Abstract
We compared the DNA sequence difference of isolates of Clonorchis sinensis from one Korean (Kimhae) and two Chinese areas (Guangxi and Shenyang). The sequences of nuclear rDNA (18S, internal transcribed spacer 1 and 2: ITS1 and ITS2) and mitochondrial DNA (cytochrome c oxidase subunit 1: cox1) were compared. A very few intraspecific nucleotide substitution of the 18S, ITS1, ITS2 and cox1 was found among three isolates of C. sinensis and a few nucleotide insertion and deletion of ITS1 were detected. The 18S, ITS1, ITS2 and cox1 sequences were highly conserved among three isolates. These findings indicated that the Korean and two Chinese isolates are similar at the DNA sequence level.
Keywords: Clonorchis sinensis, ribosomal DNA, mitochondrial DNA, China, Korea
Clonorchiasis is endemic in East Asia, and this zoonotic infection ranges from China, Hong Kong, Macao, and Korea, to Laos and Vietnam (Rim, 1986). There is a previous report that isolates of C. sinensis of Korea (Kimhae) and of China (Liaoning) showed low level intraspecific variation of DNA. The two populations are very closely related within the range of a genetic identity value of 0.998-1.0 and they have a high homology in the nucleotide sequences of the 18S rDNA, ITS2 (internal transcribed spacer 2) and cox1 (cytochrome c oxidase subunit 1) (Park and Yong, 2001). However, there had not been sequence data of ITS1 of C. sinensis. To understand whether the difference of the geographical region is related with the differences of the intraspecific variation, we compared the DNA sequences of C. sinensis from one Korean and two different Chinese isolates besides of Liaoning. DNA sequences of C. sinensis were analyzed for the nuclear rDNA (18S, ITS1 and ITS2) and mitochondrial DNA (cox1) to reveal how much the sequence variation is among the geographic isolates.
Metacercariae of C. sinensis were collected after the artificial digestion of the muscle of the freshwater fish, Psudorasbora parva in Kimhae, Korea (A), Guangxi (B) and Shenyang (C), China. The adult worms were obtained from the rat (Sprague-Dawley, 4 to 6-weeksold male) liver one month post-infection with metacercariae (A and B) or from the rabbit liver five months post-infection (C). The worms collected were then stored at -70℃ until assayed. The frozen worms were lyophilized and lysed with a lysis buffer containing 1% SDS, proteinase K (500 µg/ml), and RNase at 37℃for 2-3 hr. The DNA was extracted in phenol/chloroform and precipitated in ethanol as reported by Sambrook and Russell (2001). The PCR primers previously designed for amplification of target DNA region were used (18S, Barker and Blair, 1996; ITS1, Bowles et al., 1993; ITS2, Bowles et al., 1995; cox1, Bowles et al., 1992). Each PCR was carried out in a volume of 50 µl, as follows: 10-100 ng of the extracted genomic DNA as template, 200 µM of each dNTP, 10 pmoles of each primer and 0.5 unit of Ex Taq enzyme (TAKARA Shuzo Co., Japan). PCR amplification consisted of 40 cycles of 20 second denaturation at 95℃ 30 second annealing at 50℃ 30 second extension at 72℃ followed by a final extension at 72℃for 6 minutes. The amplified PCR products were extracted using QIAEX II Gel extraction Kit (QIAGEN Co., Germany) and ligated into a T cloning vector (pT7Blue Perfectly Blunt Cloning Kit, Novagen Co., USA). Transformation was carried by E. coli NovaBlue competent cells provided in T cloning kit. Positive recombinant clones were picked and grown overnight in 2 ml of LB broth (in the presence of 50 µg/ml ampicillin) at 37℃ and the positive plasmid DNAs were purified using a QIAprep spin plasmid kit (QIAGEN Co.). The recombinant plasmids were selected by blue/white screening using isopropyl-β-thiogalactoside and 5-bromo-4 chloro-3-indolyl-β-D-galactoside. Plasmids DNAs of white colonies were digested with BamHI and HindIII restriction enzymes at 37℃ run in 1% agarose gels and stained with ethidium bromide. DNA sequencing was performed by the dideoxy chain termination method (Sanger et al, 1977) using a Sequenase kit (ABI Prism Dye Terminator Cycle Sequencing Core Kit, Perkin Elmer) and an automated DNA sequencer (Applied Biosystems model 373A, Perkin Elmer) according to manufacture's instructions. Sequencing primers included the universal primers T3 and T7 for both directions. At least two clones were sequenced per isolate with additional clones being sequenced as necessary to resolve ambiguous sites. The aligned sequence was done using the CLUSTAL W program (European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw/) among three isolates. Alignment gaps were treated as missing data (Higgins et al., 1992). For sequence analysis, we used BLAST in National Center for Biotechnology Information, NIH, Bethesda, USA. We also calculated the fractional GC content of nucleic acid sequences using an EMBOSS GEECEE program in Sanger Institute, Cambridge, U.K. (http://analysis.molbiol.ox.ac.uk/pise_html/geecee.html).
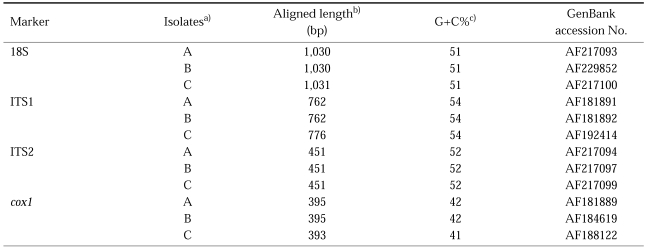
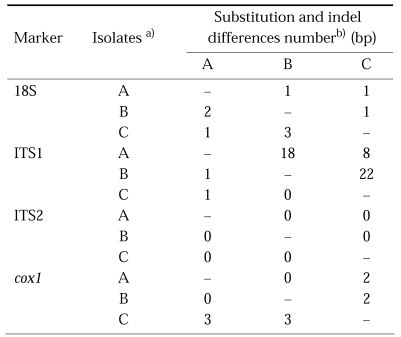
As results of sequencing, the length for each product of isolates was 1,030-1,031 bp for 18S; 762-776 bp for the ITS1; 451 bp for ITS2 and 393-395 bp for cox1 (Table 1). A G+C content was 51% (18S), 54% (ITS1), 52% (ITS2), and ranged from 41% to 42% (cox1) (Table 1). Sequences were deposited in GenBank by the accession numbers listed in Table 1. The nucleotide substitution differences among the isolates were 1-3 bp for the 18S, 1 bp for ITS1 and 3 bp for cox1. For the ITS2, no variation was detected among and within isolates from Korea and China. The nucleotide gap (insertion, deletion) differences among the isolates were 1 bp for the 18S, 8-22 bp for the ITS1 and 2 bp for cox1. For the ITS2, no gap was detected among isolates from Korea and China (data of multiple sequences alignment not shown). The ITS1 sequences showed much higher gap differences at nucleotide composition, 8-22 bp insertion and deletion at the same position among three isolates (Table 2).
Table 1.
Aligned sequence analysis for Clonorchis sinensis
a) A, isolate from Kimhae, Korea; B, isolate from Guangxi, China; C, isolate from Shenyang, China
b) Aligned length contains no missing data from Clustal W.
c) GC content was calculated from the EMBOSS GEECEE program.
Table 2.
Analysis of nucleotide differences of the each sequence among Clonorcis sinensis isolates
a) A, isolate from Kimhae, Korea; B, isolate from Guangxi, China; C, isolate from Shenyang, China
b) Below the diagonal is the nucleotide substitution number not adjusted for gaps (indel; insertion, deletion), and above the diagonal is the indel number from aligned sequence.
According to the above results, it was found that Korean and Chinese isolates of C. sinensis are remarkably similar at the DNA sequential level for ribosomal DNA and mitochondrial DNA. A few intraspecific variations in C. sinensis were found in the rDNA repeat (18S, ITS1 and ITS2) and cox1, and a few base insertion and deletions were detected in the ITS1 sequences. Extensive intraspecific sequence variations were not found between the Korean and two Chinese isolates. There were other comparable data on the intraspecific variations of ITS2 and cox1 sequence of parasitic trematodes in human. The nucleotide sequence of ITS 2 and cox1 of Opisthorchis viverrini in northeast Thailand showed intraspecific variation, that has been classified into 5 patterns, but no areaspecific pattern was observed. Nucleotide sequences in a region of the ITS2 from different areas were identical (Ando et al., 2001). Those data compared very well to the present results suggested that the geographic differences are not so significant for some DNA markers. The average sequence divergence among the Echinostoma species range from 2.2% in the ITS rDNA to about 8% for the cox1 (Morgan and Blair, 1988). DNA sequence similarities observed between the Korean and Chinese isolates of C. sinensis are indicative of a common ancestry. There is negligible intraspecific variation among one Korean and two Chinese isolates of C. sinensis.
ACKNOWLEDGEMENTS
The authors wish to thank Drs. Gab-Man Park, Kwandong University College of Medicine, Korea and Yoon Kong, Sungkyunkwan University College of Medicine, Korea for their kind donation of the two Chinese isolates.
Footnotes
This study was supported by the grant of Korea Health 21 R & D Project, Ministry of Health and Welfare, the Republic of Korea (HMP96-PJ1-PG2-M-0164).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under Accession Numbers See Table 1.
References
- 1.Ando K, Sithithaworn P, Nuchjunggreed C, et al. Nucleotide sequence of mitochondrial COI and ribosomal ITSII of Opisthorchis viverrini in Northeast Thailand. Southeast Asian J Trop Med Public Health. 2001;32:17–22. [PubMed] [Google Scholar]
- 2.Barker SC, Blair D. Molecular phylogeny of Schistosoma species supports traditional groupings within the genus. J Parasitol. 1996;82:292–298. [PubMed] [Google Scholar]
- 3.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- 4.Bowles J, Blair D, McManus DP. A molecular phylogeny of the human schistosomes. Mol Phylogenet Evol. 1995;4:103–109. doi: 10.1006/mpev.1995.1011. [DOI] [PubMed] [Google Scholar]
- 5.Bowles J, Hope M, Tiu WU, Liu X, McManus DP. Nuclear and mitochondrial genetic markers highly conserved between Chinese and Philippine Schistosoma japonicum. Acta Trop. 1993;55:217–229. doi: 10.1016/0001-706x(93)90079-q. [DOI] [PubMed] [Google Scholar]
- 6.Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL W: Improved software for multiple sequence alignment. Comput Appl Biosci. 1994;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 7.Morgan JA, Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial COI and ND1 genes for distinguishing among Echinostoma species (Trematoda) Parasitology. 1998;116:289–297. doi: 10.1017/s0031182097002217. [DOI] [PubMed] [Google Scholar]
- 8.Park GM, Yong TS. Geographical variation of the liver fluke, Clonorchis sinensis, from Korea and China based on the karyotypes, zymodeme and DNA sequences. Southeast Asian J Trop Med Public Health. 2001;32(Suppl 2):12–16. [PubMed] [Google Scholar]
- 9.Rim HJ. The current pathobiology and chemotherapy of Clonorchiasis. Korean J Parasitol. 1986;24(suppl):1–141. doi: 10.3347/kjp.1986.24.suppl.1. [DOI] [PubMed] [Google Scholar]
- 10.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. New York, USA: Cold Spring Harbor Laboratory Press; 2001. Chapter 6; pp. 6.1–6.30. [Google Scholar]