Abstract
The pro-apoptotic P2X7 receptor regulates growth of epithelial cells. The objectives of the study were to understand P2X7 gene transcription; to identify the active promoter and the transcription initiation site (TpIS); and to begin understanding regulation of P2X7 gene transcription. Experiments in vitro utilized normal and cancerous cultured human uterine cervical epithelial cells, and HEK293 cells overexpressing P2X7-luciferase reporters. Experiments in vivo used surgical specimen of normal and cancerous uterine cervix. Assays involved DNA, RNA, and protein techniques. (a) The P2X7 TpIS was localized to adenine (+1) at nt 1683 of the human P2X7 gene [GenBank Y12851]), with a TTAAA sequence at nt −32/−28 and an active promoter region within nt −158/+32. (b) P2X7 transcription was found to be regulated by two enhancers located at nt + 222/+232 and +401/+573 regions downstream of the active P2X7 promoter. (c) The putative enhancer regions formed four DNA–protein complexes. (d) P2X7 transcription was found to be controlled by hypermethylated cytosines at cytosine-phosphodiester-guanosines (CpG) that cluster or co-localize with the enhancers’ sites. (e) We identified nine CpGs as inhibitory cis elements, and three CpG sites that are hypermethylated in cultured cervical epithelial cells and in cervix epithelia in vivo. (f) In cancer cervical cells, the degree of hypermethylation of the CpG sites was greater than in the normal cervical cells. Expression of the P2X7 receptor is controlled by hypermethylated CpGs that flank transcription enhancers located within a 547-nt region downstream of the promoter.
Keywords: Cervix, Cervical, Cancer, Methylation, CpG, Enhancer, Inhibition
Introduction
Apoptosis is a regulated homeostatic process of selective cell deletion [1–3], and it controls the growth of cells [4]. Dysregulation of apoptotic cell death has been implicated in states of disease and in the neoplastic transformation [5, 6]. Among the pro-apoptotic systems that operate in various types of tissues, including epithelia, the P2X7 receptor mechanism is an important mechanism because the receptor is expressed in proliferating cells [7], and P2X7-induced apoptosis controls physiologically the growth of cells [8, 9]. Recent data showed lesser expression of the P2X7 receptor in cancer cells of epithelia derived from ectodermal, uro-genital sinus, and the distal paramesonephric duct tissues [7, 10–14], suggesting that reduced expression of the receptor may be related to the development of those cancers.
The P2X7 receptor is a membrane-bound, ligand-operated channel [15–17]. The natural ligand of the receptor is ATP [15, 16] which is present in the extracellular fluid of epithelial cells at concentrations that can activate the receptor [4, 8, 18–21]. Binding of ATP to the P2X7 receptor can activate various cell-specific signaling cascades, including the IL-1β [22], TNFα-TRAIL [23], and the p38, JNK / SAPK [24], and NF-κB cascades [25]. However, a unique effect of activation of the P2X7 receptor is formation of pores in the plasma membrane [16]. In uterine epithelial cells [8, 11, 17] and in mouse keratinocytes [9], formation of P2X7 receptor pores induces apoptosis by a mechanism that involves uncontrolled influx of Ca2+ via P2X7-pores and activation of the mitochondrial caspase-9 pathway [8, 11].
Central to the ability of ATP to induce apoptosis via the P2X7 mechanism is the degree of cellular expression of the P2X7 receptor [17]. For instance, in human uterine cervical epithelial cells [8] and in mouse keratinocytes [9], apoptosis induced by treatment with ATP or with the P2X7-specific agonist 2′,3′-0-(4-benzoylbenzoyl)-adenosine 5′-triphosphate (BzATP) was less in cancer than in normal cells, and the differences in agonist-induced apoptosis correlated with cellular expression of the receptor [11–13]. One of the mechanisms that explain the lower expression of P2X7 receptor in cancer cervical cells is greater instability of the P2X7 transcript in the cancer cells [13]. However, recent preliminary data from the author’s lab also suggested modulation of transcription in cancer cells. Until recently, little was known about the regulation of transcription of the P2X7 gene. The human P2X7 gene is localized within a 55-kb region of chromosome 12q24; it has 13 exons and it encodes a 595-amino-acid protein [26]. Previous studies reported P2X7 promoter activity within a 2-kb DNA segment of the 5′ of the gene [26, 27], but the location of the active promoter and the transcription initiation site (TpIS) were unknown. Five single nucleotide polymorphisms were identified within the 5′ 2 kb DNA segment of the gene (sites −298, −762, −838, −1140, and −1269 [relative to exon 1]), but none were associated with a specific ATP response phenotype [28]. Furthermore, previous sequencing efforts have identified numerous P2X7 variations and had proposed linkage to human disease, but at present, there is no consensus about their biological role [29].
The major objective of the present study was to better understand P2X7 gene transcription. Specific objectives were to identify the active promoter; to define the TpIS; and to begin understanding regulation of P2X7 gene transcription. An improved understanding of the regulation of P2X7 gene expression could be important for our understanding of the development of cancers because defective apoptosis may lead to cancer [30–32]. The experiments used, as a model, human epithelial uterine cervical cells. The physiological pro-apoptotic role of P2X7 in these cells was previously demonstrated [7, 8, 13]. Previous studies in cervical cells have also identified tumor suppressor genes [33–46] and apoptosis-related genes [35, 39, 43, 44, 46–49] that are regulated by changes in DNA methylation. However, until recently, little was known about the effects of DNA methylation on P2X7 gene expression.
Methods
Cell cultures
Primary cultures of human ectocervical-vaginal epithelial cells (hEVEC), a well-characterized model of the normal human ectocervical epithelium, were generated from discarded normal ectocervical-vaginal tissues [4]. Human cervical epithelial cancer cell lines (Caski, HeLa, Siha, and HT3), and human embryonic kidney 293 cells (HEK293), which lack endogenous expression of P2X7 [11], were obtained from the ATCC. Cell culture conditions were described [4, 11].
Elucidation of the promoter region
HindIII-tailed sense primers and the NcoI-tailed antisense primers (InVitrogen, Carlsbad CA) that were used for the elucidation of the promoter region of the human P2X7 gene (GenBank Y12851) are shown in Table 1. The corresponding cDNA fragments were synthesized by PCR using human genomic DNA. The PCR fragments were digested with HindIII and NcoI and ligated into pGL3 enhancer vector (Promega, Madison, WI) with HindIII and NcoI sites using Rapid DNA Ligation Kit (Roche, Indianapolis, IN). Reporters in this experiment and in all the experiments described below were confirmed by sequencing.
Table 1.
Primers for elucidation of the promoter region
| HindIII-tailed sense | ||||||||||||
| −1664 | TTT | TTA | AGC | TTA | AGA | TGT | GAA | GCC | AGG | ATC | G | |
| −1179 | TTT | TTA | AGC | TTG | GAT | CAA | GCC | AGC | TGTA | |||
| −698 | TTT | TTA | AGC | TTG | GTG | GTG | TCC | CTC | ACT | GAA | T | |
| −399 | TTT | TTA | AGC | TTG | GGG | CTG | AAT | AAA | GGG | TTG | T | |
| −204 | TTT | TTA | AGC | TTA | ATG | CCC | ATC | CTC | TGA | ACA | C | |
| −158 | TTT | TTA | AGC | TTG | CCA | GCT | GGG | GTG | AGG | TCA | TCT | G |
| −122 | TTT | TTA | AGC | TTT | AGG | ACT | TGG | CGC | TTC | TTG | T | |
| −73 | TTT | TTA | AGC | TTA | GGG | CCC | GCC | CCA | ACT | CTG | CAG | |
| −53 | TTT | TTA | AGC | TTA | GTC | ATT | GGA | GGA | GCT | TGA | AGT | TA |
| NcoI-tailed antisense | ||||||||||||
| −380 | TTT | TTC | CAT | GGA | CAA | CCC | TTT | ATT | CAG | CCC | C | |
| +32 | TTT | TTC | CAT | GGC | ACA | GCA | AGC | CCC | CTC | CCA | GTA | |
| +91 | TTT | TTC | CAT | GGG | GTG | ACA | GCC | TCC | CTC | CCT | GCG | CG |
| +221 | TTT | TTC | CAT | GGC | TTA | CCA | AAC | GTA | GGA | AAA | ||
| +232 | TTT | TTC | CAT | GGC | CCA | GAT | CCC | ACT | TAC | CAA | A | |
| +337 | TTT | TTC | CAT | GGA | GAG | CAC | GTC | TCA | GAT | TCG | AA | |
| +402 | TTT | TTC | CAT | GGG | CTG | CAG | CCT | GGC | ACC | GTT | TC | |
| +470 | TTT | TTC | CAT | GG | TGC | GCG | CCC | TGG | CGG | GC | ||
| +503 | TTT | TTC | CAT | GGC | CTG | CGC | TTT | CCT | ACC | TTC | CC | |
| +573 | TTT | TTC | CAT | GGT | CAG | ATG | CCA | GCA | TGA | TCA | CCA | GGC |
Segments are numbered relative to the transcription initiation start site
Control plasmids containing the pGL3 luciferase enhancer vector (5064 nt) or test plasmids (P2X7-luciferase) were transfected into subconfluent cultured HEK293 cells that were plated 14 h earlier in six-well plates at a density of 2.5 × 105 cells per well. The culture medium was replaced with fresh medium plus 100 μl serum-free medium per well, containing 2.5 μl Fugene 6 (Roche) and 750 ng DNA of the control or test vectors. For luciferase activity determinations cells were cotransfected with Renilla luciferase and 30 ng of the pRL-CMV vector (Promega) [13]. At the completion of incubations cells were harvested and maintained in lyses buffer for 24 h (Promega); Firefly and Renilla luciferase activities were measured consecutively by using Dual-Luciferase Reporter Assay System (Promega), and luciferase activity was determined in terms of Fluc/Rluc. For determinations of changes in P2X7 and Firefly luciferase (Fluc) mRNA, cells were lysed followed by RNA extraction. P2X7 and Fluc mRNA levels were determined by real-time PCR (qPCR) relative to cytokeratin-18 (CK-18) or GAPDH, and expressed in terms of the threshold cycle of fluorescence (Ct), using our described methodology [12, 13]. Primers for P2X7, Fluc, CK-18 and GAPDH were described [7, 11–13].
Elucidation of the transcription initiation start site
A modified 5′ rapid amplification of cDNA ends method (5′-RACE) was used. Primers (InVitrogen) are shown in Tables 2 and 3. Cloning, by reverse transcription (RT), was carried out at 55°C (total of 20 μg RNA per reaction), using Invitrogen SuperScript™ III Reverse Transcriptase (InVitrogen). Biotin-labeled primers were used for RT to produce cDNA, and the biotin-labeled cDNA was combined with Dynabeads in order to concentrate the cDNA product and facilitate its purification. To this aim, the RT reaction was mixed with 40 μl Dynabeads M-280 streptavidin (Invitrogen) at 25°C for 30 min; beads were washed in buffer containing 10 mM Tris pH 7.5, 1 mM EDTA, and 2 M NaCl, and the beads-attached cDNA was tailed using Terminal Transferase (Promega) with dATP at 37°C for 20 min and at 70°C for 10 min. PCR was done with a primer pair of anchor TTT and the gene-specific nested primers. A second PCR was done with the anchor primer and the nested HindIII antisense primer using the first PCR products diluted 100-fold. The resulting cDNA fragment was cloned into pGL3 enhancer vector with HindIII site, and confirmed by sequencing. The primary TpIS was identified from the sequence, beginning with adenine, and designated as site +1 (nt 1683 of the published human P2X7 gene sequence [GenBank Y12851]).
Table 2.
Primers for elucidation of the transcription Initiation start site (TpIS)
| Anchor HindIII-tailed dT | |
| GGA CCA AGC TTA TCG ATG TCG ACT TTT TTT TTT TTT TTT V | |
| HindIII-including anchor | |
| GGA CCA AGC TTA TCG ATG TCG AC | |
| Antisense primers | |
| Biotin-labeled (located at Exon-3) | GCT CTT GGC CTT CTG TTT TG |
| Nested (located at Exon-2) | GGT GTA GTC TGC GGT GTC AA |
| nested HindIII-tailed (located at Exon-2) | CCG CTA AGC TTG CTT GTC ACT CAC CAG AGC A |
Table 3.
Primers for elucidation of TATA-like Sites
| Mutation | Primers |
| CATT/GTAA (nt −1 to +3) | Forward—CAG TAC GTT TGT AAT TGC AGT TAC TG |
| Reverse—CAG TAA CTG CAA TTA CAA ACG TAC TG | |
| TA/CC (nt −31 to −30) | Forward—GAG CTT GAA GTC CAA GAC TCC TG |
| Reverse—CAG GAG TCT TGG ACT TCA AGC TC | |
| AGGG/TATA (nt −73 to −70) | Forward—GCC ACT GCC TAT ACC CGC CCC A |
| Reverse—TGG GGC GGG TAT AGG CAG TGG C | |
| TT/CC (nt −102 to −101) | Forward—TGG CGC TTC TTG TCC ATC ACA GC |
| Reverse—GCT GTG ATG GAC AAG AAG CGC CA |
Oligonucleotides-directed mutagenesis of regions of interest within the promoter region [13] utilized the pGL3 enhancer vector reporter containing fragment −158 to +32 and the PCR method. Regions of interest and mutations were as follows: nt −1 to +3 (CATT/GTAA); nt −31 to −30 (TA/CC); nt −73 to −70 (AGGG/TATA); and nt −102 to −101 (TT/CC). Primers are shown in Tables 2 and 3.
Mutagenesis of CpG sites
Mutations in the CpG sites within the 547-nt region downstream of the P2X7 promoter were as follows: +211/+212 (CG/AA), +257/+258 (CG/TT), +278/+279 (CG/TT), +319/+320 (CG/AT), +330/+331 (CG/AT), +424/+425 (CG/TT), +461/+464 (CGCG/ATTA), +453/+454 (CG/TT), +475/+476 (CG/TT), +498/+499 (CG/TT), and +548/+549 (CG/TT). Primers are shown in Table 4. cDNA fragments composed of the P2X7 active promoter attached with the downstream 547-nt region containing the wild-type or mutant sequences were inserted into a luciferase vector and transfected into HEK293 cells. Promoter activity was determined in terms of luciferase activity.
Table 4.
Primers for CpG mutagenesis experiments
| Segment (CpG site) | Mutation | Primers |
|---|---|---|
| −158/+221 | ||
| (+211/+212) | CG/AA | Reverse—TTT TTC CAT GGC TTA CCA AAT TTA GGA AAA G |
| −158/+337 | ||
| (+257/+258) | CG/TT | Forward—ATC TCT GCA GTG GCT TAC AGC ACA |
| Reverse—TGT GCT GTA AGC CAC TGC AGA GAT | ||
| (+278/+279) | CG/TT | Forward—AAG CCC CAG TTG GCA GCT TCA G |
| Reverse—CTG AAG CTG CCA ACT GGG GCT T | ||
| (+319/+320) | CG/AT | Reverse—TTT TTC CAT GGA GAG CAC GTC TCA GAT TAT AAA |
| (+330/+331) | CG/AT | Reverse—TTT TTC CAT GGA GAG CAA TTC TCA GAT TCG AA |
| −158/+470 | ||
| (+424/+425) | CG/TT | Forward—CAC AGG ACA AGT TGG ATT CCT |
| Reverse—AGG AAT CCA ACT TGT CCT GTG | ||
| (+453/+454) | CG/TT | Reverse—Nco)TTT TTC CAT GGC CCT GCG CGC CCT GGA AGG C |
| (+461/+464) | CGCG/ATTA | Reverse—TTT TTC CAT GGC CCT GTA ATC CCT GGC GGG C |
| −158/+573 | ||
| (+475/+476) | CG/TT | Forward—GCA GGG TTT GCC TGG GGA AGG TAG |
| Reverse—CTA CCT TCC CCA GGC AAA CCC TGC | ||
| (+498/+499) | CG/TT | Forward—GTA GGA AAG TTC AGG GCA ACA C |
| Reverse—GTG TTG CCC TGA ACT TTC CTA C | ||
| (+548/+549) | CG/TT | Reverse—Nco)TTT TCC ATG GTC AGA TGC CAG CAT GAT CAC CAG GAA TGC C |
P2X7-receptor immunostaining
The previously described method [7, 12] utilized rabbit polyclonal anti-P2X7 receptor antibody (primary, from Alomone Laboratories, Jerusalem, Israel) [9] and goat anti-rabbit Alexa Fluro 594 (secondary, from Invitrogen) [9]. Immunofluorescence was captured in a fluorescence microscope Nikon Eclipse 80i (Nikon, Melville NY) [11]. Image analysis of the immunofluorescence data was done as described [11] and expressed in terms of average pixel density per cell.
Apoptosis assays
Apoptosis was quantified by using Roche Cell Death Detection ELISA Kit (Roche) [11].
In vitro methylation assays
Control (pGL3 luciferase enhancer vector) or test plasmids (P2X7-promoter-luciferase) were methylated in vitro prior to transfections by incubations with the CpG-Methylase M.SssI (which adds a methyl group in cytosine residues) according to the manufacturer’s protocol (New England Biolabs, Ipswich, MA).
Electrophoretic mobility shift assay
A pGL3 enhancer Vector with an insert of the P2X7 promoter was used as the template, and assays were performed using LightShift® Chemiluminescent EMSA Kit (Pierce, Rockford, IL). Briefly, biotin-labeled fragments were amplified by PCR with one of the primers listed in Table 5. After amplification, P2X7 promoter fragments were separated on 1.5% agarose gel and extracted using the Agarose Gel Extraction Kit (QIAGEN). The binding reaction was performed for 30 min at room temperature using the electrophoretic mobility shift assay (EMSA) kit system in a total volume of 20 μL containing 2 μl binding buffer, 0.5 μg of HeLaScribe Nuclear Extract (Promega), 40 fmol biotin-labeled oligonucleotide, and 1 μl poly (dI-dC). DNA–protein complexes formed were fractionated by electrophoresis over 4% polyacrylamide gels in 1× Tris–boric acid–EDTA buffer. Gels were electrophoretically transferred at 100 V for 1 h on ice to a positively charged nylon membrane and immediately cross-linked with a UV transilluminator. Streptavidin-horseradish peroxidase conjugate and the LightShift Chemiluminescent Substrate (Pierce) were used to detect the biotin end-labeled DNA, and the nylon membranes were exposed to X-ray film for 3 min for detection of possible DNA–protein binding reactions.
Table 5.
Primers for the electrophoretic mobility shift assays
| Fragment | Forward (biotin-labeled at 5′) | Reverse | PCR product size |
|---|---|---|---|
| F401-475 | GCAGAGAGAAGCCACAGGA | GCCCTGCGCGCCCTGGCG | 75 bp |
| F401-530 | GCAGAGAGAAGCCACAGGA | CCTGCGCTTTCCTACCTTCCC | 103 bp |
| F401-573 | GCAGAGAGAAGCCACAGGA | TCAGATGCCATCATGATCACC | 173 bp |
| F217-237 | GTAAGTGGGATCTGGGGAGGA | TCCTCCCCAGATCCCACTTAC | 21 bp |
Human tissues
Discarded human uterine tissues were used for DNA methylation analysis. Tissues were obtained from the Human Tissue Procurement Facility of University Hospital CASE Medical Center, Case Western Reserve University, Cleveland Ohio, according to approved research protocols 12-03-50 and 03-90-300 by the Institutional Review Board. Cross sections of cervical segments were obtained from paraffin embedded blocks that were prepared by the Department of Pathology initially to establish the patient’s diagnosis. For the assays, additional parallel 10-μm sections were cut and slides were made according to standard procedures. For each case, an H&E stained slide was used to identify normal or cancerous cervical epithelial regions. Tissue epithelial fragments were obtained from parallel regions of non-stained slides by microdissection. The tissue material was dispersed into 100 μl digestion buffer (50 mM Tris pH 8.5, 0.5% Tween 20, 200 μg/ml proteinase-K), incubated at 42°C overnight, and proteinase-K was inactivated by incubation at 80°C for 10 min. The data presented in this paper are considered preliminary proof-of-concept, and no formal Power Analysis was performed to determine the number of tissues to be included in testing any specific hypotheses. No attempt was made to categorize results by patients’ ethnic origin or race.
DNA methylation analysis
Assays used the method of combined bisulfite restriction analysis (COBRA) which employs restriction enzyme digestion to reveal methylation-dependent sequence differences in PCR products of sodium bisulfite-treated DNA [50]. Briefly, bisulfite-treated, genomic DNA unmethylated cytosines are converted to thymidines whereas methylated cytosine residuals are retained as cytosines. DNA segments of interest are amplified using PCR primers that do not contain CpG dinucleotides so that the amplification step does not discriminate between templates by their original methylation status. The PCR products are digested by restriction enzymes that recognize sequences containing CpG. Cleavage occurs if the CpG sequence has been retained during the bisulfite conversion according to the methylated status of the cytosine residue. The digested PCR products are resolved and separated by gel electrophoresis and stained with ethidium bromide.
Cloning of CpG-rich regions downstream of the promoter Patterns of methylation were determined within a 547-bp region of nt +26/+573 (relative to the subsequently defined TpIS, nt + 1; Fig. 1). The region of interest was arbitrarily divided to three segments with partial overlaps, designated segment 1 (nt +26/+247), segment 2 (nt +223/+399), and segment 3 (nt +352/+573). Primers are shown in Table 6. PCR conditions (annealing temperature) and the restriction enzyme used were as follows: segment 1 56°C (MaeII); Segment-2 56°C (MaeII); segment 3 59°C (BstUI).
Fig. 1.
Sequence of the 5′ region of the human P2X7, containing the active promoter (white symbols on black background, nt −158/+32); a 547-nt CpG-rich region (underlined, nt +26/+573) downstream of the promoter; Exon 1 (underlined and italics, nt +92/+216); and the proximal part of intron-1 (distal to Exon 1, underlined, beginning at nt +217). Nucleotides were numbered relative to the Transcription Initiation Start Site (TpIS; +1; nt 1683 according to GenBank Y12851). TpISs- and TATA-like sequences within the active promoter bases/regions are bolded and doubly underlined. CpG dinucleotides are bolded and doubly underlined. Vertical thick empty arrows point to MaeII-sensitive CpG sites (nt +193/+194, +211/+212, and +330/+331). The vertical thick-filled arrow points to a BstUI-sensitive CpG–CpG site (nt +461/+462 and +463/+464). For DNA methylation experiments the 547-nt CpG-rich region was subdivided into segment 1 (nt +26/+247), segment 2 (nt +223/+399), and segment 3 (nt +352/+573)
Table 6.
Primers for experiments using segments 1–3
| Segment 1 | Forward—TGT TGT GGT TTT GTT AGG AAG AGT A |
| Reverse—AAA AAT CTA AAT CCT CCC CAA ATC | |
| Segment 2 | Forward—GGA TTT GGG GAG GAT TTAGATT-3 |
| Reverse—CAA CCT AAC ACC GTT TCCTCTT-3 | |
| Segment 3 | Forward—GGG AGG GAG GAA GTA GTA GTA GGT A |
| Reverse—TCA AAT ACC AAC ATA ATC ACC AAA C |
Evaluation of DNA Methylation Genomic DNA was extracted from cultured cells and from human tissues using DNA purification kit (Promega). To convert CpG non-associated cytosines to uracil (and thymidines), 1 μg of genomic DNA was denatured with 2 M NaOH at 37°C for 10 min; 30 μl of 10 mM hydroquinone were added and the solution was incubated with 3 M sodium bisulfite (pH 5) at 53°C for 16 h in darkness. After treatment, DNA was purified by DNA clean up kit (Promega). The solution was incubated with 2 M NaOH at room temperature and precipitated with 100% ethanol, washed with 70% ethanol, and resuspended in 20 μl of distilled water. The bisulfite-treated DNA was amplified by PCR (95°C 5 min/95°C 30 s/55–59°C 45 s/and 72°C 45 s, 37 cycles). PCR products were digested with restriction enzymes and separated in 6% polyacrylamide gel at 4 W for 3 h and the amplified products were checked for accuracy by sequencing. Positive control for the DNA methylation experiments was human placental genomic DNA treated with the CpG-Methylase M.SssI and the negative control was water. DNA methylation was determined from the polyacrylamide gel pictures in terms of samples showing de novo appearance of a low MW band, corresponding to a cleaved band at a previously methylated site. The degree of methylation was determined by densitometry in terms of the intensity of the cleaved fragment (lower MW band) relative to the density of the non-methylated plus the methylated bands (lower plus higher MW bands). Densitometry was done as described [11].
To confirm the efficacy of the bisulfite method for converting genomic DNA cytosines (but not methylcytosines) to uracils, bisulfite-treated genomic DNA was amplified with primers for Segments 1 and 2. PCR products were purified with PCR purification kit (QIAGEN) and cloned into vector pCR II-TOPO (Invitrogen). Subclones were transformed into E. coli bacteria type BL21 and cultured overnight at 37°C. Plasmid-containing PCR products were extracted with miniprep kit (Promega) and sequenced. The results (not shown) confirmed that all genomic DNA cytosines converted to uracils, except methylcytosines.
Data analysis
Data were analyzed using GraphPad Instat (GraphPad Software Inc., San Diego, CA). Significance of differences between groups was estimated by t test, or by one-way or two-way ANOVA with Tukey–Kramer Multiple Comparisons post test analysis.
Supplies
All chemicals, unless specified otherwise, were obtained from Sigma Chemicals (St. Louis, MO).
Results
Elucidation of the active promoter region
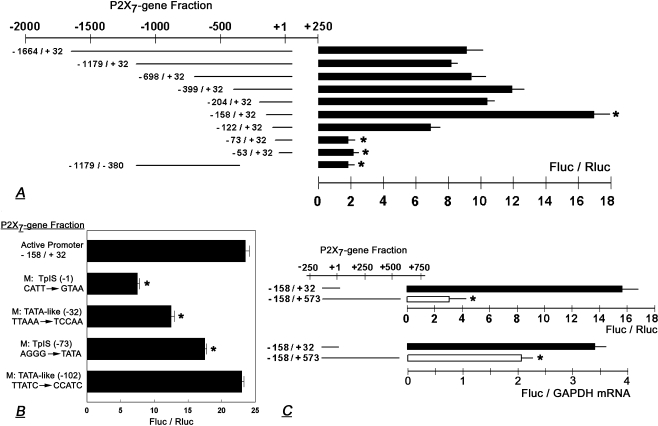
To define the active promoter region and the TpIS, a series of cDNA fragments were generated encompassing a 1.7-kb DNA segment at the 5′ region of the human P2X7 gene. Nucleotides were numbered relative to the subsequently elucidated TpIS (+1), which corresponds with nt 1683 of the human P2X7 gene (GenBank Y12851). Initial experiments included cDNA fragments ranging from −1664/+32 to −53/+32 nt (Figs. 1 and 2a). The cDNA fragments were inserted into a luciferase vector, and the P2X7-luciferase reporter was transfected into HEK293 cells which lack endogenous expression of the P2X7. Significant promoter activity was found in fragments ranging from −1664/+32 to −158/+32 nt, while shorter fragments lacked significant promoter activity (Fig. 2a). A cDNA fragment of −1179/−380 nt lacked significant promoter activity, suggesting that there is little promoter activity upstream of nt −380. Since maximal promoter activity was found in experiments using the −158/+32 nt fragment (Fig. 2a), the data suggested location of the active promoter of the human P2X7 gene in the −158/+32 nt region.
Fig. 2.
a–b. Elucidation of the P2X7 Active Promoter Region. a cDNA fragments corresponding to regions within a 1.7-kb DNA segment of the 5′ region of the human P2X7 gene were inserted into luciferase vector; the P2X7-luciferase reporters were transfected into HEK293 and P2X7 promoter activity was determined in terms of changes in luciferase activity (Fluc/Rluc). Data (means ± SD, three to five experiments in triplicates) were normalized (=1) to Fluc/Rluc recorded in cells transfected with empty vector. *p < 0.01 compared to the rest. b Confirmation of P2X7 TpIS. Two potential TpISs and their related TATA-like regions were mutated and effects on P2X7 transcription were determined as in a (means±SD, three to five experiments in triplicates). *p < 0.01 compared to −158/+32. c P2X7 transcription is modulated by effectors downstream of the active promoter. P2X7 −158/+32 or −158/+573 luciferase reporters were transfected into HEK293 cells and P2X7 promoter activity was determined in terms of changes in luciferase activity (Fluc/Rluc, upper panel) or in terms of changes in Fluc/GAPDH mRNA (lower panel). Shown are means (±SD) of one to three experiments in triplicates. Data of Fluc/GAPDH mRNA were normalized (=0) to those recorded in cells transfected with empty vector. *p < 0.01
The TpIS was elucidated using the modified 5′ RACE method. By sequencing, two possible TpISs were found upstream of the subsequently defined TpIS: adenine bases at +1 nt and −73 nt (Fig. 1). To determine which of the two is important, the TpIS-corresponding regions were mutated as (−1) CATT to GTAA, and (−73) AGGG to TATA (Fig. 2b). Fragments carrying the mutated regions were inserted into the luciferase vector and transfected into HEK293 cells. The results showed 70% luciferase activity in the (−73)AGGG→TATA construct but only 30% luciferase activity in the (−1)CATT→GTAA construct (Fig. 2b). Sequence analysis also suggested two TATA-like sequences located upstream of the subsequently defined TpIS: TTAAA at −32 nt, and TTATC at −102 nt (Fig. 1). Mutation analysis revealed 50% luciferase activity in the (−32)TTAAA→TCCAA construct and no change in luciferase activity in the (−102)TTATC→CCATC construct (Fig. 2b). Collectively, these data suggest a functionally important TpIS at site +1 nt and a TATA-like sequence TTAAA at site −32 nt.
Regions distal to the TpIS inhibit transcription
Sequence analysis of the human P2X7 gene (GenBank Y12851) downstream of the active promoter revealed an unusually high concentration of cytosine-phosphodiester-guanosine (CpG) dinucleotides sites in the +26 to +573 nt region (Fig. 1). This region of 547 nt contains 20 CpG sites, in contrast to the −158/+32 190-bp active promoter region, which contains only 4 CpG sites (Fig. 1). Since changes in methylation of cytosines within CpG sites can modulate gene function [51], we tested the hypothesis that changes in the methylation status of CpG sites in the region downstream of the active promoter can regulate P2X7 transcription.
A construct containing the fragment −158/+573 resulted in less transcription compared to the −158/+32 construct (Fig. 2c). To ascertain that the changes in Fluc/Rluc shown in Fig. 2c (upper panel) are not artifactual, the experiment was repeated using as endpoint changes in Fluc/GPDH mRNA. The data were similar (Fig. 2c, lower panel) indicating that cis regulatory elements contained within the +33 to +573 nt region downstream of the active P2X7 promoter inhibit transcription.
Aza-dC effects on P2X7 mRNA levels
To determine whether changes in DNA methylation modulate P2X7 gene transcription, cultured cervical cells were treated with the de-methylation agent 5-aza-2′-deoxycytidine (Aza-dC), and effects on P2X7 mRNA steady-state levels were measured. For experiments, cells were treated with 1 μM Aza-dC, which in preliminary experiments exerted near maximal effects (not shown).
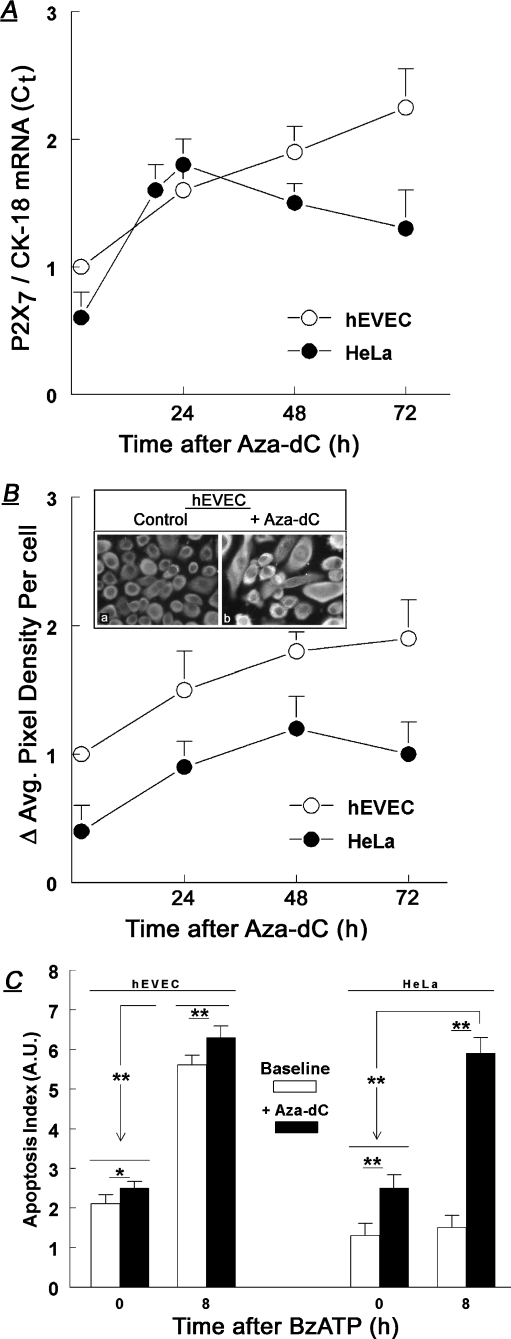
Baseline levels of P2X7 mRNA steady-state levels (relative to CK-18) were higher in the normal hEVEC than in the HeLa cancer cervical epithelial cells (Fig. 3a), confirming previous reports [7, 11]. Treatment with Aza-dC increased P2X7 mRNA both in hEVEC and in HeLa cells (Fig. 3a). The effect was time-dependent, and increases in P2X7 mRNA were observed already 18–24 h after treatment (Fig. 3a). In hEVEC cells, levels of P2X7 mRNA continued to increase; in HeLa cells P2X7 mRNA levels plateaued after 24 h and began to decrease afterwards, but remained elevated compared to baseline for at least 72 h after the start of treatment (Fig. 3a).
Fig. 3.
Effects of treatments with Aza-dC (1 μM) on steady-state levels of P2X7 mRNA (a), P2X7 receptor protein levels (b), and BzATP-induced apoptosis (in arbitrary units [A.U.]) (c). Means ± SD of three to six experiments in triplicates. Insert in b shows immunofluorescence data (×20). Data in a and b were normalized (=1) to levels in hEVEC cells at t = 0. The time-related increases in P2X7 mRNA (a) and P2X7 receptor protein levels (b) for both hEVEC and HeLa cells were significant (p < 0.01). Protein levels of P2X7 remained higher (p < 0.01) in hEVEC than in HeLa cells throughout the length of the experiment. In c, treatments with Aza-dC were followed by 100 μM BzATP (added for 8 additional hours). The degree of apoptosis (in arbitrary units [A.U.]) was normalized to levels determined in non-treated cells. In c, *p < 0.05; **p < 0.01
Aza-dC effects on P2X7 protein levels
Aza-dC effects on P2X7 protein were determined in terms of changes in cellular immunoreactivity to the anti P2X7 antibody. In non-treated hEVEC cells, staining with the anti P2X7 receptor antibody revealed homogenous P2X7 immunoreactivity (Fig. 3b, insert), and treatment with 1 μM Aza-dC for 48 h increased the P2X7 immunoreactivity significantly (Fig. 3b). A similar effect was seen in HeLa cells (not shown). Image analysis of P2X7 immunofluorescence density in Aza-dC-treated cells revealed a twofold increase after 48 h of treatment in P2X7 immunoreactivity for both the hEVEC and HeLa cells (Fig. 3b).
Aza-dC effects on baseline and P2X7-mediated apoptosis
Baseline apoptosis, which is induced paracrinologically by ATP secreted by the cultured cells [8], was twofold higher in hEVEC cells than in HeLa cells (Fig. 3c), confirming previous reports [8, 11]. Treatment with the P2X7-receptor-specific agonist BzATP increased apoptosis 2.5-fold in hEVEC cells but it had no significant effect in HeLa cells (Fig. 3c). Treatment with Aza-dC increased mildly baseline apoptosis in hEVEC cells, and significantly (twofold) baseline apoptosis in HeLa cells (Fig. 3c). In hEVEC cells pretreated with Aza-dC, co-treatment with BzATP resulted in greater apoptosis than in cells treated only with BzATP, but the effect was mild (Fig. 3c). In contrast, in Aza-dC-pretreated HeLa cells co-treatment with BzATP resulted in 2.5-fold greater apoptosis than in cells treated only with BzATP, and the degree of apoptosis was nearly that observed in hEVEC cells under similar conditions (Fig. 3c). These data indicate that Aza-dC sensitized normal and cancer cervical cells to the pro-apoptotic effect of BzATP, probably by upregulating the expression of P2X7 mRNA (Fig. 3a) and protein receptor (Fig. 3b).
To determine whether apoptosis per se can stimulate an increase in P2X7 mRNA, hEVEC and HeLa cells were incubated in serum-free medium for 14 h. The rationale was that serum deprivation increases apoptosis of cultured cervical cells [8]. The results (not shown) indicated that P2X7 mRNA steady-state levels in hEVEC and in HeLa cells were similar in serum-deprived and in serum-exposed cells.
Modulation of DNA methylation in HEK293 cells
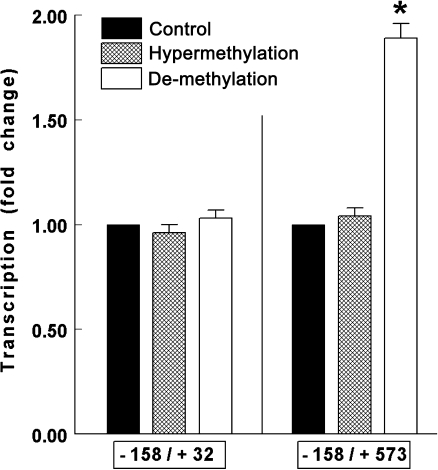
Effects of changes in the methylation status on P2X7 transcription were tested more directly in HEK293 cells transfected with the luciferase P2X7 −158/+32 or the −158/+573 reporters. Hypermethylation assays involved incubation of the test plasmids with the CpG-Methylase M.SssI prior to transfections, and de-methylation assays were done by treating transfected cells with Aza-dC. Hypermethylation had no significant effects on luciferase mRNA levels in HEK293 cells transfected either with the luciferase P2X7 −158/+32 or luciferase −158/+573 reporters (Fig. 4). De-methylation, induced by treatment with Aza-dC had no significant effect on luciferase mRNA levels in cells expressing the −158/+32 nt construct. However, it increased luciferase mRNA levels twofold in cells expressing the −158/+573-nt construct (Fig. 4).
Fig. 4.
Effects of hypermethylation and de-methylation on changes in transcription in HEK293 cells transfected with the luciferase −158/+32 or −158/+573 reporters (means±SD of two experiments in triplicates). Hypermethylation assays were done by incubating the test plasmids prior to transfections with the CpG-Methylase M.SssI and changes in transcription were determined in terms of changes in luciferase activity (Fluc/Rluc). De-methylation assays were done by treating transfected cells with Aza-dC (1 μM for 48 h). Changes in transcription were determined in terms of changes in Fluc/GAPDH mRNA levels. Data were normalized (=1) to levels in control cells. *p < 0.01
The specificity of the effects shown above was determined by measuring effects of de-methylation and hypermethylation on CK-18 and GAPDH mRNA steady-state levels. Both in hEVEC and in HeLa cells, treatment with Aza-dC had no significant effects on the steady-state levels of CK-18 and GAPDH mRNA levels (not shown). Also, in HEK293 cells transfected with either the luciferase P2X7 −158/+32 or the −158/+573 reporters, de-methylation and hypermethylation assays did not affect significantly steady-state levels of GAPDH mRNA (not shown).
Collectively, the data in Figs. 3 and 4 suggest that regions downstream of the active P2X7 promoter regulate transcription by modulation of DNA methylation.
CpGs downstream of the promoter inhibit transcription
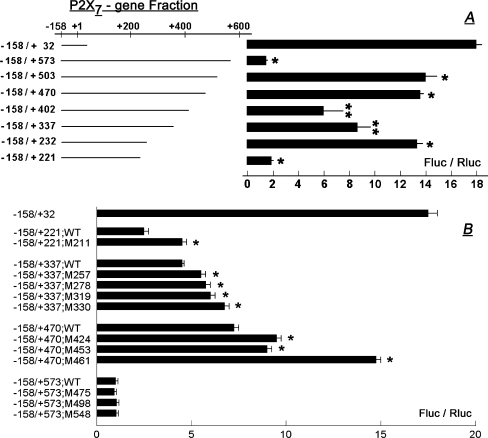
To understand which CpG sites in the 547-nt region downstream of the promoter play a role in the regulation of P2X7 transcription, cDNA fragments were constructed containing the active promoter (nt −158/+32) attached with one of different segments of the 547-nt region downstream of the promoter (Fig. 5a). The cDNA fragments were inserted into a luciferase vector; the P2X7-luciferase reporter was transfected into HEK293 cells, and P2X7 transcription was determined in terms of luciferase activity.
Fig. 5.
Elucidation of transcription regulatory cis elements within a CpG-rich 547-nt region downstream of the P2X7 promoter. a cDNA fragments were constructed containing the P2X7 active promoter (nt −158/+32) attached with one of the shown segments of the 547-nt region downstream of the promoter (Fig. 1). b Effects of mutations in the CpG sites within the 547-nt region downstream of the promoter on P2X7 transcription. WT wild-type. Mutations are described in the section of “Methods” section. For both a and b cDNA fragments composed of the P2X7 active promoter attached with one of the shown segments were inserted into a luciferase vector and transfected into HEK293 cells. Promoter activity was determined in terms of changes in luciferase activity (Fluc/Rluc, means ± SD, of two experiments in triplicates). In a *p < 0.01 compared to −158/+32; **p < 0.05–0.01 compared to −158/+232. In b *p < 0.01 compared to the wild-type sequence in each case
Luciferase activity in the fragment −158/+221 was low, about 10% compared to that of the active promoter alone (nt −158/+32; Fig. 5a). Luciferase activity in the fragment −158/+232 was higher, about 75% compared to the active promoter (Fig. 5a). Luciferase activities in fragments −158/+337 and −158/+402 were lower by 25% and 50%, respectively than in fragment −158/+232 (Fig. 5a). Luciferase activity in fragments −158/+470 and −158/+503 was similar to that in fragment −158/+232 (Fig. 5a). Luciferase activity in fragment −158/+573 was low, about 10% of that in fragment −158/+32 (the active promoter; Fig. 5a).
To determine if CpG sites in the 547-nt region downstream of the promoter affect P2X7 transcription, selected CpG sites were mutated (Fig. 5b; Table 4), and the effects on transcription were tested. The following mutations resulted in a significant increase in luciferase activity: +211/+212 (CG/AA), +257/+258 (CG/TT), +278/+279 (CG/TT), +319/+320 (CG/AT), +330/+331 (CG/AT), +424/+425 (CG/TT), +453/+454 (CG/TT), and +461/+464 (CGCG/ATTA; Fig. 5b). These data suggest that CpGs +211/+212, +257/+258, +278/+279, +319/+320, +330/+331, +424/+425, +453/+454, and the bi-CpG complex +461/+464 inhibit P2X7 transcription.
CpG sites downstream of the active promoter are constitutively hypermethylated in cervical cells
The data in the cultured host HEK293 cells suggested that CpG sites in the CpG-rich 547-nt region downstream of the active promoter can modulate P2X7 transcription through changes in their methylation status. To confirm the discovery and to understand its biological relevance, the degree of methylation of cytosines within selected CpG sites in the 547-nt region was determined in cultured normal and cancerous human uterine cervical epithelial cells, and in normal and cancerous human cervix epithelial tissues. The specific objective was to correlate cytosines methylation status with cell histology (normal versus cancer).
The control experiments are shown in Fig. 6a and b. The first experiment tested effects of treatment with Aza-dC on the methylation status of cytosines within CpG sites of the 547-nt region. cDNA segments of interest (segments 1–3) were generated from Aza-dC-treated HeLa cells and analyzed by the genomic DNA bisulfite conversion method followed by gene-specific PCR and restriction enzyme cutting. The amplified segments were cut with restriction enzymes to detect potential methylation-sensitive cytosines at CpG sites. Sites +193/+194 and or +211/+212 (segment 1), and +330/+331 (segment 2) were cut with MaeII; sites +461/+462 and or +463/+464 (segment 3) were cut with BstUI (Fig. 1). The employed method could not differentiate between sites +193/+194 and +211/+212 due to their close proximity which could not be resolved by gel electrophoresis; or between sites +461/+462 and +463/+464 because either could be the target of the BstUI.
Fig. 6.
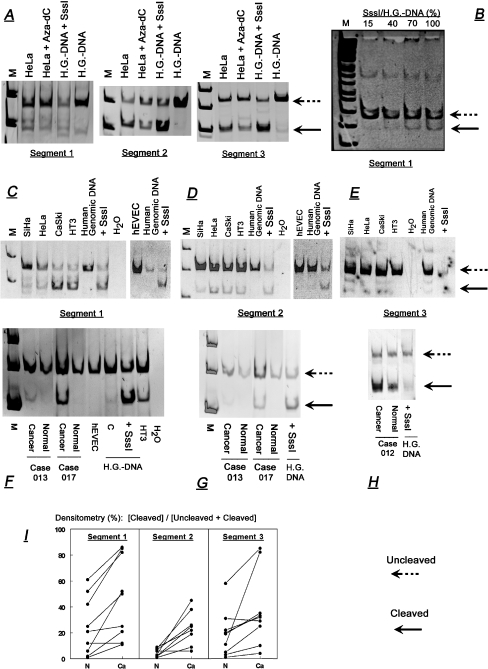
a Effects of treatment with Aza-dC in HeLa cells (1 μM, 48 h) on methylation of cytosines in CpG sites +193/+194 (and/or +211/+212), +330/+331, and +461/+462 (and/or +463/+464), within segments 1, 2, and 3 respectively of a 547-nt region downstream of the P2X7 active promoter (Fig. 1). Methylation of cytosines in CpG sites was determined in terms of cleavage at CpG sites using the genomic DNA bisulfite conversion method followed by gene-specific PCR and restriction enzyme cutting. Left-pointing arrows show bands corresponding to uncleaved (broken lines) and cleaved fractions (continuous lines) at the CpG sites. M markers. Controls were aliquots of human placental genomic DNA (H.G.-DNA) treated in vitro with the CpG-methylase SssI. b Aliquots of human placental genomic DNA were mixed with different molar concentrations of SssI and the degree of cleavage at CpG sites +193/+194 (and/or +211/+212) within segment 1 was determined as in a. Data were normalized to the effect (100%) obtained in a reaction mixture containing 1 μg DNA. c–h Methylation status of cytosines in CpG sites in cultured human epithelial uterine cervical cells (c–e) and in human uterine cervix tissues in vivo (f–h). Experiments were done as in a, and data on the degree of the cleaved fractions (in terms of the ratio of densitometry of the cleaved, versus the uncleaved plus cleaved bands [%]) are summarized in Table 1. i Degree of cleavage at CpG sites in tissues of human cervix. Data were compiled from ten sets of paired cervical specimens, including in each case normal and squamous cell carcinoma tissues. Available for analysis were nine cases for segments 1 and 3, and eight cases for segment 2. Experiments and data evaluation were done as above. Lines connect paired tissues (Normal [N] and Cancer [Ca]) from the same patient. Data are explained in the section of “Results” section
Figure 6a shows that treatment with Aza-dC decreased cleavage at CpG sites in all three segments. Densitometry of the data in Fig. 6a showed that the degree of cleavage at CpG sites (defined in terms of the ratio of densitometry of the cleaved band, relative to the uncleaved plus cleaved bands [%]) decreased in segment 1 from 22% to 5%; in segment 2 from 65% to 43%; and in segment 3 from 48% to 22%. These data suggest that Aza-dC induced de-methylation of cytosines within CpG sites at the 547-bp region, and confirmed the validity of the method that was used.
The positive control for the experiment in Fig. 6a was human placental genomic DNA treated in vitro with the CpG-methylase SssI. Aliquots of placental DNA were mixed with different amounts of SssI and the degree of methylation was determined in terms of cleavage at CpG sites, as in Fig. 6a. The degree of methylation in the absence of methylase SssI was small (Fig. 6a), but it increased in a dose-related manner (Fig. 6b), relative to the amount of Sss1 versus placental DNA in the reaction mixture.
A similar method was used to evaluate the methylation status of cytosines within those CpG sites in cultured normal and cancer human cervical cells (Fig. 6c–e), and in normal and cancer human cervix epithelial tissues (Fig. 6f–i). The results in cultured cervical cells showed bands corresponding to fractions cleaved at those CpG sites (Fig. 6c–e). The results also revealed greater degree of cleavage in cultured cancer cells than in cultured normal epithelial cervical cells (Fig. 6c–e). Similar experiments were done on human cervix epithelial tissues using specimens obtained by microdissection from uterine tissue cross sections. Experiments utilized paired specimens from ten patients, including, in each case, normal and squamous cell carcinoma tissues. Sufficient amounts of tissues were available in nine cases for segments 1 and 3 and in eight cases for segment 2. The results showed bands corresponding to fractions cleaved at the above CpG sites both in cancer and in normal tissues, with a similar tendency of greater degree of cleavage in cancer tissues than in normal tissues (Fig. 6f,h).
Semi-quantitative analysis of the data in cultured cervical cells showed a five- to tenfold-higher degree of cleaved fractions in all three segments in cancer cells than in normal cells (Table 7), suggesting greater degree of hypermethylation of cytosines at the tested CpG sites in the cancer cells than in the normal cervical cells. Similar trends were obtained in cervix epithelial tissues in vivo. Data analysis in those tissues used the densitometry ratios of [cleaved]/[uncleaved plus cleaved] fractions in normal and cancer tissues obtained from the same patient. The results showed greater degree of cleaved fractions in cancer cases than in the corresponding normal tissues in eight of nine cases of segment 1 (p < 0.05); in seven of eight cases of segment 2 (p < 0.01), and in eight of nine cases of segment 3 (p < 0.05p; Fig. 6i). Collectively, the data in Fig. 6c–l suggest that in cultured cervical cells and in the cervix in vivo cytosines within CpG sites +193/+194 (and or +211/+212), +330/+331, and +461/+462 (and or +463/+464) in the 547-bp region downstream of the active P2X7 promoter are hypermethylated to a greater degree in cancer cells than in normal epithelial cells.
Table 7.
The degree of the cleaved fractions (in terms of the ratio of densitometry of the cleaved versus the uncleaved plus cleaved bands [%]) at CpG sites +193/+194 (and/or +211/+212; segment 1); +330/+331 (segment 2); and +461/+462 (and/or +463+464; segment3) downstream the P2X7 active promoter in cultured cancer and normal cervical cells (means ± SD of three to four experiments). *p < 0.01
| Segment 1 | Segment 2 | Segment 3 | |
|---|---|---|---|
| Cancer cells | |||
| HeLa | 24 ± 63 | 23 ± 4 | 23 ± 2 |
| CaSki | 55 ± 23 | 38 ± 13 | 20 ± 4 |
| HT3 | 53 ± 17 | 35 ± 10 | 22 ± 4 |
| SiHa | 7 ± 3 | 12 ± 5 | 25 ± 5 |
| Normal cells (hEVEC) | 2 ± 1* | 1 ± 2* | 13 ± 3* |
DNA–protein binding within putative enhancer regions
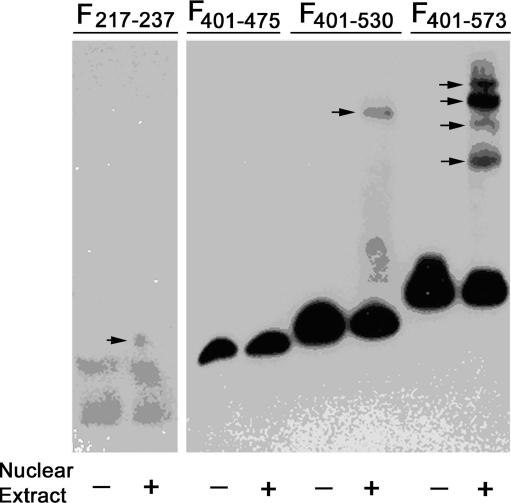
One of the mechanisms by which cells regulate gene expression is via transcription modulating factors that bind in defined recognition sites. The data in Figs. 2 and 5 suggest that the transcription inhibitory effects of the hypermethylated CpGs are associated with their proximity to regions that may have enhancer activity. To begin understanding the molecular mechanism of P2X7 transcription regulation we tested the hypothesis that the putative enhancer regions nt + 222/+232 and +401/+573 in the 547-bp region downstream of the active P2X7 promoter contain protein binding sites. To this aim, electrophoretic mobility shift assays (EMSA) were used to detect DNA–protein complexes. Using four amplified fragments (Table 5) we found one shifted band in the +217/+237 fragment; one shifted band in the +401/+530 fragment; and four shifted bands in the +401/+573 fragment (Fig. 7). These data indicate the presence of DNA–protein complexes in the +217/+237 and the +476/+573 regions.
Fig. 7.
Elucidation of DNA–protein binding within the 547-nt region downstream of the P2X7 promoter. cDNA fragments were constructed containing the indicated P2X7 gene segments (Table 5). Electrophoretic mobility shift assays (EMSA) were used to detect DNA–protein complexes. Right-pointing arrows indicate shifted bands. The experiment was repeated twice
Discussion
The present study reports, for the first time, the TpIS of the P2X7 gene, and defines its active promoter region. We also discovered that a CpG-rich 547-nt region downstream of the active promoter regulates transcription: regions at nt +222/+232 and +401/+573 downstream of the active P2X7 promoter contain enhancer cis elements, while methylated CpGs located in the +33/+573 region downstream of the active P2X7 promoter exert an inhibitory effect on transcription.
The data that a CpG-rich 547-nt region downstream of the active promoter inhibits transcription are novel in terms of epigenetic (methylation) control of transcription by cis-inhibitory elements located downstream of an active promoter. Regulation of transcription by methylation of CpGs is a known epigenetic mechanism that modulates gene function [52]. However, in most genes, methylation involves the promoter, where the addition of methyl groups to the 5′ of cytosines can alter the appearance of the DNA groove to which the DNA-binding proteins bind [52]. This can modulate the spatial conformation of transcription factors recognition sites [53], or interfere with the recruitment of proteins that function as adaptors between DNA and chromatin-modifying enzymes [54, 55]. The net effect would be modulation of the transcription machinery [56].
Although the hypothesis that regions outside an active promoter can regulate transcription by modulation of DNA methylation is not new [51, 52, 57–59], until recently, there have been no reports of studies that tested it directly. The present data show, for the first time, direction-dependent regulation of promoter activity by methylated CpGs located downstream of an active promoter. The conclusion that the inhibition of P2X7 transcription imparted by CpGs within the 547-nt region involves effects of hypermethylated CpGs is supported by the data that CpGs located in the +33/+573 region are constitutively methylated; that methylation exerts an inhibitory effect on transcription; and that de-methylation increases transcription. The data also identified specific CpG sites that determine the inhibitory effect. Mutagenesis of CpGs +211/+212, +257/+258, +278/+279, +319/+320, +330/+331, +424/+423, +453/+454, and the bi-CpG complex +461/+464 increased transcription, and each of these CpGs inhibits transcription when methylated. Moreover, the data of bisulfite restriction experiments in cultured cervical cells, and in cervix epithelial tissues in vivo, showed that cytosine within CpG sites +193/+194 (and / or +211/+212), +330/+331, and +461/+462 (and/or +463+464) are endogenously methylated, and that the degree of methylation was greater in cancer cells than in normal cervical cells. From a functionality point of view, the data in segment 1 are relevant only to CpG +211/+212 since CpG +193/+194 had only minimal effect on transcription. Also, the effect in segment 3 involves mainly the bi-CpG +461/+464 complex because mutagenesis of both +461/+462 and +463/+464 CpGs produced the greatest effect (Fig. 5b). It is possible that in addition to CpGs +211/+212, +330/+331, and +461/+464, cytosines at additional CpG sites within the 547-nt region downstream of the promoter are also hypermethylated in cancer cells than in normal cervical cells. However, the lack of commercially available restriction enzymes to target additional CpG sites in this unique DNA segment made it impossible to extend the study to other CpG sites. An alternative method, cloning and sequencing of representative DNA segments could yield meaningful results only if single cells are processed, but in the context of the present study, this would have become an impractical approach given the large number of cells/tissues that were needed to be processed vis-à-vis the large cell variability in cultured and intact tissues.
Interestingly, the differences in methylation between the cancer and normal cervical cells correlated inversely with levels of P2X7 expression in these cells. P2X7 mRNA and protein receptor levels are lower in cancer cervical cells than in normal cells, both in culture and in vivo [7, 11–13], while CpG sites +211/+212, +330/+331, and +461/+464 were hypermethylated in the cancer cervical cells compared to the normal cells (present data). In other systems, hypermethylation of genes has been associated with repression of tumor suppressor genes [60], and has been implicated in cancer development and metastasis [51, 56, 61, 62]. DNA hypermethylation in the promoter [51] or in 5′ non-coding or coding regions [52, 57, 58] can lead to defects in DNA repair, cell cycle control, cell adherence, and apoptosis. Therefore, the correlation in cervical cancer cells between hypermethylation of cytosines in CpG sites downstream of the P2X7 promoter and the reduced expression of P2X7 mRNA and protein, together with the finding that those CpG sites exert an inhibitory effect on P2X7 transcription suggest a novel mechanism of P2X7 transcription. Accordingly, hypermethylated CpG sites +211/+212, +330/+331, and +461/+464, and perhaps additional hypermethylated CpGs within the 547-nt region downstream of the promoter, are direction-dependent inhibitory cis elements of P2X7 transcription.
The molecular mechanism by which hypermethylated CpGs within the 547-nt region downstream of the P2X7 promoter inhibit transcription is, at present, unknown. One possibility is by modulation of the spatial conformation of transcription factors recognition sites within the putative enhancer regions (present data). This conclusion is supported by the EMSA data which suggested the presence of four DNA–protein complexes in the +217/+237, in the +476/+530, and in the +531/+573 regions, and by the methylation assays which suggested that those regions are flanked by hypermethylated CpGs (Fig. 8). The possibility that one or more of the four DNA–protein complexes reflect transcription factors recognition sites is supported by bioinformatics analysis of the P2X7 gene [63] which revealed an association between the locations of the putative enhancer regions with factors known to regulate transcription of other genes. The sequence of the putative enhancer region +222/+232 matched with binding sites for p300, Elk-1, and E47. p300 promotes transcription by acetylating histones and integrating signaling from enhancer and promoter regions [64]; Elk-1 is regulated by phosphorylation in response to activation of mitogen-activated protein kinase (MAPK) pathways [65]; while E47 is a member of the E2 protein family encoded by the E2A gene. E47 regulates cell development and differentiation, and repression or absence of E47 activity has been implicated in cancer development [66]. Bioinformatics analysis of the sequence of the putative enhancer region +401/+573 also revealed putative binding sites for p300, Elk-1, and E47, as well for EIIaE, E2F, and p53. EIIaE and E2F are also members of the E protein family; both control cell cycle progression [67–69], and over-expression of E2F-1 can activate apoptosis [70]. The p53 tumor suppressor transcription factor controls expression of genes involved in the regulation of cell cycle progression and cell death [71], and there is an association between activation of the P2X7 receptor and the p53 apoptotic pathway [72], and between increased expression of the P2X7 receptor and p53 protein levels [73]. Additional studies are needed to determine whether the experimentally found four DNA–protein complexes correspond with any of the above suggested transcription factors.
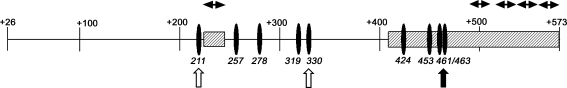
Fig. 8.
Schema of the CpG-rich 547-nt DNA region (+26/+573) downstream of the active promoter of the P2X7 gene. Filled ellipses denote CpG sites that were found experimentally to inhibit P2X7 transcription. Upwards pointing arrows denote CpG sites that were found experimentally to be hypermethylated in cultured cervical cells and in cervix epithelial tissues in vivo. Hatched squares are cis regions that were found experimentally to possess transcription enhancer activity. Horizontal bi-directional arrows show putative sites within the cis-enhancer regions that were found to form DNA–protein complexes
The present data showed that the putative enhancer +222/+232 and +401/+573 regions co-localize with, or are flanked by, constitutively methylated CpGs. The association of inhibitory CpGs with binding sites of putative enhancers suggests a novel mechanism, such that hypermethylated CpGs inhibit P2X7 transcription by modulating the interaction of enhancer transcription factors with their cognate DNA-binding domains.
Based on these findings, we propose a novel model of P2X7 transcription regulation (Fig. 8). Transcription of the P2X7 receptor is regulated by cis-enhancer elements located in nt +222/+232 and +401/+573 downstream of the active P2X7 promoter, which contain binding sites for transcription factors. Transcription of the P2X7 receptor is negatively controlled by methylated CpG sites +193/+194 (and/or +211/+212), +330/+331, and +461/+462 (and/or +463+464) that flank the enhancers (Fig. 8). Since transcription in cells expressing the −158/+573-nt construct was lower compared to that in cells expressing the active promoter (nt −158/+32), the data suggest that under baseline conditions, the effect of the putative inhibitors surpasses that of the putative enhancer(s).
The present data and previous studies in the field provide better understanding of the physiological and biological roles of the P2X7. The P2X7 receptor is a pro-apoptotic mechanism that controls cell growth, but cells have devised mechanisms to regulate expression and activity of the P2X7 receptor and to avoid death. First, enhancers of transcription downstream of the P2X7 promoter are controlled by neighboring CpG sites in a hypermethylated state (present data). Second, P2X7 transcripts can be degraded post-transcriptionally by the actions of micro-RNAs [13]. Third, cells produce non-functional variants of the P2X7 receptor [11, 74, 75] that can oligomerize with the functional full-length receptor [11]. In epithelial cells, P2X7-mediated apoptosis involves formation of pores that are composed of oligomers of P2X7 molecules [11, 76] and pannexins [77, 78], but co-expression of variants, e.g., the P2X7-j [11] would favor formation of non-active complexes of the receptor [11]. Together, these data indicate that under physiological conditions, the P2X7 mechanism is tightly controlled and regulated. The data also indicate that in several types of epithelia, e.g., the cervix, the expression of P2X7 in cancer cells is reduced [7, 11–13], suggesting that the abrogated P2X7-mediated apoptosis is associated with the development of cancer. At present, it is unknown whether the changes that lead to reduced expression of the P2X7 receptor, e.g., hypermethylation of the CpGs in cancer cells, are secondary to the cancer process; or whether they precede the carcinogenic stimuli such that the abrogated apoptosis predisposes cells to the development of cancer.
In summary, the present data suggest that P2X7 transcription is regulated by two groups of direction-dependent cis regulatory enhancers located within a 547-nt region downstream of the active promoter (segments +222/+232 and +403/+573), and is controlled by the actions of CpGs that co-localize with or cluster the enhancer sites. The experiments identified nine CpGs as inhibitory cis elements, and suggest that their effects depend on the degree of hypermethylation of the respective cytosines. Data in cervical cells elucidated three CpG sites, +211/+212, +330/+331, and +461/+464 that are hypermethylated in cultured cells and in vivo, with greater degree of hypermethylation in cancer cells than in the normal cervical cells.
Conclusions
P2X7-dependent apoptosis is an important mechanism that controls the growth of epithelial cells.
Reduced expression of the P2X7 receptor is associated with abrogated apoptosis, and it may predispose cells to the development of cancer.
P2X7 transcription is regulated by two groups of enhancers located within a 547-nt region (+26/+573) downstream of the active promoter.
P2X7 transcription was found to be controlled by the action of hypermethylated cytosines at CpG sites that cluster or co-localize with the enhancers’ sites.
Low expression of the P2X7 receptor in epithelial cancer cells, e.g., uterine cervical cancer cells is determined, in part, by the inhibition of the action of transcription enhancers, located within a 547-nt region downstream of the promoter, through the action of flanking hypermethylated CpGs.
Acknowledgment
The study was supported in part by NIH grant AG15955 and by an unrestricted grant by CytoCore Inc. to GIG.
Disclosure CytoCore Inc. funded a small part of the study but it has no financial interest in the study. Dr. Gorodeski was a paid consultant to CytoCore Inc. and he holds restricted stocks of CytoCore. The ties between Dr. Gorodeski and CytoCore were severed in March 2008 and Dr. Gorodeski has no financial interest in the study. None of the other authors had any ties with CytoCore. Neither Dr. Gorodeski, nor any of the other authors have other financial or non-financial competing interests. None of the authors hold any patents related to the study. University Hospital CASE Medical Center and Case Western Reserve University have a financial interest in the study.
Footnotes
Lingyin Zhou and Liping Luo are equal first authors.
References
- 1.Wyllie AH, Kerr JFR, Currie AR (1980) Cell death: the significance of apoptosis. Int Rev Cytol 68:251–306 [DOI] [PubMed]
- 2.Ellis HM, Yuan J, Horvitz HR (1991) Mechanisms and functions of cell death. Annul Rev Cell Biol 7:663–698 [DOI] [PubMed]
- 3.Fawthrop DJ, Boobis AR, Davies DS (1991) Mechanisms of cell death. Arch Toxicol 65:437–444 [DOI] [PubMed]
- 4.Wang Q, Wang L, Feng YH, Li X, Zeng R, Gorodeski GI (2004) P2X7-receptor mediated apoptosis of human cervical epithelial cells. Am J Physiol 287:C1349–C1358 [DOI] [PubMed]
- 5.Soti C, Sreedhar AS, Csermely P (2003) Apoptosis, necrosis and cellular senescence: chaperone occupancy as a potential switch. Aging Cell 2:39–45 [DOI] [PubMed]
- 6.Renvoize C, Biola A, Pallardy M, Breard J (1998) Apoptosis: identification of dying cells. Cell Biol Toxicol 14:111–120 [DOI] [PubMed]
- 7.Li X, Zhou L, Feng YH, Abdul-Karim F, Gorodeski GI (2006) The P2X7 receptor: a novel biomarker of uterine epithelial cancers. Cancer Epidemiol Biomark Prev 15:1–8 [DOI] [PMC free article] [PubMed]
- 8.Wang L, Feng YH, Gorodeski GI (2005) EGF facilitates epinephrine inhibition of P2X7-receptor mediated pore formation and apoptosis: a novel signaling network. Endocrinology 146:164–174 [DOI] [PMC free article] [PubMed]
- 9.Fu W, McCormick T, Qi X, Luo L, Zhou L, Li X, Wang BC, Gibbons HE, Abdul-Karim FW, Gorodeski GI (2009) Activation of P2X7-mediated apoptosis inhibits DMBA/TPA-induced formation of skin papillomas and cancer in mice. BMC Cancer 9:114 [DOI] [PMC free article] [PubMed]
- 10.Greig AV, Linge C, Healy V, Lim P, Clayton E, Rustin MH, McGrouther DA, Burnstock G (2003) Expression of purinergic receptors in non-melanoma skin cancers and their functional roles in A431 cells. J Invest Dermatol 121:315–327 [DOI] [PubMed]
- 11.Feng YH, Li X, Wang L, Zhou L, Gorodeski GI (2006) A truncated P2X7 receptor variant (P2X7-j) endogenously expressed in cervical cancer cells antagonizes the full-length P2X7 receptor through hetero-oligomerization. J Biol Chem 281:17228–17237 [DOI] [PMC free article] [PubMed]
- 12.Li X, Qi X, Zhou L, Catera D, Rote NS, Potashkin J, Abdul-Karim FW, Gorodeski GI (2007) Decreased expression of P2X7 in endometrial epithelial pre-cancerous and cancer cells. Gynecol Oncol 106:233–243 [DOI] [PMC free article] [PubMed]
- 13.Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, Gorodeski GI (2008) Micro-RNAs miR-186 and miR-150 downregulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem 283:28274–28286 [DOI] [PMC free article] [PubMed]
- 14.Li X, Qi X, Zhou L, Fu W, Abdul-Karim FW, MacLennan G, Gorodeski GI (2009) P2X7 receptor expression is decreased in epithelial cancer cells of ectodermal, uro-genital sinus, and distal paramesonephric-duct origin. Purinergic Signalling, in press. doi:10.1007/s11302-009-9161-3 [DOI] [PMC free article] [PubMed]
- 15.Dubyak GR, el-Moatassim C (1993) Signal transduction via P2-purinergic receptors for extracellular ATP and other nucleotides. Am J Physiol 265:C577–C606 [DOI] [PubMed]
- 16.Ralevic V, Burnstock G (1998) Receptors for purines and pyrimidines. Pharmacol Rev 50:413–492 [PubMed]
- 17.North RA (2002) Molecular physiology of P2X receptors. Physiol Rev 82:1013–1067 [DOI] [PubMed]
- 18.Sperlágh B, Haskó G, Németh Z, Vizi ES (1998) ATP released by LPS increases nitric oxide production in raw 264.7 macrophage cell line via P2Z/P2X7 receptors. Neurochem Int 33:209–215 [DOI] [PubMed]
- 19.Grahames CBA, Michel AD, Chessell IP, Humphrey DPA (1999) Pharmacological characterization of ATP- and LPS-induced IL-1β release in human monocytes. Br J Pharmacol 127:1915–1921 [DOI] [PMC free article] [PubMed]
- 20.Henriksen KL, Novak I (2003) Effect of ATP on intracellular pH in pancreatic ducts involves P2X7 receptors. Cell Physiol Biochem 13:93–102 [DOI] [PubMed]
- 21.Loomis WH, Namiki S, Ostrom RS, Insel PA, Junger WG (2003) Hypertonic stress increases T-cell Interleukin-2 expression through a mechanism that involves ATP release, P2 Receptor, and p38 MAPK activation. J Biol Chem 278:4590–4596 [DOI] [PubMed]
- 22.Ferrari D, Chiozzi P, Falzoni S, Dal Susino M, Melchiorri L, Baricordi OR, Di Virgilio F (1997) Extracellular ATP triggers IL-1β release by activating the purinergic P2Z receptor of human macrophages. J Immunol 159:1451–1458 [PubMed]
- 23.Aggarwal BB, Rath PC (1999) TNF-induced signaling in apoptosis. J Clin Immunol 19:350–364 [DOI] [PubMed]
- 24.Humphreys BJ, Rice J, Kertesy SB, Dubyak GR (2000) Stress-activated protein kinase/JNK activation and apoptotic induction by the macrophage P2X7 nucleotide receptor. J Biol Chem 275:26792–26798 [DOI] [PubMed]
- 25.Ferrari D, Wesselborg S, Bauer MK, Schulze-Osthoff K (1997) Extracellular ATP activates transcription factor NF-κB through the P2Z purinoreceptor by selectively targeting NF-κB p65. J Cell Biol 139:1635–1643 [DOI] [PMC free article] [PubMed]
- 26.Buell GN, Talabot F, Gos A, Lorenz J, Lai E, Morris MA, Antonarakis SE (1998) Gene structure and chromosomal localization of the human P2X7 receptor. Recept Channels 5:347–354 [PubMed]
- 27.Li CM, Campbell SJ, Kumararatne DS, Bellamy R, Ruwende C, McAdam KP, Hill AV, Lammas DA (2002) Association of a polymorphism in the P2X7 gene with tuberculosis in a Gambian population. J Infect Dis 186:1458–1462 [DOI] [PubMed]
- 28.Li CM, Campbell SJ, Kumararatne DS, Hill AVS, Lammas DA (2002) Response heterogeneity of human macrophages to ATP is associated with P2X7 receptor expression but not to polymorphisms in the P2RX7 promoter. FEBS Letters 531:127–131 [DOI] [PubMed]
- 29.Denlinger LC, Coursin DB, Schell K, Angelini G, Green DN, Guadarrama AG, Halsey J, Prabhu U, Hogan KJ, Bertics PJ (2006) Human P2X7 pore function predicts allele linkage disequilibrium. Mol Diagnostics Gen 52:995–1004 [DOI] [PubMed]
- 30.Gasser S, Raulet D (2006) The DNA damage response, immunity and cancer. Semin Cancer Biol 16:344–347 [DOI] [PubMed]
- 31.Kujoth GC, Leeuwenburgh C, Prolla TA (2006) Mitochondrial DNA mutations and apoptosis in mammalian aging. Cancer Res 66:7386–7389 [DOI] [PubMed]
- 32.Rodriguez-Nieto S, Zhivotovsky B (2006) Role of alterations in the apoptotic machinery in sensitivity of cancer cells to treatment. Curr Pharm Des 12:4411–25 [DOI] [PubMed]
- 33.Gustafson KS, Furth EE, Heitjan DF, Fansler ZB, Clark DP (2004) DNA methylation profiling of cervical squamous intraepithelial lesions using liquid-based cytology specimens: an approach that utilizes receiver-operating characteristic analysis. Cancer Cytopathol 102:259–268 [DOI] [PubMed]
- 34.Li J, Zhang Z, Bidder M, Funk MC, Nguyen L, Goodfellow PJ, Rader JS (2005) IGSF4 promoter methylation and expression silencing in human cervical cancer. Gynecol Oncol 96:150–158 [DOI] [PubMed]
- 35.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB (2005) Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst 97:273–282 [DOI] [PubMed]
- 36.Sova P, Feng Q, Geiss G, Wood T, Strauss R, Rudolf V, Lieber A, Kiviat N (2006) Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomark Prev 15:114–123 [DOI] [PubMed]
- 37.Bhattacharjee B, Sengupta S (2006) CpG methylation of HPV 16 LCR at E2 binding site proximal to P97 is associated with cervical cancer in presence of intact E2. Virology 354:280–285 [DOI] [PubMed]
- 38.Jeong DH, Youm MY, Kim YN, Lee KB, Sung MS, Yoon HK, Kim KT (2006) Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer 16:1234–1240 [DOI] [PubMed]
- 39.Kang S, Kim JW, Kang GH, Lee S, Park NH, Song YS, Park SY, Kang SB, Lee HP (2006) Comparison of DNA hypermethylation patterns in different types of uterine cancers: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. Int J Cancer 118:2168–2171 [DOI] [PubMed]
- 40.Kitkumthorn N, Yanatatsanajit P, Kiatpongsan S, Phokaew C, Triratanachat S, Trivijitsilp P, Termrungruanglert W, Tresukosol D, Niruthisard S, Mutirangura A (2006) Cyclin A1 promoter hypermethylation in human papillomavirus-associated cervical cancer. BMC Cancer 6:55 [DOI] [PMC free article] [PubMed]
- 41.Narayan G, Goparaju C, Arias-Pulido H, Kaufmann AM, Schneider A, Dürst M, Mansukhani M, Pothuri B, Murty VV (2006) Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. BMC Molecular Cancer 5:16 [DOI] [PMC free article] [PubMed]
- 42.Okino K, Nagai H, Nakayama H, Doi D, Yoneyama K, Konishi H, Takeshita T (2006) Inactivation of Crk SH3 domain-binding guanine nucleotide-releasing factor (C3G) in cervical squamous cell carcinoma. Int J Gynecol Cancer 16:763–71 [DOI] [PubMed]
- 43.Wisman GBA, Nijhuis ER, Hoque MO, Reesink-Peters N, Koning AJ, Volders HH, Buikema HJ, Boezen HM, Hollema H, Schuuring E, Sidransky D, van der Zee AGJ (2006) Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer 119:1908–1914 [DOI] [PubMed]
- 44.Yang HJ, Liu VWS, Wang Y, Tsang PCK, Ngan HYS (2006) Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer 6:212 [DOI] [PMC free article] [PubMed]
- 45.Lai HC, Lin YW, Chang CC, Wang HC, Chu TW, Yu MH, Chu TY (2007) Hypermethylation of two consecutive tumor suppressor genes, BLU and RASSF1A, located at 3p21.3 in cervical neoplasias. Gynecol Oncol 104:629–635 [DOI] [PubMed]
- 46.Zhang Z, Joh K, Yatsuki H, Zhao W, Soejima H, Higashimoto K, Noguchi M, Yokoyama M, Iwasaka T, Mukai T (2007) Retinoic acid receptor β2 is epigenetically silenced either by DNA methylation or repressive histone modifications at the promoter in cervical cancer cells. Cancer Lett 247:318–327 [DOI] [PubMed]
- 47.Dong SM, Pai SI, Rha SH, Hildesheim A, Kurman RJ, Schwartz PE, Mortel R, McGowan L, Greenberg MD, Barnes WA, Sidransky D (2002) Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomark Prev 11:3–6 [PubMed]
- 48.Ivanova T, Petrenko A, Gritsko T, Vinokourova S, Eshilev E, Kobzeva V, Kisseljov F, Kisseljova N (2002) Methylation and silencing of the retinoic acid receptor-β2 gene in cervical cancer. BMC Cancer 2:4 [DOI] [PMC free article] [PubMed]
- 49.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV (2003) Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer 2:24 [DOI] [PMC free article] [PubMed]
- 50.Xiong Z, Laird PW (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acid Res 25:2532–2534 [DOI] [PMC free article] [PubMed]
- 51.Chen T, Li E (2006) Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol 301:179–201 [DOI] [PubMed]
- 52.Jones PA, Takai D (2001) The role of DNA methylation in mammalian epigenetics. Science 293:1068–1070 [DOI] [PubMed]
- 53.Comb M, Goodman HM (1990) CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res 18:3975–3982 [DOI] [PMC free article] [PubMed]
- 54.Hendrich B, Tweedie S (2003) The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet 19:269–277 [DOI] [PubMed]
- 55.Wade PA (2001) Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene 20:3166–3173 [DOI] [PubMed]
- 56.Bird AP, Wolffe AP (1999) Methylation-induced repression: belts, braces, and chromatin. Cell 99:451–444 [DOI] [PubMed]
- 57.Youssef EM, Chen XQ, Higuchi E, Kondo Y, Garcia-Manero G, Lotan R, Issa JPJ (2004) Hypermethylation and silencing of the putative tumor suppressor tazarotene-induced gene 1 in human cancers. Cancer Res 64:2411–2417 [DOI] [PubMed]
- 58.Youssef EM, Estecio MR, Issa JP (2004) Methylation and regulation of expression of different retinoic acid receptor beta isoforms in human colon cancer. Cancer Biol Ther 3:82–86 [DOI] [PubMed]
- 59.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JPJ (2005) MGMT promoter methylation and field defect in sporadic colorectal cancer. J Nat Cancer Inst 97:1330–1338 [DOI] [PubMed]
- 60.Tryndyak V, Kovalchuk O, Pogribny IP (2006) Identification of differentially methylated sites within unmethylated DNA domains in normal and cancer cells. Anal Biochem 356:202–207 [DOI] [PubMed]
- 61.Plass C, Soloway PD (2002) DNA methylation, imprinting and cancer. Eur J Hum Genet 10:6–16 [DOI] [PubMed]
- 62.Villagra A, Gutierrez J, Paredes R, Sierra J, Puchi M, Imschenetzky M, Wijnen Av A, Lian J, Stein G, Stein J, Montecino M (2002) Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J Cell Biochem 85:112–122 [DOI] [PubMed]
- 63.Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31:3651–3653 [DOI] [PMC free article] [PubMed]
- 64.Li M, Luo J, Brooks CL, Gu W (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 277:50607–50611 [DOI] [PubMed]
- 65.Yang SH, Sharrocks AD (2006) Convergence of the SUMO and MAPK pathways on the ETS-domain transcription factor Elk-1. Biochem Soc Symp 73:121–129 [DOI] [PubMed]
- 66.Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L (2008) E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol 181:5885–5894 [DOI] [PMC free article] [PubMed]
- 67.Mathis DJ, Elkaim R, Kédinger C, Sassone-Corsi P, Chambon P (1981) Specific in vitro initiation of transcription on the adenovirus type 2 early and late EII transcription units. Proc Natl Acad Sci USA 78:7383–7387 [DOI] [PMC free article] [PubMed]
- 68.Hamel PA, Gill RM, Phillips RA, Gallie BL (1992) Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol 12:3431–3438 [DOI] [PMC free article] [PubMed]
- 69.Zheng N, Fraenkel E, Pabo CO, Pavletich NP (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F–DP. Genes Dev 13:666–674 [DOI] [PMC free article] [PubMed]
- 70.Pardee AB, Li CJ, Reddy GP (2004) Regulation in S phase by E2F. Cell Cycle 3:1091–1094 [PubMed]
- 71.Laptenko O, Prives C (2006) Transcriptional regulation by p53: one protein, many Possibilities. Cell Death Differ 13:951–961 [DOI] [PubMed]
- 72.Schulze-Lohoff E, Hugo C, Rost S, Arnold S, Gruber A, Brüne B, Sterzel RB (1998) Extracellular ATP causes apoptosis and necrosis of cultured mesangial cells via P2Z/P2X7 receptors. Am J Physiol 275:F962–F971 [DOI] [PubMed]
- 73.Turner CM, Tam FWK, Lai PC, Tarzi RM, Burnstock G, Pusey CD, Cook HT, Unwin RJ (2007) Increased expression of the pro-apoptotic ATP-sensitive P2X7 receptor in experimental and human glomerulonephritis. Nephrol Dial Transplant 22:386–395 [DOI] [PubMed]
- 74.Cheewatrakoolpong B, Gilchrest H, Anthes JC, Greenfeder S (2005) Identification and characterization of splice variants of the human P2X7 ATP channel. Biochem Biophys Res Comm 332:17–27 [DOI] [PubMed]
- 75.Georgiou JG, Skarratt KK, Fuller SJ, Martin CJ, Christopherson RI, Wiley JS, Sluyter R (2005) Human epidermal and monocyte-derived langerhans cells express functional P2X receptors. J. Invest Dermatol 125:482–490 [DOI] [PubMed]
- 76.Kim M, Spelta V, Sim J, North RA, Surprenant A (2001) Differential assembly of rat purinergic P2X7 receptor in immune cells of the brain and periphery. J Biol Chem 276:23262–23267 [DOI] [PubMed]
- 77.Locovei S, Scemes E, Qiu F, Spray DC, Dahl G (2007) Pannexin1 is part of the pore forming unit of the P2X7R death complex. FEBS Lett 581:483–488 [DOI] [PMC free article] [PubMed]
- 78.Iglesias R, Locovei S, Roque A, Alberto AP, Dahl G, Spray DC, Scemes E (2008) P2X7 receptor-pannexin1 complex: pharmacology and signaling. Am J Physiol 295:C752–C760 [DOI] [PMC free article] [PubMed]