels of AO+ cells in the branchial arches (Fig. 5e). By 36 hpf, the number of detectable AO+ cells was negligible in both control (Fig. 5c) and MO-injected embryos (Fig. 5f). Identical patterns of elevated staining were observed for each of the three specific MO, whereas no elevated staining was seen in embryos injected with the control MO (data not shown).
Fig. 5.
Knockdown of p2rx3.1 results in increased cell death in the head of embryos. Epifluorescent microscopy (×100) of both control (panels a–c) and morphant (d–f) embryos revealed acridine orange positive cells (seen as bright dots) in the head at 16 hpf (a and d) and 24 hpf (b and e), but not 36 hpf (c and f). Increased cell death in the morphant embryos are highlighted by the arrows in d and e
Development of epibranchial ganglionic neurons requires p2rx3.1 function
Previous studies have demonstrated that formation of the peripheral sensory neurons comprising the cranial sensory ganglia (CSG) is dependent upon interactions between the cranial neural crest, the endoderm of the branchial arches and the lateral ectoderm [46, 47]. Given our findings that lateral ectodermal cells express p2rx3.1 and that branchial arch neural crest cell differentiation is perturbed in morphant embryos, we hypothesized that CSG development would be perturbed in the absence of p2rx3.1 function.
To visualize the cranial sensory neurons, we utilized the Tg(p2rx3.2:eGFP)sl1 line, in which nearly all of the peripheral sensory neurons that make up gV and the epibranchial ganglia are eGFP+ ([21] and Fig. 6a–c). eGFP+ cranial sensory neurons first appear as the relevant ganglia are forming and the reporter expression is still present well after the ganglia have formed and established their axonal connections. This allows us to visualize all stages of CSG formation.
Fig. 6.
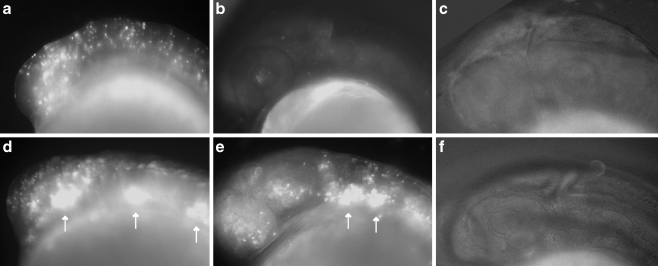
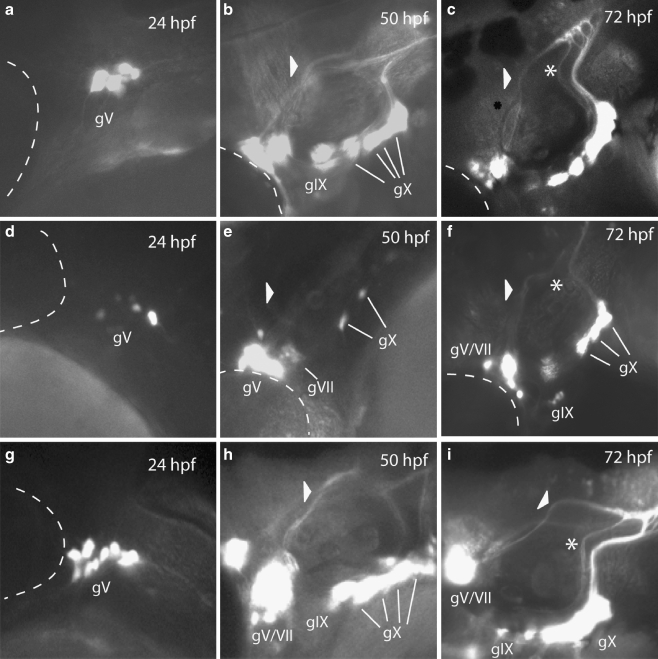
Loss of p2rx3.1 function results in disrupted epibranchial ganglia formation. Epifluorescent micrographs (×100) of eGFP expression in control (a–c), MO-start injected (d–f) and MO-e10i10 (4 ng)/rP2X3 (25 ng) RNA co-injected (g–i) Tg(p2xr3.2:eGFP)sl1 embryos selected times post-fertilization. Panels a, d, and g show the trigeminal ganglia (gV) at 24 hpf, panels b, e, and h show gV and the epibranchial ganglia at 50 hpf, and panels c, f, and i show the heads of 72-hpf embryos. The triangles in panels b, e, and h mark the sensory afferent of gVII; asterisks in panels c, f, and i mark the afferent projection of gIX. The dotted line indicates the margins of the eye, gV trigeminal ganglia, gVII facial ganglia, gIX glossopharyngeal ganglia, gX vagal ganglia
Between 24 and 72 hpf, the gV and epibranchial ganglia in control Tg(p2rx3.2:eGFP)sl1 larvae form, coalesce, and establish their central axonal projections (Fig. 6a–c). In contrast, knockdown of p2rx3.1 function in Tg(p2rx3.2:eGFP)sl1 embryos (Fig. 6d–f) resulted in the malformation of all labeled ganglia in over half of MO-injected larvae (MO-start, n = 238/411; MO-e10i10, n = 55/80), with a spectrum of phenotypes visible: the observed defects ranged from a slight decrease in the total number of neurons in any one ganglia to near-total loss of eGFP+ neurons in one or more ganglia. We did not see any ectopic expression of the transgene in MO-injected animals (n > 1,000).
In all morphant embryos/larvae examined, gV was the least affected with similar numbers of eGFP+ neurons present in both control and MO-injected larvae. However, at 24 hpf, the neurons of the gV were not as compacted when compared to control animals (Fig. 6a and d). This phenotyope fails to persist as by 50 hpf, the gV in morphants were indistinguishable from controls (Fig. 6b and e) and there were no detectable alterations in the central projections of these neurons with regards to either the time at which they appeared or in their projection pathways (Fig. 6c and f). These findings suggest that p2rx3.1-mediated signaling only has a minor, and possibly transient, role in the normal development of gV.
A different set of results were obtained upon examination of the epibranchial ganglia. Neuronal loss within the facial ganglia (gVII) was detected in some, but not all, morphants. However, it can be difficult to determine absolute numbers of gV and gVII neurons in 3 dpf embryos due to the proximity of the ganglia to one another. Irrespective of cell numbers, the central afferents of these neurons are easily visible and act as reliable indicators of their presence (arrowheads in Fig. 6b and c). In p2rx3.1 morphant larvae, gVII is developmentally delayed (Fig. 6e). In 50-hpf control embryos, gVII have already formed and are sending their projections into the hindbrain (arrowhead in Fig. 6b), whereas at this stage in morphants, gVII appears to be still undergoing coalescence and lacks central afferent projections (arrowhead in Fig. 6e). By 72 hpf, when gVII neurons in the control larvae have already established their central connections within the hindbrain, gVII afferents in morphant embryos have formed but appear to still be pathfinding in the CNS (arrowhead in Fig. 6f). The other epibranchial ganglia also appeared to be affected.
The eGFP+ neurons that constitute the glossopharyngeal ganglia (gIX) are normally visible by 50 hpf in control animals (Fig. 6b), but in morphant embryos, there were no discernible transgene-expressing gIX neurons at this time point (Fig. 6e). However, by 75 hpf (Fig. 6f), there was a small number of eGFP+ neurons near the normal location of gIX which appeared to be displaced ventrally. The central afferents of gIX were also perturbed in p2rx3.1 morphants. By 72 hpf in control larvae, gIX central afferents follow a defined path to the hindbrain as shown by the asterisk in Fig. 7c. In contrast, this central projection is missing in morphant larvae (Fig. 7f, where the asterisk marks the expected position of the afferent nerve), which is consistent with the lack of a defined gIX in these animals.
Fig. 7.
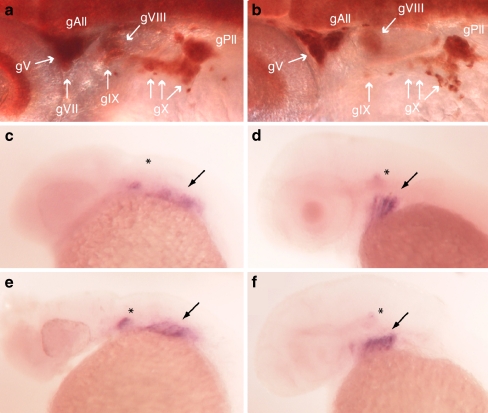
Disruption of p2rx3.1-mediated signaling perturbs epibranchial gangliogenesis. Panel a HuC immunoreactivity (brown) showing neurons in a 54 hpf wild-type embryo. b MO-start morphant 54-hpf embryo. gV trigeminal ganglia, gVII facial ganglia, gIX glossopharyngeal ganglia, gX vagal ganglia, gAll and gPll anterior and posterior lateral line ganglia. foxi1 mRNA expression (purple) in 24-hpf wild-type (c) or morphant (e) embryos, and in 48-hpf wild-type (d) or morphant (f) embryos. Asterisks mark otic placode expression, arrows point to branchial arch expression
The vagal ganglia (gX) were also affected by p2rx3.1 MO injection. At 50 hpf, there is a dramatic reduction in the number of eGFP+ neurons in gX (Fig. 6e) compared to control larvae (Fig. 6b). The number of gX eGFP+ neurons in morphants increases by 75 hpf (Fig. 6f) but still remains less than observed in controls (Fig. 6c). Unlike gVII and gIX, the gX central projections do not appear to be affected by p2rx3.1 knockdown.
All observed CSG defects in p2rx3.1 morphant embryos were rescued by co-injection of rP2X3 RNA {disrupted ganglia in MO-start alone, n = 238/411 (58%); MO-start plus P2X3 RNA, n = 59/217 (27%): for M0-i9e10 alone, n = 528/800 (66%); M0-i9e10 plus P2X3 RNA, n = 40/187 (24%): differences in the numbers of embryos displaying ganglia disruption in the MO alone versus MO plus RNA were significant (p < 0.001) by the Chi-square test}. An example of a rescue of MO-e10i10 is shown in Fig. 6g–i.
The decreased number of epibranchial eGFP+ neurons we observed in p2rx3.1 morphant embryos could be a reflection of defective ectodermal signaling, as many of the peripheral sensory neurons are generated from ectodermal-placode-derived precursors that are induced by the overlying ectoderm [48]. Such a defect could result in either a decrease in the number of mature neurons produced by the placodes, or in the formation of post-mitotic neurons that are not differentiated and do not express p2rx3.2 and thus are present but not labeled in the transgenic embryos. To determine if the former was occurring in the MO-injected larvae, we examined expression of the mRNA-binding protein HuC, which is a marker of post-mitotic neurons [49]. At 52 hpf in wild-type control larvae, immunostaining of HuC+ cells in the branchial arch region revealed normally shaped and positioned epibranchial ganglia (n = 45/45) (Fig. 7a). However, in MO-injected larvae, a distinct loss of HuC+ cells was observed within gVII, gIX and gX. Larvae showing disruption or loss of HuC+ staining at the position of the epibranchial ganglia (Fig. 7b) were detected when either MOstart (n = 21/37) or M0-i9e10 (n = 17/31) were injected. These findings suggest that perturbation of purinergic signaling via knockdown of the P2X3.1 receptor subunit resulted in decreased neurogenesis involving the epibranchial sensory ganglia.
The specification and differentiation of cranial sensory neurons from ectodermal cells occurs via a stepwise process in which placodal precursor cells differentiate sequentially from neural progenitors, to neural precursors, then differentiating post-mitotic neurons and finally, mature neurons [48]. The loss of HuC staining in p2rx3.1 morphants suggested that the last two steps in this process were perturbed. To determine if the lack of functional P2X3.1 receptors in cranial ectodermal cells was interfering with their differentiation into neurogenic ectoderm, we investigated the expression of the neurogenic placode marker foxi1 [50]. Analysis of foxi1 expression revealed expression in the branchial arches at both 24 hpf (MO-start n = 48/48) and 48 hpf (n = 37/37; Fig. 7c–f), suggesting that elements of the ectoderm undergo normal differentiation into neuroectoderm and that the actions of purinergic signaling on neurogenesis via P2X receptors occurs downstream of foxi1 function and upstream of HuC expression.
Discussion
Little is known about the functional roles that P2X receptor-mediated ATP signaling may have during embryogenesis. A previous study from this lab showed that a number of P2X receptor subunit genes were expressed during embryogenesis in the zebrafish, primarily in neurons [20]. In a subsequent report, one subunit gene, p2rx3.1, was found to be transiently expressed in non-neural cells in the head of 12–16-hpf zebrafish embryos [21], suggesting that ATP signaling may be involved in some aspect of craniofacial development during early embryogenesis. Here, we report that the non-neural p2rx3.1 expression is in ventrolateral cells that are tfap2a-positive but sox10/foxd3-negative, thus identifying the cranial ectoderm as the source of p2rx3.1 expression. Data obtained from MO knockdown experiments demonstrate that purinergic signaling via P2X3.1 receptors in the embryonic head serves two important functions: first, it is required for a subpopulation of cranial neural crest cells (CNCC) to achieve both proper patterning and terminal differentiation into chondrocytes, and second, it is necessary for the production of mature peripheral sensory neurons from the epibranchial neuroectodermal placodes.
Craniofacial development is dependent upon multiple, complex, and hierarchical interactions between combinations of cells derived from the three embryonic germ cell layers. These cellular interactions include both cell–cell contacts (juxtacrine) and extracellular signaling (paracrine) mechanisms [13, 15]. It is through such signaling that the progenitor cells in the germ layers are enabled to undergo the processes of specification and morphogenesis, both of which are required for formation of the anatomically complex vertebrate head [51].
Expression of p2rx3.1 is in a population of cells that are appropriately positioned to affect these processes in the branchial arch region. There is strong evidence that the specification and differentiation of cranial neural crest cells is influenced by the ectoderm, and vice-versa (for a recent review, see [14]). This is partially due to cell–cell contacts that occur as the neural crest cells migrate through the embryo and come into contact with the three germ layers. In addition, it has also been demonstrated that diffusible factors from the ectoderm act upon the neural crest and that the presence of the neural crest is required for those factors to be expressed by the ectoderm [14]. It is therefore feasible to postulate that the release of ATP into the extracellular milieu occurs in the head of the developing embryo influences ectoderm function via activation of the p2rx3.1 gene product.
This hypothesis is supported by the MO-mediated knockdown experiments reported here. In morphant embryos, the most apparent phenotype observed was malformation of the pharyngeal skeleton. The disruption of cartilage formation observed could arise from actions at one or more of the primary steps in the formation of these cartilages: the migration of non-differentiated CNCC into the branchial arches, the condensation of the pre-chondrogenic CNCC into the branchial masses and/or chondrogenesis and proliferation [52]. The dlx2a expression results demonstrate that the pre-chondrogenic CNCC are migrating into their proper positions in the arches in the morphants. In addition, the migratory behavior of melanophores (another CNCC derivative) does not appear to be altered in morphant embryos (data not shown). Thus, reduction in P2X3.1 signaling does not alter the ability of the CNCC to migrate or begin the processes of specification (as shown by dlx2a expression) and differentiation (Alcian Blue staining). However, the dlx2a results suggested, and the fli1:eGFP-derived data confirmed, that the branchial neural crest masses were malformed. The elevated AO staining suggested this was at least partially due to increased cell death in the branchial arches of morphant embryos. Other published studies [51] have shown that such malformed masses lead to altered jaw cartilages and the p2rx3.1 morphant larvae exhibit this phenotype. Taken together, our results suggest that ATP signaling, via P2X3.1 receptors in the ectoderm, plays an essential role in extrinsically influencing chondrogenesis programs of neural crest cells.
The perturbations in epibranchial ganglia development indicate that ATP signaling affects intrinsic ectodermal function as well. Ectodermal cells become either neural or epidermal. The specification of ectoderm to a neural fate is called neural induction [48, 53] and increased intracellular Ca2+ levels have been implicated as playing a pivotal role in this process [54, 55]. P2X receptors can influence intracellular Ca2+ concentrations either directly [12] or indirectly via depolarization-induced activation of calcium channels. Once induction occurs and the ectodermal placodes are formed, the stepwise process of neurogenesis begins [56–58]. Cells first delaminate and migrate away from the ectoderm. At this stage, they begin to differentiate and express pro-neural genes. As these neural progenitors continue their migration towards their final position in the appropriate ganglia, they become neuroblasts and finally differentiate into mature neurons. Because we observe normal expression of foxi1 in p2rx3.1 morphant embryos, we conclude that lack of P2X3.1-mediated signaling does not interfere with induction of the epibranchial placodes from ectoderm. However, a loss of HuC+ cells in these same embryos implies an alteration in the competency of placodally derived neuroblasts to undergo their terminal differentiation.
The data presented in this paper demonstrates that loss of P2X3.1-mediated signaling leads to craniofacial defects in zebrafish. These findings are in contrast to mice, where ablation of P2rx3 has not been reported to lead to craniofacial defects [59]. This disparity cannot be explained by differences in gene expression patterns, as the patterns of the two p2rx3 paralogs in zebrafish together mimic that reported for the single P2X3 subunit in mouse embryos [60, 61]. In both species, these genes are expressed in nearly all CSG neurons during neurogenesis and continue to be expressed throughout the rest of embryonic development. In addition, p2rx3 genes in both species are also expressed transiently in non-neuronal cells in the ventral cranial region: p2rx3.1 by ectoderm in zebrafish during 12–16 hpf, and P2rx3 in sub-epidermal regions of the branchial arches in the mouse between E9.5 and E10.5 [60].
So how can the interspecies differences in the outcomes of signaling perturbation be accounted for? There are two potential explanations. The first hypothesis is derived from the fact that P2X receptors are oligomeric and, oftentimes, hetero-oligomeric in nature [62]. This hypothesis proposes that there is only one receptor subunit, p2rx3.1, expressed in zebrafish ectoderm whereas there is more than one P2X receptor subunit present in mouse ectoderm. Thus, loss of a single subunit ablates function in the zebrafish whereas the presence of an additional subunit in mouse is compensatory and allows for normal ectodermal function. The lack of data detailing the embryonic expression patterns of the majority of mammalian subunit genes prevents us from determining if this hypothesis is possible. A second explanation can arise from the differences in the temporal expression of the two genes. In mouse, p2rx3 expression is first reported occurring at embryonic day 9.5 [60, 61]. At this stage in development, the cranial neural crest cells have already migrated into the branchial arches, and the neuroectodermal placodes have already formed and are in the process of producing neuroblasts and differentiated neurons. In contrast, p2rx3.1 expression in zebrafish (at 12–16 hpf) is occurring prior to both neural crest migration into the arches and neural induction in the epibranchial placodes [63]. Therefore, the difference in temporal expression patterns could allow for the loss of P2X-mediated signaling to have major impacts on the ability of the zebrafish ectoderm to both produce neurons and affect chondrogenic processes in the branchial arches, while having a much lesser effect on mouse neuroectodermal function. Which of the two putative mechanisms is operative will require further study of P2X receptor gene expression during mouse embryogenesis.
In conclusion, the data presented here show, for the first time, an essential functional role for P2X receptor-mediated signaling during vertebrate embryogenesis. Further studies will be needed to investigate the mechanism(s) by which loss of receptor signaling in ectoderm results in defects in chondrogenesis and epibranchial gangliogenesis.
Acknowledgments
We would like to thank Jasmina Mandzukic for help with fish husbandry, and Dr. Steve Johnson (Washington Univ. St. Louis) for his great insight and help on all things zebrafish. The zn8 antibody developed by Dr. W. Trevarrow was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by the NIH grants NS50261 (MMV), NS051140 (SK), and GM008306 (SK and AL).
References
- 1.Burnstock G, Knight GE (2004) Cellular distribution and functions of P2 receptor subtypes in different systems. Int Rev Cytol 240:31–304. doi:10.1016/S0074-7696(04)40002-3 [DOI] [PubMed]
- 2.Burnstock G (1996) P2 purinoceptors: historical perspective and classification. Ciba Found Symp 198:1–28 discussion 29-34 [DOI] [PubMed]
- 3.Masse K, Bhamra S, Eason R, Dale N, Jones EA (2007) Purine-mediated signalling triggers eye development. Nature 449:1058–1062. doi:10.1038/nature06189 [DOI] [PubMed]
- 4.Pearson RA, Dale N, Llaudet E, Mobbs P (2005) ATP released via gap junction hemichannels from the pigment epithelium regulates neural retinal progenitor proliferation. Neuron 46:731–744. doi:10.1016/j.neuron.2005.04.024 [DOI] [PubMed]
- 5.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR (2004) Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron 43:647–661. doi:10.1016/j.neuron.2004.08.015 [DOI] [PubMed]
- 6.Burnstock G (2007) Purine and pyrimidine receptors. Cell Mol Life Sci 64:1471–1483. doi:10.1007/s00018-007-6497-0 [DOI] [PMC free article] [PubMed]
- 7.Burnstock G (2008) Unresolved issues and controversies in purinergic signalling. J Physiol 586:3307–3312. doi:10.1113/jphysiol.2008.155903 [DOI] [PMC free article] [PubMed]
- 8.Dale N (2008) Dynamic ATP signalling and neural development. J Physiol 586:2429–2436. doi:10.1113/jphysiol.2008.152207 [DOI] [PMC free article] [PubMed]
- 9.Zimmermann H (2006) Nucleotide signaling in nervous system development. Pflugers Arch 452:573–588. doi:10.1007/s00424-006-0067-4 [DOI] [PubMed]
- 10.Egan TM, Samways DS, Li Z (2006) Biophysics of P2X receptors. Pflugers Arch 452:501–512. doi:10.1007/s00424-006-0078-1 [DOI] [PubMed]
- 11.North RA (2002) Molecular physiology of P2X receptors. Physiol Rev 82:1013–1067 [DOI] [PubMed]
- 12.Egan TM, Khakh BS (2004) Contribution of calcium ions to P2X channel responses. J Neurosci 24:3413–3420. doi:10.1523/JNEUROSCI.5429-03.2004 [DOI] [PMC free article] [PubMed]
- 13.Hall BK, Miyake T (2000) Craniofacial development of avian and rodent embryos. Methods Mol Biol 135:127–137 [DOI] [PubMed]
- 14.Knight RD, Schilling TF (2006) Cranial neural crest and development of the head skeleton. Adv Exp Med Biol 589:120–133. doi:10.1007/978-0-387-46954-6_7 [DOI] [PubMed]
- 15.Kuratani S (2005) Craniofacial development and the evolution of the vertebrates: the old problems on a new background. Zoolog Sci 22:1–19. doi:10.2108/zsj.22.1 [DOI] [PubMed]
- 16.Schilling TF (1997) Genetic analysis of craniofacial development in the vertebrate embryo. Bioessays 19:459–468. doi:10.1002/bies.950190605 [DOI] [PubMed]
- 17.Bronner-Fraser M (1994) Neural crest cell formation and migration in the developing embryo. FASEB J 8:699–706 [DOI] [PubMed]
- 18.Holder N, Klein R (1999) Eph receptors and ephrins: effectors of morphogenesis. Development 126:2033–2044 [DOI] [PubMed]
- 19.Trainor PA, Krumlauf R (2000) Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci 1:116–124. doi:10.1038/35039056 [DOI] [PubMed]
- 20.Kucenas S, Li Z, Cox JA, Egan TM, Voigt MM (2003) Molecular characterization of the zebrafish P2X receptor subunit gene family. Neuroscience 121:935–945. doi:10.1016/S0306-4522(03)00566-9 [DOI] [PubMed]
- 21.Kucenas S, Soto F, Cox JA, Voigt MM (2006) Selective labeling of central and peripheral sensory neurons in the developing zebrafish using P2X(3) receptor subunit transgenes. Neuroscience 138:641–652. doi:10.1016/j.neuroscience.2005.11.058 [DOI] [PubMed]
- 22.Lawson ND, Weinstein BM (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248:307–318. doi:10.1006/dbio.2002.0711 [DOI] [PubMed]
- 23.Westerfield M (2000) The zebrafish book. University of Oregon, Eugene
- 24.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310 [DOI] [PubMed]
- 25.Lewis C, Neidhart S, Holy C, North RA, Buell G, Surprenant A (1995) Coexpression of P2X2 and P2X3 receptors subunits can account for ATP-gated currents in sensory neurons. Nature 377:432–435. doi:10.1038/377432a0 [DOI] [PubMed]
- 26.Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi:10.1016/0003-2697(87)90021-2 [DOI] [PubMed]
- 27.Dutton KA et al (2001) Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128:4113–4125 [DOI] [PubMed]
- 28.Pohl BS, Knochel W (2001) Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech Dev 103:93–106. doi:10.1016/S0925-4773(01)00334-3 [DOI] [PubMed]
- 29.Knight RD et al (2003) lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130:5755–5768. doi:10.1242/dev.00575 [DOI] [PubMed]
- 30.Baker CV, Bronner-Fraser M (2000) Establishing neuronal identity in vertebrate neurogenic placodes. Development 127:3045–3056 [DOI] [PubMed]
- 31.Knight RD, Javidan Y, Nelson S, Zhang T, Schilling T (2004) Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev Dyn 229:87–98. doi:10.1002/dvdy.10494 [DOI] [PubMed]
- 32.Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF (2005) AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development 132:3127–3138. doi:10.1242/dev.01879 [DOI] [PubMed]
- 33.Li W, Cornell RA (2007) Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol 304:338–354. doi:10.1016/j.ydbio.2006.12.042 [DOI] [PMC free article] [PubMed]
- 34.Ekker SC (2000) Morphants: a new systematic vertebrate functional genomics approach. Yeast 17:302–306. doi:10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-# [DOI] [PMC free article] [PubMed]
- 35.Egan TM, Haines WR, Voigt MM (1998) A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J Neurosci 18:2350–2359 [DOI] [PMC free article] [PubMed]
- 36.Torres GE, Egan TM, Voigt MM (1999) Identification of a domain involved in ATP-gated ionotropic receptor subunit assembly. J Biol Chem 274:22359–22365. doi:10.1074/jbc.274.32.22359 [DOI] [PubMed]
- 37.Boue-Grabot E, Akimenko MA, Seguela P (2000) Unique functional properties of a sensory neuronal P2X ATP-gated channel from zebrafish. J Neurochem 75:1600–1607. doi:10.1046/j.1471-4159.2000.0751600.x [DOI] [PubMed]
- 38.Egan TM, Cox JA, Voigt MM (2000) Molecular cloning and functional characterization of the zebrafish ATP-gated ionotropic receptor P2X(3) subunit. FEBS Lett 475:287–290. doi:10.1016/S0014-5793(00)01685-9 [DOI] [PubMed]
- 39.Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC (1998) The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol 203:245–263. doi:10.1006/dbio.1998.9016 [DOI] [PubMed]
- 40.Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB (2000) sucker encodes a zebrafish endothelin-1 required for ventral pharyngeal arch development. Development 127:3815–3828 [DOI] [PubMed]
- 41.Richman JM, Lee SH (2003) About face: signals and genes controlling jaw patterning and identity in vertebrates. Bioessays 25:554–568. doi:10.1002/bies.10288 [DOI] [PubMed]
- 42.Hall BK, Miyake T (2000) All for one and one for all: condensations and the initiation of skeletal development. Bioessays 22:138–147. doi:10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4 [DOI] [PubMed]
- 43.Furutani-Seiki M et al (1996) Neural degeneration mutants in the zebrafish, Danio rerio. Development 123:229–239 [DOI] [PubMed]
- 44.Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW (2004) Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development 131:1463–1477. doi:10.1242/dev.01033 [DOI] [PubMed]
- 45.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC (2007) p53 activation by knockdown technologies. PLoS Genet 3:e78. doi:10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed]
- 46.Holzschuh J et al (2005) Requirements for endoderm and BMP signaling in sensory neurogenesis in zebrafish. Development 132:3731–3742. doi:10.1242/dev.01936 [DOI] [PubMed]
- 47.Nechiporuk A, Linbo T, Raible DW (2005) Endoderm-derived Fgf3 is necessary and sufficient for inducing neurogenesis in the epibranchial placodes in zebrafish. Development 132:3717–3730. doi:10.1242/dev.01876 [DOI] [PubMed]
- 48.Baker CV, Bronner-Fraser M (2001) Vertebrate cranial placodes I. Embryonic induction. Dev Biol 232:1–61. doi:10.1006/dbio.2001.0156 [DOI] [PubMed]
- 49.Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N (1996) Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett 216:109–112. doi:10.1016/0304-3940(96)13021-4 [DOI] [PubMed]
- 50.Lee SA, Shen EL, Fiser A, Sali A, Guo S (2003) The zebrafish forkhead transcription factor Foxi1 specifies epibranchial placode-derived sensory neurons. Development 130:2669–2679. doi:10.1242/dev.00502 [DOI] [PubMed]
- 51.Kimmel CB, Miller CT, Moens CB (2001) Specification and morphogenesis of the zebrafish larval head skeleton. Dev Biol 233:239–257. doi:10.1006/dbio.2001.0201 [DOI] [PubMed]
- 52.Yelick PC, Schilling TF (2002) Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med 13:308–322. doi:10.1177/154411130201300402 [DOI] [PubMed]
- 53.Schlosser G (2006) Induction and specification of cranial placodes. Dev Biol 294:303–351. doi:10.1016/j.ydbio.2006.03.009 [DOI] [PubMed]
- 54.Moreau M, Leclerc C (2004) The choice between epidermal and neural fate: a matter of calcium. Int J Dev Biol 48:75–84. doi:10.1387/ijdb.15272372 [DOI] [PubMed]
- 55.Webb SE, Moreau M, Leclerc C, Miller AL (2005) Calcium transients and neural induction in vertebrates. Cell Calcium 37:375–385. doi:10.1016/j.ceca.2005.01.005 [DOI] [PubMed]
- 56.D'Amico-Martel A (1982) Temporal patterns of neurogenesis in avian cranial sensory and autonomic ganglia. Am J Anat 163:351–372. doi:10.1002/aja.1001630407 [DOI] [PubMed]
- 57.Northcutt RG, Brandle K (1995) Development of branchiomeric and lateral line nerves in the axolotl. J Comp Neurol 355:427–454. doi:10.1002/cne.903550309 [DOI] [PubMed]
- 58.Zou D, Silvius D, Fritzsch B, Xu PX (2004) Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 131:5561–5572. doi:10.1242/dev.01437 [DOI] [PMC free article] [PubMed]
- 59.Cockayne DA et al (2005) P2X2 knockout mice and P2X2/P2X3 double knockout mice reveal a role for the P2X2 receptor subunit in mediating multiple sensory effects of ATP. J Physiol 567:621–369 [DOI] [PMC free article] [PubMed]
- 60.Boldogkoi Z, Schutz B, Sallach J, Zimmer A (2002) P2X(3) receptor expression at early stage of mouse embryogenesis. Mech Dev 118:255–260. doi:10.1016/S0925-4773(02)00280-0 [DOI] [PubMed]
- 61.Cheung KK, Burnstock G (2002) Localization of P2X(3) receptors and coexpression with P2X(2) receptors during rat embryonic neurogenesis. J Comp Neurol 443:368–382. doi:10.1002/cne.10123 [DOI] [PubMed]
- 62.Khakh BS et al (2001) International union of pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacol Rev 53:107–118 [PubMed]
- 63.Sun SK, Dee CT, Tripathi VB, Rengifo A, Hirst CS, Scotting PJ (2007) Epibranchial and otic placodes are induced by a common Fgf signal, but their subsequent development is independent. Dev Biol 303:675–686. doi:10.1016/j.ydbio.2006.12.008 [DOI] [PubMed]