Abstract
Mathematical neuronal models are normally expressed using differential equations. The Parker-Sochacki method is a new technique for the numerical integration of differential equations applicable to many neuronal models. Using this method, the solution order can be adapted according to the local conditions at each time step, enabling adaptive error control without changing the integration timestep. The method has been limited to polynomial equations, but we present division and power operations that expand its scope. We apply the Parker-Sochacki method to the Izhikevich ‘simple’ model and a Hodgkin-Huxley type neuron, comparing the results with those obtained using the Runge-Kutta and Bulirsch-Stoer methods. Benchmark simulations demonstrate an improved speed/accuracy trade-off for the method relative to these established techniques.
Keywords: Parker-Sochacki, Spiking neural network, Numerical integration, Izhikevich, Hodgkin-Huxley
Introduction
Spiking neural network simulations are a flexible and powerful method for investigating the behaviour of neuronal systems. Spiking neuron models can be described mathematically as hybrid systems (Brette et al. 2007), with continuous evolution of the state variables punctuated by discrete synaptic and/or firing events. The continuous part of the system is generally described by a set of differential equations, and running a simulation involves repeatedly solving these equations using analytical or numerical integration methods.
The Parker-Sochacki (PS) method is a new technique for numerically integrating differential equations. PS computes iterative Talyor series expansions, enabling extraordinary integration accuracy in practical simulation time. The method is broadly applicable in computational modelling but has so far been largely overlooked in the biological sciences.
In this article, we explore the Parker-Sochacki method by applying it to two neuronal models: the Izhikevich ‘simple’ model (Izhikevich 2003), and a Hodgkin-Huxley neuron described in Brette et al. (2007). Benchmark simulations based on those established in Brette et al. (2007) are employed to compare the PS method with the established Runge-Kutta and Bulirsch-Stoer methods.
The Parker-Sochacki method
Most neuronal models can be expressed as initial value ordinary differential equations (ODEs) of the form
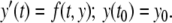 |
1 |
Picard’s method of successive approximations was designed to prove the existence of solutions to such equations. The method uses an equivalent integral form for Eq. (1)
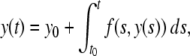 |
2 |
whose solution can be obtained as the limit of a sequence of functions yn(t) given by the following recurrence relation
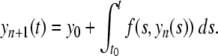 |
3 |
Provided f(t,y) satisfies the Lipschitz condition locally, this sequence is guaranteed to converge locally to y. However, the iterates become increasingly hard to compute, limiting the practicality of the method in this general form.
Parker and Sochacki (1996), considered a form of Eq. (1), with t0 = 0 and polynomial f. Note that the first condition is insignificant, since systems of the form of Eq. (1) can always be translated to the origin with a change of independent variable t →t + t0. Parker and Sochacki showed that polynomial f resulted in Picard iterates that were also polynomial. Furthermore, if yn(t) is truncated to degree n at each iteration, then the n-th Picard iterate is identical to the degree n Maclaurin Polynomial for y(t). Using a truncated Picard iteration to compute the Maclaurin series for a polynomial ODE was termed the Modified Picard Method in Parker and Sochacki (1996), but we follow Rudmin (1998) in calling it the Parker-Sochacki method.
For a system of ODEs with all polynomial right hand sides, the PS method can be used to compute the Maclaurin series for each variable to any degree desired, thus enabling arbitrarily accurate solutions for the ODE system within the regions of convergence of the series approximations. Parker and Sochacki (1996) went on to demonstrate that a broad class of analytical ODEs can be converted into polynomial form via variable substitutions, thus rendering them solvable via the PS method. The method was subsequently extended to partial differential equations (Parker and Sochacki 2000).
Rudmin (1998) established the practical utility of the PS method by using it to solve the N-body problem in celestial mechanics. Pruett et al. (2003) developed an adaptive time-stepping version of the method for the same problem. Carothers et al. (2005) built on the algorithmic work of Rudmin to derive an efficient, algebraic PS method using Cauchy products to solve for higher order terms.
Application
To apply the PS method to a polynomial ODE system, we first define Maclaurin series for each model variable
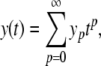 |
4 |
with y0 = y(0), y1 = y′(0), 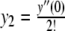 and so on. Now, because the Maclaurin series is polynomial, we can write down a series for the first derivative in terms of the original series
and so on. Now, because the Maclaurin series is polynomial, we can write down a series for the first derivative in terms of the original series
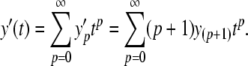 |
5 |
Equating terms, we have y′p = (p + 1)y(p + 1). Rearranging for coefficients in the original series, we arrive at a relation that lies at the heart of the PS method
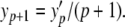 |
6 |
The basis of the method is to use the model differential equations to replace y′p with an expression in terms of the model variables. This is best illustrated through examples.
Example 1 Consider the linear system
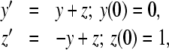 |
7 |
Here, y′p = yp + zp, z′p = − yp + zp and the PS solution is
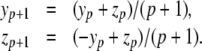 |
8 |
Thus, each coefficient of the Maclaurin series can be computed using the previous coefficients and we can easily obtain solutions of arbitrary order. This is the general principle of the PS method.
Example 2 To demonstrate how to handle constants and higher order terms, we consider
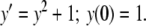 |
9 |
The series for y and y′ are as defined above, but an additional series is also defined for y2.
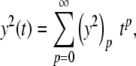 |
10 |
with coefficients generated using Cauchy products,
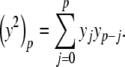 |
11 |
Since we can obtain the value of y2 given y, we refer to y2 as a derived variable, while y is a basic variable. The PS solution to Eq. (9) is given by
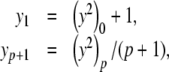 |
12 |
with p ≥ 1 and 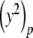 given by Eq. (11). Note that the constant term appears in the initial step but not in the subsequent iterations.
given by Eq. (11). Note that the constant term appears in the initial step but not in the subsequent iterations.
Simulations
In numerical simulations, the PS method is applied at each time step to solve for the system variables using initial conditions given by the solution at the previous time step. Thus, for a step size 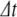 , the variables are updated using truncated series approximations (up to order n), as follows:
, the variables are updated using truncated series approximations (up to order n), as follows:
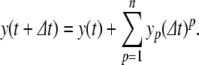 |
13 |
In a “clock-driven” simulation with fixed time step, it is always possible to rescale the system such that the effective step size becomes equal to one. Thus,
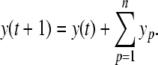 |
14 |
Adaptive order processing
One of the advantages of the Parker-Sochacki method is that the order of the Maclaurin series approximations depends only on the number of iterations, and can therefore be adapted according to the local conditions at each time step (Pruett et al. 2003; Carothers et al. 2005).
The PS solution is a sum over terms 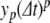 , which approximate the local truncation error for variable y on iteration p. However, with floating point numbers, rounding means that the actual change in solution at each iteration will only be approximately equal to
, which approximate the local truncation error for variable y on iteration p. However, with floating point numbers, rounding means that the actual change in solution at each iteration will only be approximately equal to 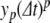 . Taking this into account, we apply adaptive error control by incrementally calculating the solution and halting the iterations when the absolute changes in value of all variables are less than or equal to some error tolerance value, ε.
. Taking this into account, we apply adaptive error control by incrementally calculating the solution and halting the iterations when the absolute changes in value of all variables are less than or equal to some error tolerance value, ε.
Variable substitutions
Equations containing exponential and trigonometric functions can often be converted into a form solvable by PS via the substitution of variables (Parker and Sochacki 1996, 2000; Carothers et al. 2005). We illustrate the method using a simple example relevant to neuronal modelling.
Example 3 Consider the system
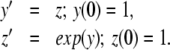 |
15 |
In order to transform the system, we let x = exp(y). Like y2 in Example 3, x here is a derived variable, while y and z are basic variables. Since the derivative of an exponential function is equal to the function itself, x′ = y′x, and the system can be rewritten:
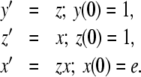 |
16 |
Power series operations
Application of the Parker-Sochacki method can be viewed in terms of power series operations, and the examples above demonstrate all of the operations required to solve any polynomial system of ODEs. Addition and subtraction operations are applied in term-wise fashion, while the Cauchy product performs multiplication. Since integer powers can be obtained using multiplication (y3 = y2y, y4 = y2y2 etc.), addition, subtraction and multiplication operations are sufficient to solve polynomial equations.
Knuth (1997) describes further power series operations that can be used to apply the Parker-Sochacki method to non-polynomial equations. First, we consider division. If we take two variables x,y, expressed as power series, and define a new variable to represent the quotient z = x/y, then using the Cauchy product we can write
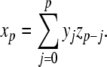 |
17 |
By rearrangement, we have (for 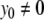 ):
):
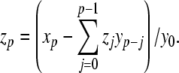 |
18 |
Just as the Cauchy product permits variable multiplication in ODEs solvable by the PS method, this formula adds division to the list of permissible operations. Thus, the ODEs need not be strictly polynomial as suggested in prior works. Rather, PS can be applied to any equation composed only of numbers, variables, and the four basic arithmetic operations (addition, subtraction, multiplication and division), with higher powers handled through iterative multiplication.
Alternatively, Knuth (1997) presents a formula, due to Euler, for raising series to powers directly. We consider only positive integer powers here. Briefly, if y0 = 1, then the coefficients of z = yα are given by z0 = 1 and (for p > 0)
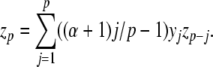 |
19 |
For series with 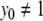 ,
, 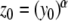 and (for p > 0)
and (for p > 0)
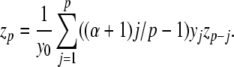 |
20 |
If the ((α + 1)j/p − 1) terms are pre-calculated, this method uses 2p multiplications and a single division to calculate the p-th coefficient. Thus, Euler’s power method can provide a computational saving over iterative multiplication only if more than two Cauchy products are required to calculate the power, i.e. α > 4.
As presented, both division and general power operations run the risk of encountering division by zero. We will return to the issue of division by zero in quotient calculations in the context of the Hodgkin-Huxley neuron model in Section 4. For Euler’s power method, we can circumvent the issue. If ym is the first non-zero coefficient in y, we define a new series x, with xp = yp + m. Next, we take w = xα and calculate the coefficients using Eq. (20). The series z has a number of leading zeros equal to the number of leading zeros in y, multiplied by the power (mα). Finally, zp + mα = wp.
The presented methods for performing power series division and power operations are not new (though we are not aware of a prior description of the technique for handling leading zeros in the power calculations). However, their incorporation into the Parker-Sochacki method is both novel and powerful, significantly expanding the method’s scope.
The Izhikevich model
The Izhikevich model (Izhikevich 2003, 2007) is a two variable, phenomenological neuron model, featuring a quadratic membrane potential, 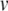 , and a linear recovery variable, u. The model is interesting because it has simple equations yet is capable of a rich dynamic repertoire (Izhikevich 2004, 2007). The model can act as either an integrator or a resonator and can exhibit adaptation or support bursting. Indeed, this is claimed to be the simplest model capable of spiking, bursting, and being either an integrator or a resonator (Izhikevich 2007).
, and a linear recovery variable, u. The model is interesting because it has simple equations yet is capable of a rich dynamic repertoire (Izhikevich 2004, 2007). The model can act as either an integrator or a resonator and can exhibit adaptation or support bursting. Indeed, this is claimed to be the simplest model capable of spiking, bursting, and being either an integrator or a resonator (Izhikevich 2007).
Subthreshold behaviour and the upstroke of the action potential can be represented as follows:
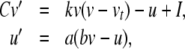 |
21 |
where 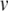 is the membrane potential minus the resting potential
is the membrane potential minus the resting potential 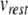 (
(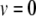 at rest),
at rest), 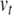 is the threshold potential, C is the membrane capacitance, a is the rate constant of the recovery variable, k and b are scaling constants, and I is the total (inward) input current from sources other than
is the threshold potential, C is the membrane capacitance, a is the rate constant of the recovery variable, k and b are scaling constants, and I is the total (inward) input current from sources other than 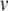 and u. Assuming the threshold potential is greater than the resting potential
and u. Assuming the threshold potential is greater than the resting potential 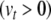 , then when
, then when 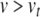 , the quadratic expression in Eq. (21) will be positive, and
, the quadratic expression in Eq. (21) will be positive, and 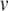 will tend to escape towards infinity. This escape process models the action potential upstroke. The action potential downstroke is modelled using an instantaneous reset of the membrane potential, plus a stepping of the recovery variable:
will tend to escape towards infinity. This escape process models the action potential upstroke. The action potential downstroke is modelled using an instantaneous reset of the membrane potential, plus a stepping of the recovery variable:
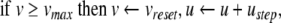 |
22 |
where 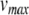 is the action potential peak,
is the action potential peak, 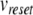 is the post-spike reset potential, and ustep is used to model post-spike adaptation effects. Spike times are taken as the times when Eq. (22) is applied.
is the post-spike reset potential, and ustep is used to model post-spike adaptation effects. Spike times are taken as the times when Eq. (22) is applied.
In our benchmark network simulations, synaptic interactions were modelled using a conductance-based formalism (Vogels and Abbott 2005; Brette et al. 2007). With the addition of fast excitatory (η) and inhibitory (γ) conductance-based synaptic currents, Eq. (21) becomes
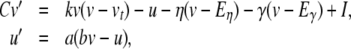 |
23 |
where η and γ are the total excitatory/inhibitory conductances, and Eη, Eγ are corresponding reversal potentials. The conductance values are stepped by incoming synaptic events of matching type, and decay exponentially with time
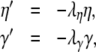 |
24 |
where the λ parameters are decay rate constants.
The Parker-Sochacki solution
In this section, we develop an efficient PS solution for the Izhikevich model system Eqs. (22), (23), (24). Most calculations in the PS method require a fixed number of floating point operations at each iteration, but Cauchy products require a number of operations that scales linearly with the number of iterations. Consequently, we seek to minimise the use of Cauchy products in designing an efficient algorithm.
A straightforward solution based on Eq. (23) would require three Cauchy products: one to compute 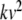 , one for
, one for 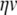 , and another for
, and another for 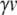 . Noting that these products contain the common factor
. Noting that these products contain the common factor 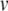 , we rearrange the membrane equation such that only one Cauchy product is required
, we rearrange the membrane equation such that only one Cauchy product is required
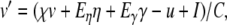 |
25 |
where 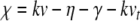 . Thus,
. Thus, 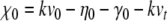 and (for p > 0)
and (for p > 0) 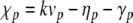 . Then the term
. Then the term 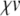 is given by a Cauchy product:
is given by a Cauchy product:
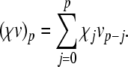 |
26 |
Using this construction, an efficient Parker-Sochacki solution for the Izhikevich model can be written down as:
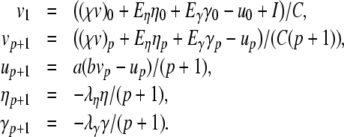 |
27 |
We can pre-calculate 1/C, 1/(p + 1) and 1/C(p + 1) and solve using only add, subtract and multiply floating point operations.
Calculating exact spike times
In clock-driven simulations, spike times are normally restricted to discrete time samples, offering limited spike timing accuracy despite accurate integration. Furthermore, Eq. (22) implies that discretisation of spike times will dramatically affect the subsequent accuracy of the solution. Specifically, the membrane potential increases rapidly during an action potential upstroke and, because of the voltage-dependence of the recovery variable, spike timing discretisation will tend to result in significant errors in the values of both 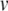 and u prior to the application of Eq. (22). When Eq. (22) is applied,
and u prior to the application of Eq. (22). When Eq. (22) is applied, 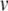 is reset to a fixed value regardless of its prior state, but u is stepped and thus depends on its prior value. Thus, errors in the value of u are propagated through to the post-spike state. This propagation of errors can be minimised by applying Eq. (22) at the correct times.
is reset to a fixed value regardless of its prior state, but u is stepped and thus depends on its prior value. Thus, errors in the value of u are propagated through to the post-spike state. This propagation of errors can be minimised by applying Eq. (22) at the correct times.
We now show how to calculate precise spike times for the Izhikevich model despite using large time steps in simulation. To establish our method, we note the following:
The Maclaurin series solution for
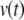 is a polynomial.
is a polynomial.Via a shift in
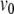 , locating a voltage threshold crossing can be posed as a polynomial root-finding problem.
, locating a voltage threshold crossing can be posed as a polynomial root-finding problem.Having found a supra-threshold voltage value at a discrete time point, we know that the threshold crossing must have occurred during the preceding time step.
Because of the escape process used to model the action potential upstroke, the membrane voltage will be monotonically increasing close to the threshold crossing/root.
Given these conditions, it is clear that we can efficiently solve this root finding problem using the Newton-Raphson method with pre-calculated polynomial coefficients.
For original step size 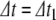 , this root-finding process returns a value
, this root-finding process returns a value 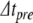 , in
, in 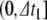 , reflecting the spike time within the local time step, and we solve for u, η, γ at
, reflecting the spike time within the local time step, and we solve for u, η, γ at 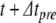 . Next, Eq. (22) is applied to model the action potential downstroke and post-spike adaptation effects. Finally, an additional time step is run using the post-spike variable values as initial conditions and a step size
. Next, Eq. (22) is applied to model the action potential downstroke and post-spike adaptation effects. Finally, an additional time step is run using the post-spike variable values as initial conditions and a step size 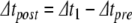 . This returns the solution to time
. This returns the solution to time 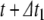 .
.
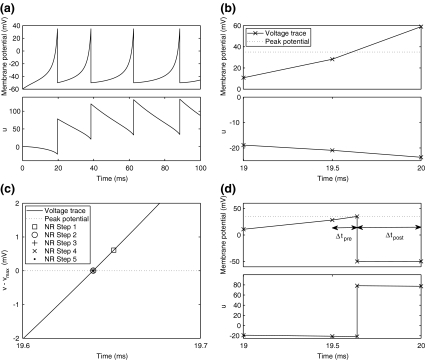
Figure 1 illustrates the application of this algorithm to a single neuron under constant current injection, with fixed time step 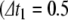 ms). In Fig. 1(a), the cell fires four times in 100 ms, with spike-rate adaptation due to the recovery variable. Figure 1(b) zooms in on the first spike, where a peak voltage crossing occurs between the 19.5 ms and 20 ms time samples. Figure 1(c) illustrates the use of the Newton-Raphson method to find the exact spike time, and the post-spike reduced step up to 20 ms is depicted in Fig. 1(d).
ms). In Fig. 1(a), the cell fires four times in 100 ms, with spike-rate adaptation due to the recovery variable. Figure 1(b) zooms in on the first spike, where a peak voltage crossing occurs between the 19.5 ms and 20 ms time samples. Figure 1(c) illustrates the use of the Newton-Raphson method to find the exact spike time, and the post-spike reduced step up to 20 ms is depicted in Fig. 1(d).
Fig. 1.
Calculating exact spike times for the Izhikevich neuron. (a) Membrane potential (top) and recovery variable (bottom) traces over 100 ms under current injection. The true membrane potential is recovered using 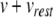 . (b) Twentieth millisecond of simulation using 0.5 ms standard time steps. A peak voltage threshold crossing is detected after solving for
. (b) Twentieth millisecond of simulation using 0.5 ms standard time steps. A peak voltage threshold crossing is detected after solving for 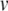 on the second step shown. (c) Locating the threshold crossing using the Newton-Raphson (NR) method for root-finding. First,
on the second step shown. (c) Locating the threshold crossing using the Newton-Raphson (NR) method for root-finding. First, 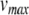 is subtracted from
is subtracted from 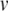 so that the threshold crossing becomes a root of the polynomial. Next, the Newton-Raphson method finds the root through iterative refinement. The results after steps 3–5 of this process are all contained within the circle used to plot the result from step 2, and convergence is obtained after 5 steps. (d) Post-spike reset and continuation. First, we solve for u at the spike time (around 19.64 ms). Next, Eq. (22) is applied to update the post-spike values of
so that the threshold crossing becomes a root of the polynomial. Next, the Newton-Raphson method finds the root through iterative refinement. The results after steps 3–5 of this process are all contained within the circle used to plot the result from step 2, and convergence is obtained after 5 steps. (d) Post-spike reset and continuation. First, we solve for u at the spike time (around 19.64 ms). Next, Eq. (22) is applied to update the post-spike values of 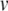 and u. Finally, we integrate over a reduced time step (
and u. Finally, we integrate over a reduced time step (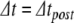 ) to arrive at the correct solution at the 20 ms time point
) to arrive at the correct solution at the 20 ms time point
An adaptive order algorithm
With adaptive order processing, the complete algorithm for one Izhikevich neuron over a single time step is as follows:
Run Eq. (27) to order n where error checking succeeds
Use Eq. 13 to get a new value for
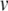
- If
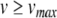
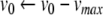
- Apply the Newton-Raphson method to find (
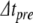 )
) 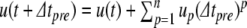
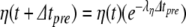
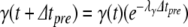
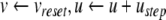
- Run a reduced time step with
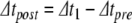
This algorithm omits synaptic events. We have developed a system for scheduling and delivering events at arbitrary, continuous time points despite using a fixed global time step, 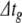 . Provided that synaptic transmission delays are always longer than
. Provided that synaptic transmission delays are always longer than 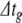 , we can calculate in advance whether a neuron receives any synaptic events during the time interval
, we can calculate in advance whether a neuron receives any synaptic events during the time interval 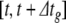 (Morrison et al. 2007). If events are to be delivered, we move through the global step via local substeps separated by synaptic events, with each substep being processed using the algorithm presented above.
(Morrison et al. 2007). If events are to be delivered, we move through the global step via local substeps separated by synaptic events, with each substep being processed using the algorithm presented above.
A Hodgkin-Huxley model
The Hodgkin and Huxley (1952) (HH) model of the squid giant axon has been arguably the most influential work in the field of computational neuroscience, and their conductance-based modelling framework remains widely employed. HH model equations are more complex than those of the Izhikevich neuron, and they are not generally expressed in a form to which the Parker-Sochacki method can be directly applied. In this section, we show how to apply the power series operations and variable substitutions described in Sections 2.5 and 2.4, to produce a PS solution algorithm.
The particular HH model neuron considered here was described in Brette et al. (2007), as a modification of a hippocampal cell model described by Traub and Miles (1991). With conductance-based synapses, the equations are,
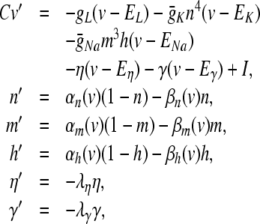 |
28 |
where 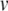 is the membrane potential (in mV), n, m, h are gating variables for the voltage-gated sodium (m, h) and potassium (n) currents, and η and γ are excitatory and inhibitory synaptic conductances, respectively. The gating variables evolve according to voltage-dependent rate constants,
is the membrane potential (in mV), n, m, h are gating variables for the voltage-gated sodium (m, h) and potassium (n) currents, and η and γ are excitatory and inhibitory synaptic conductances, respectively. The gating variables evolve according to voltage-dependent rate constants,
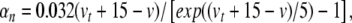 |
29 |
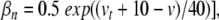 |
30 |
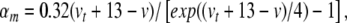 |
31 |
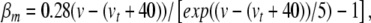 |
32 |
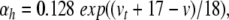 |
33 |
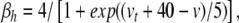 |
34 |
where 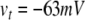 sets the threshold (Brette et al. 2007).
sets the threshold (Brette et al. 2007).
The Parker-Sochacki solution
Since Eqs. 29–34 feature exponential functions, variable substitutions are required before PS can be applied. First, we let a = βn and b = αh. As described in Section 2.4, the new equations are as follows:
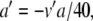 |
35 |
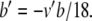 |
36 |
Equation 34 takes the form of a Boltzmann function. Now, letting 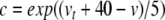 , we can write
, we can write
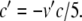 |
37 |
Applying this substitution, we have βh = 4/(c + 1), and h′ = b(1 − h) − 4h/(c + 1). Carothers et al. (2005) showed that the substitution z = 1/y yields an equation of the form z′ = − y′z2, and this substitution can be employed to convert βh, and hence h′, into polynomial form. However, a simpler solution is obtained via series division using Eq. (18). In this application, we let d = h/(c + 1), and use
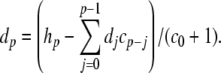 |
38 |
Then, h′ = b(1 − h) − 4d. Note, there is no danger of encountering division by zero here since the denominator in Eq. (38) is of the form (exp(x) + 1), which is always positive.
Equations 29, 31, 32 can all be written in the form x/(exp(x)−1), multiplied by some scaling constant. As with (34), we begin here by substituting for the exponential terms in the denominators of these equations. Thus, letting 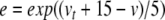 ,
, 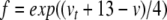 , and
, and 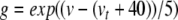 , we have
, we have
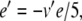 |
39 |
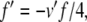 |
40 |
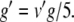 |
41 |
Next we introduce variables for the quotient terms. Thus, 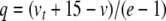 ,
, 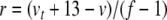 ,
, 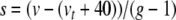 , with coefficients given by:
, with coefficients given by:
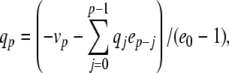 |
42 |
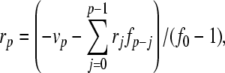 |
43 |
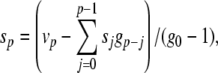 |
44 |
yielding αn = 0.032q, αm = 0.32 r, and βm = 0.28 s. There is a danger of division by zero here since the denominators in Eqs. (42)–(44) follow (exp(x) − 1), which equals zero when x = 0. Furthermore, this condition is encountered within the normal voltage range for the model neuron. In examining the stability of the PS method around these singular points, we found that the Taylor series expansions diverged, causing the PS method to fail. To examine whether this problem was specific to the series division operation, an alternative formulation was testing using substitutions of the form z = 1/x, z′ = − x′z2 (Carothers et al. 2005). The same failures were observed here. To solve this problem, code was added to first detect series divergence and then substitute in an alternative integration method to repeat the failed step. This system was used in the HH model benchmarking simulations in Section 5.2.
As for the Izhikevich model, we simplify the membrane potential equation by grouping all the terms multiplied by 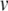 , and defining
, and defining 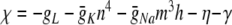 . Thus,
. Thus, 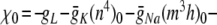 η0 − γ0 and (for p > 0)
η0 − γ0 and (for p > 0) 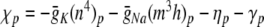 . Similarly, ψ = − (αn + βn) = − (0.032q + a), ξ = − (αm + βm) = − (0.32 r + 0.28 s).
. Similarly, ψ = − (αn + βn) = − (0.032q + a), ξ = − (αm + βm) = − (0.32 r + 0.28 s).
Equation (28) can now be re-written as:
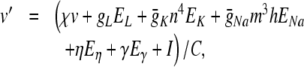 |
45 |
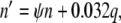 |
46 |
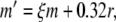 |
47 |
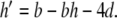 |
48 |
The complete PS solution is listed below. Since no powers greater than four are calculated, Cauchy products are used rather than the Euler power operation. Cauchy products will comprise the major computational cost of the solution method, especially in cases where a high-order solution is required. Equation 50 uses one Cauchy product to obtain 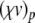 . Two Cauchy products are needed to obtain (n4)p (via an intermediate n2 term). Three Cauchy products are needed for (m3h)p, via intermediate m2 and m3 terms. Equations 51 to 65 each require one Cauchy product to solve (in slightly modified form for the quotient variables). Thus, a total of 19 Cauchy products are required at each iteration to solve this HH model using the PS method.
. Two Cauchy products are needed to obtain (n4)p (via an intermediate n2 term). Three Cauchy products are needed for (m3h)p, via intermediate m2 and m3 terms. Equations 51 to 65 each require one Cauchy product to solve (in slightly modified form for the quotient variables). Thus, a total of 19 Cauchy products are required at each iteration to solve this HH model using the PS method.
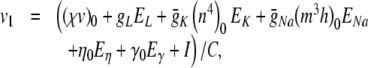 |
49 |
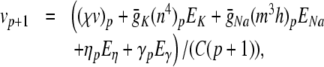 |
50 |
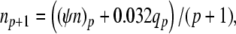 |
51 |
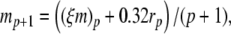 |
52 |
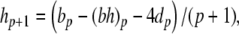 |
53 |
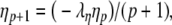 |
54 |
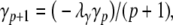 |
55 |
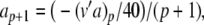 |
56 |
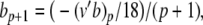 |
57 |
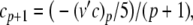 |
58 |
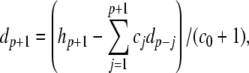 |
59 |
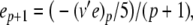 |
60 |
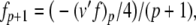 |
61 |
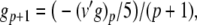 |
62 |
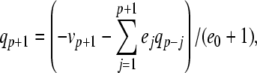 |
63 |
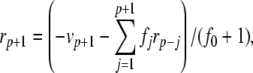 |
64 |
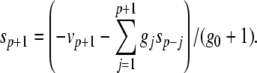 |
65 |
With adaptive order processing, the complete algorithm for one HH neuron over a single time step is:
For the non-power derived variables (a − s), we have the option of either updating using Eq. (13) or Eq. (14), or using the definition of the variable to recalculate its value at each step. For example, for the variable c, we can use
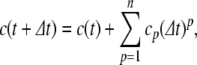 |
or
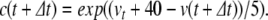 |
Using the latter method, the variable is guaranteed to match its definition at each time step. We term this method tethering, and any variable so updated a tethered variable. In preliminary testing, it was found that the stability of the PS solution was improved by tethering all the variables involved in quotient calculations (c, d, e, f, g, q, r, s), but that tethering a and b produced no improvement in the solution.
Results
In this section we assess the speed and accuracy of our adaptive PS algorithms by running benchmark simulations for the Izhikevich and Hodgkin-Huxley neuron models. The results are compared to those obtained using the 4th-order Runge-Kutta (RK) and Bulirsch-Stoer (BS) methods.
The 4th-order Runge-Kutta method is one of the most commonly used numerical integration methods (Press et al. 1992). The method offers moderate accuracy at moderate computational cost. For each equation, the derivative is evaluated four times per step: once at the start point, twice at trial midpoints, and once at a trial endpoint. These results are then combined in such a way that the first, second and third order error terms cancel. Thus, the solution agrees with the Taylor series expansion up to the 4th-degree term. Derivative evaluations are the major computational cost of RK.
The Bulirsch-Stoer method is another popular method. For smooth ODEs without singular points inside the integration interval, BS is described by Press et al. (1992) as the best known way to obtain high-accuracy solutions to ODEs with minimal computational effort. The method combines the (second order) modified midpoint method with the technique of Richardson extrapolation. In a single BS step, a sequence of crossings of the step is made with an increasing number n of modified midpoint substeps. Following Press et al. (1992), we use the sequence nk = 2k, where k is the crossing number. After each crossing, a rational function extrapolation is carried out to approximate the solution that would be obtained if the step size were zero. The extrapolation algorithm also returns error estimates. If the latter are acceptable, we terminate the sequence and move to the next step. If not, we continue with the next crossing. For a given step size, BS can be expected to be more accurate but also more computationally expensive than RK.
Both BS and PS can apply adaptive error control without adaptive time stepping. To examine this process, adaptive stepping was not implemented for any of the methods. For PS, adaptive order processing was implemented as described in Section 2.3. Equivalent adaptive error control, based on change in the iterative solution, was employed for BS. PS was limited to a maximum of 200-th order, while BS was limited to a maximum of 50 crossings.
Simulations were run in the MATLAB 7.5 environment, with algorithms written in C and compiled as mex files. Code for the Runge-Kutta and Bulirsch-Stoer methods was adapted from the routines provided in Press et al. (1992) by removing adaptive time stepping. The machines used to run the simulations featured 2.2 GHz AMD Opteron processors, and at least 4 GB of memory. Simulation code will be provided in the ModelDB database.1 Major routines are listed in Appendix A.
Izhikevich model
Two types of simulation were used on the Izhikevich model. In the first type, cells were driven by current injection only. In the second, recurrent synaptic interactions were also modelled. These different simulations enabled us to separate the computational costs of integration and synaptic processing.
Current injection simulations
The current injection model featured 1000 neurons but no functional synapses. All cells had identical parameters, set by fitting the model to the HH neuron in Brette et al. (2007). Details of the cellular model and fitting process are given in Appendix B. All simulations were of one second duration. In one series of experiments, all model cells were driven by a constant, depolarising injection current sufficient to make them fire once within the simulation time period. In another, the cells were driven to fire ten times. These are the one- and ten-spike simulations, respectively. In each case, we applied nc = 15 different error tolerance conditions. For BS and PS we used a global step size of 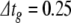 ms, and systematically varied the error tolerance, ε. All three methods were stable (no solution divergence to infinity) at this step size. For condition cn, ε = 1e−(n + 1). For RK, we varied the error tolerance indirectly by changing the global step size. For c1..15,
ms, and systematically varied the error tolerance, ε. All three methods were stable (no solution divergence to infinity) at this step size. For condition cn, ε = 1e−(n + 1). For RK, we varied the error tolerance indirectly by changing the global step size. For c1..15, 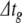 for RK was set to 1/4, 1/6, 1/8, 1/10, 1/20, 1/40, 1/60, 1/80, 1/100, 1/200, 1/400, 1/600, 1/800, 1/1000, 1/2000 ms, respectively. For time averaging, all simulations were repeated ten times. All solution algorithms included calculations for exact spike times using the Newton-Raphson method as described in Section 3.2. For RK and BS this required additional integration steps to evaluate
for RK was set to 1/4, 1/6, 1/8, 1/10, 1/20, 1/40, 1/60, 1/80, 1/100, 1/200, 1/400, 1/600, 1/800, 1/1000, 1/2000 ms, respectively. For time averaging, all simulations were repeated ten times. All solution algorithms included calculations for exact spike times using the Newton-Raphson method as described in Section 3.2. For RK and BS this required additional integration steps to evaluate 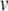 and v′ at different time points.
and v′ at different time points.
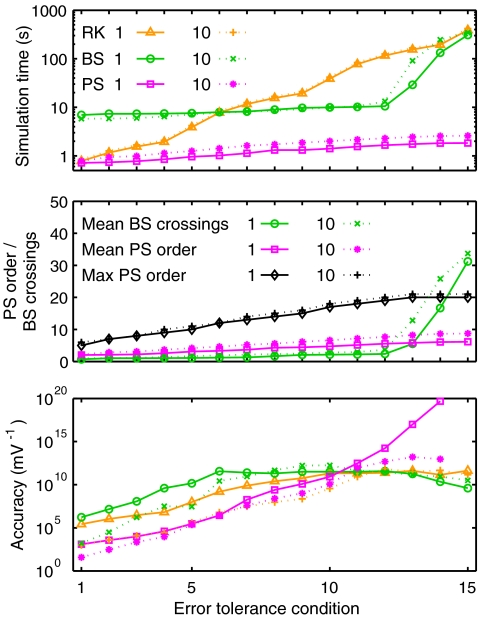
Figure 2 shows the results from these simulations. In Fig. 2 (top) simulation time is plotted as a function of c. In the one-spike simulations (solid lines), PS was the fastest method for all c, with simulation times monotonically increasing from 0.72 ±0.01 s (c1) to 1.83 ±0.02 s (c15). RK times rose from 0.78 ±0.01 s to 388 ±0.7 s for c1..15. BS times increased gradually from 7 to 11 s across c1..12 but rose steeply at tighter error tolerances to 309 ±1 s in condition 15. Results from the ten-spike simulations were similar. PS was again the fastest method in all conditions. However, times here were greater than the one-spike results in equivalent conditions, with gain ranging from 1.09 (c1) to 1.44 (c7). RK times were slightly greater than for the equivalent one-spike simulations for all c, with gain factors ranging from 1.072 (c6) to 1.085 (c2). BS times were reduced relative to the one-spike results in the first few conditions, but were greater in the last four conditions.
Fig. 2.
Izhikevich model current injection results. (Top) Mean simulation time for 1 s simulations with varying error tolerance conditions. (Middle) Adaptive processing statistics. Plots show the mean (over a simulation) number of crossings used by the BS method per step, and the mean and maximum order of the PS method. (Bottom) Simulation accuracy taken as the reciprocal of absolute voltage divergence between test and reference traces. Line styles as in top panel
In order to explain the variation in simulation times, Fig. 2 (middle) shows how adaptive processing for BS and PS varied with error tolerance by plotting representative statistics for each method. The number of BS crossings was low for c1..12 but rose steeply for c > 12. This increase reflects error tolerance failures. In both one- and ten-spike simulations there were no failures for c1..12, but for c13,14,15 there were, respectively, 21, 475, 1666 failures per cell in the one-spike simulations and 176, 1088, and 1850 failures per cell in the ten-spike simulations. In contrast, PS never failed to achieve the specified error tolerances. The mean order of the PS method increased gradually with increasing c, and the maximum order across all conditions was 20 in one- and 21 in ten-spike simulations. The mean PS order was greater in the ten- than one-spike simulations and the gain was very similar to the simulation time gain, ranging from 1.10 (c1) to 1.43 (c7).
To quantify the accuracy of the simulation output, we created reference solutions against which to test all other solutions. Since the PS Taylor series were convergent in these simulations, reference solutions were obtained by running PS to complete numerical convergence (ε = 0). There were no tolerance failures and reference simulation times were 1.84 ±0.02 s for one spike and 2.59 ±0.01 s for ten. Mean order was 6.14 and 8.73, respectively, and maximum order was 20 and 21 (as for c15). Simulation error was calculated as the mean absolute membrane voltage divergence between test and reference traces. Figure 2 (bottom) plots simulation accuracy, taken as the reciprocal of the error.
Despite using the same error tolerance conditions, BS was many times more accurate than PS for c1..10 in both one- and ten-spike simulations. In the one-spike simulations, RK was also more accurate than PS for c1..10. RK was less accurate in the ten-spike simulations, but was still more accurate than PS for c1..6. However, both BS and RK accuracy plots plateaued at low tolerances, with peak values always between 1e11 and 1e13. Indeed, BS showed reduced accuracy at the lowest tolerances where it exhibited failures. In contrast, PS showed progressive accuracy gains with decreasing tolerance until in condition 15 measured accuracy was infinite since the reference and test traces were identical for both one- and ten-spike simulations.
The reference PS simulations achieved double precision accuracy in the state variables and yet were faster than the fastest BS simulations. Furthermore, the reference runs were only 2.35 and 3.07 slower than the fastest RK simulations in the one- and ten-spike simulations, respectively.
Recurrent network model simulations
The network model here was based on Benchmark 1 from Brette et al. (2007), which was inspired by an earlier model (Vogels and Abbott 2005). The network featured 4000 neurons (80% excitatory, 20% inhibitory; parameters as above). All cells were randomly (2% probability) connected, and ns = 5 different network configurations were created.
All simulations were of one second duration. To generate recurrent activity, random stimulation was applied for the first 50 ms, as described by Brette et al. (2007). This initial stimulation was provided here by constant current injection, and each cell was independently assigned a random current value in [0, 200] pA. For each network configuration, ni = 10 different patterns of initial stimulation were applied, and each pairing of network configuration and input pattern defines a single experiment (ne = ns ×ni = 50).
In the absence of numerical errors, all simulations from the same experiment should have produced identical output. Repeated experiments therefore allowed us to examine the speed/accuracy trade-off for each integration method.
Given the results from the current injection simulations, we selected three representative error tolerance conditions to apply here. Specifically, it was specified that c1,2,3 here would be identical to c1,9,15 from the current injection simulations. Thus, for BS and PS, ε = 1e-2, 1e-10, 1e-16, and for RK, 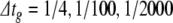 . As in the current injection simulations, reference solutions were created using PS with ε = 0.
. As in the current injection simulations, reference solutions were created using PS with ε = 0.
Single experiment results
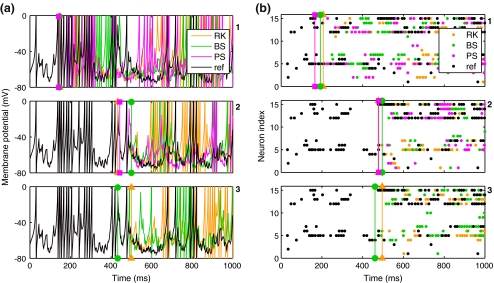
In this section, we characterise the outputs from a single experiment, using the reference solution to assess accuracy. Figure 3(a) shows membrane potential traces from a single neuron, with results from conditions 1–3 arranged in separate panels, top to bottom and integration methods represented using different colours. For comparison, the trace from the reference solution for this experiment is plotted as a black line in each panel. The reference trace was drawn last so that it would obscure the coloured traces when they were in agreement. Thus, working left to right in a single panel, the appearance of a coloured line is a visual indicator of divergence between the reference solution and the test solution from the method represented by that colour.
Fig. 3.
Izhikevich model recurrent network results: recordings from repeated simulations using the same network model and initial random inputs but varying the integration method and error tolerance/time step size. (a) Membrane potential traces from single neuron. (Top to bottom) Conditions 1–3. In each plot, the colour represents the integration method, with an additional reference trace drawn in black. The reference trace was drawn last. Thus, the appearance of coloured lines indicates divergence of test solutions from the reference. (b) Population raster plot of the first twenty cells in the same network model as in (a); layout and colours as in (a). The single cell traced in (a) appears here as neuron 5. As in (a), these plots are overlain by a reference solution in black. Thus, the appearance of coloured dots indicates divergence
A quantitative measure of trace divergence (accuracy) was obtained by recording the time point at which each test trace first differed from the reference trace by more than 1 mV. These divergence points are indicated by vertical lines in Fig. 3(a). For c1, the divergence times for all three methods were between 140 and 150 ms. For c2, divergence times were later for all methods, at 433 ms (RK), 443 ms (PS), and 500 ms (BS). In the final condition, the BS and RK results were reversed relative to the previous condition with RK diverging at 500 ms, and BS diverging earlier at 433 ms. In contrast, the PS test solution agreed with the reference solution over the full one second simulation.
Figure 3(b) shows population raster plots from the first 15 cells in the same network. Once again, the reference solution is plotted in each panel to give a visual indicator of agreement. Raster divergence (vertical lines) was taken as the time point at which a test spike time first differed from the corresponding reference spike time by more than 1 ms. Raster divergence times were generally later than trace divergence times but followed the same trends. This indicates that solution divergence for this model is a network phenomenon rather than a single cell characteristic; as expected given the highly recurrent network activity here.
Both voltage traces and spike time results from each method generally converged towards the reference solution as the error tolerance decreased. The only exception to this rule was the BS result from condition 3, which was worse than condition 2. As above, BS exhibited error tolerance failures at the lowest tolerance, and reduced accuracy probably results from roundoff errors with many crossings (Press et al. 1992). Only the PS solution from condition 3 showed agreement with the reference solution that extended beyond the time limit of the simulation. RK and BS showed neither complete agreement with the reference solution, nor agreement between their own test solutions.
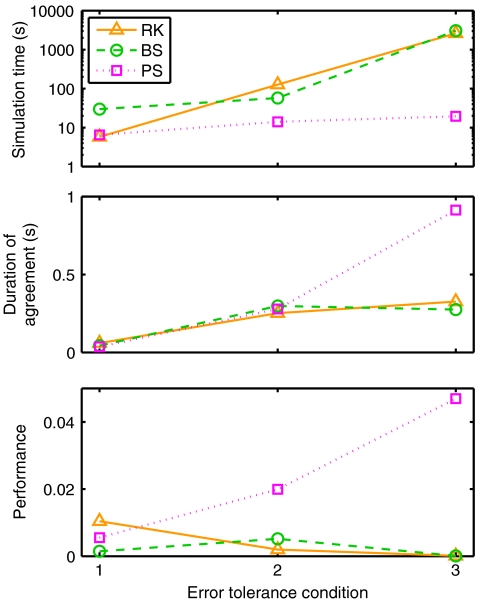
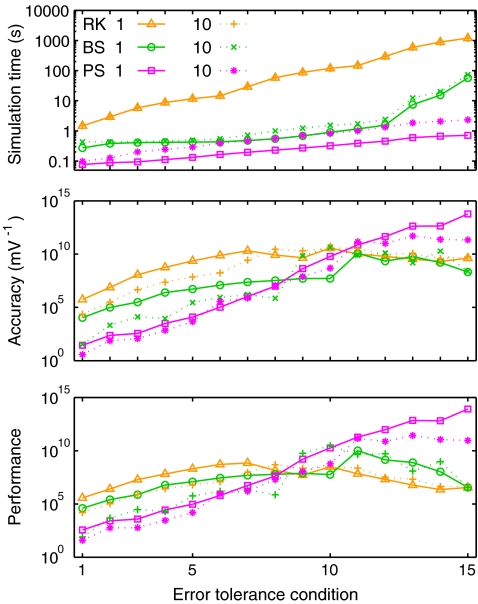
Overall performance results
In this section, we examine the overall accuracy of each integration method and derive a performance measure based on both speed and accuracy. In the single experiment results, simple heuristic measures of trace and raster plot divergence were employed to assess accuracy. While those measures were sufficient to highlight important trends in the data, a stricter measure of global solution divergence was obtained here by comparing time-ordered sequences of spikes from different simulations in the same experiment. Spike sequences consisted of {spike time, neuron index} pairs, and the duration of global solution agreement was taken as the time of the last spike at which the test and reference sequence neuron indices were identical. Assuming the reference simulations were at least as accurate as the test simulations, the duration of agreement provides a measure of solution accuracy.
Figure 4 (top) plots simulation time for each method as a function of the error tolerance condition. The general pattern of results here was qualitatively similar to the current injection results in Fig. 2(a). The mean number of spikes across all experiments was 7.66 ±0.4, so we compare with the ten-spike current injection results. RK times were between 6.07 (c2) and 6.82 (c1) times larger than in the equivalent current injection simulation. PS times were between 7.51 (c2,3) and 8.31 (c1) times larger, while BS times were between 5.09 (c1) and 8.9 (c3) times larger. These increased times, despite reduced firing rate, reflect the introduction of synaptic interactions, and the fact that the recurrent network had four times as many cells. Furthermore, trace recordings were output to file during the recurrent simulations but not during the current injection simulations. To examine the cost of synaptic interactions more directly, additional simulations were run with smaller recurrent networks of 1000 cells, without file recordings, and with firing rate adjusted to approximately ten spikes per cell via weight scaling. In these smaller simulations, RK times were between 0.97 (c2,3) and 1.03 (c1) times larger than in the equivalent current injection simulation. PS times were between 1.24 (c1) and 1.32 (c3) times larger, while BS times were between 1.27 (c1) and 1.96 (c3) times larger. Thus, the additional costs of synaptic interactions was noticeable for BS and PS, but not RK.
Fig. 4.
Izhikevich model recurrent network results: performance summary. (Top) Mean simulation time. (Middle) Simulation accuracy: mean duration over which the output from simulation runs agree with reference solutions in terms of the exact spike sequences (see text for further details). (Bottom) Performance (accuracy/time)
Figure 4 (middle) plots the mean duration of agreement for each method. These results are broadly similar to those obtained in the single experiment results. RK accuracy increased by a large amount moving from c1 to c2, and by a smaller amount from c2 to c3, reaching a maximal value of 0.33 s. BS accuracy peaked at 0.30 s for c2 and decreased at c3. PS accuracy increased progressively as the error tolerance was tightened. In condition 3, most, but not all, PS simulations agreed with the reference solution over the full duration of the simulation. Thus, unlike in the current injection simulations, tiny numerical differences between the simulations with ε = 1e-16 and ε = 0 were sufficient here to cause network state divergence in some cases within a one second simulation time period.
Morrison et al. (2007) have argued that in order to arrive at a relevant measure of the performance of an integration method, simulation time should be analysed as a function of the integration error. Following this principle, overall performance was assessed as a function of both speed and accuracy by dividing the duration of agreement by simulation time to yield a dimensionless performance measure. For example, a performance score of 1 would be obtained by a method running at real-time speed and showing complete agreement with the reference solution. Figure 4 (bottom) plots mean performance. By this measure, RK was the best performing method for c1, while PS performed second best for c1 and much better than the other methods for c2,3.
The reference PS simulations once again achieved double precision accuracy in the state variables over the simulation period and took 19.50 ±0.7 s to run. This was faster than the fastest BS simulations, and 3.39 times slower than the fastest RK simulations.
Hodgkin-Huxley model
For the Hodgin-Huxley model, current injection simulations were used to compare methods in the absence of synaptic processing. Ten identical cells were modelled for more accurate time calculations; parameters are listed in Appendix B. As for the Izhikevich model, one- and ten-spike simulations were run and 15 error tolerance conditions were applied. Preliminary testing showed solution divergence to infinity with step sizes larger than 0.01 ms for RK, and 0.1 ms for BS/PS. Consequently, 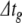 was set to 0.1 ms for all BS and PS simulations, and 0.01 for RK in condition 1. Error tolerances for BS and PS were identical to those used in Section 5.1.1. For RK,
was set to 0.1 ms for all BS and PS simulations, and 0.01 for RK in condition 1. Error tolerances for BS and PS were identical to those used in Section 5.1.1. For RK, 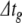 was reduced in the same manner as for the Izhikevich model, but from a lower starting point. Thus, for c15,
was reduced in the same manner as for the Izhikevich model, but from a lower starting point. Thus, for c15, 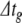 for RK was 1/80,000 ms, or 12.5 ns.
for RK was 1/80,000 ms, or 12.5 ns.
For the PS method, an adaptive algorithm with failure detection was used as described in Section 4.1, with a replacement BS step being run when PS failure was detected.
Unlike the Izhikevich model simulations, the PS method sometimes failed to achieve the specified error tolerances here, giving no guarantee of accuracy. For this reason, we conducted an analysis of within-method agreement across conditions using the c15 results from each method as reference solutions. Voltage traces were recorded from every simulation at 1 ms sampling steps, and the mean absolute difference between test and reference traces was recorded. All methods showed initial convergence towards their own reference as the error tolerance was reduced, suggesting that they were becoming more accurate. The best result in both one- and ten-spike simulations came from PS (c14) at 2.39e-13 and 1.12e-12 mV, respectively. RK and BS achieved divergences never less than 1e-10 and 1e-9 mV, respectively.
Consequently, general reference solutions for each experiment were produced using PS with ε = 0. Treating all other voltage traces as test solutions, the mean absolute voltage difference between reference and test traces was calculated, and the inverse of this value was taken as an accuracy measure for the test solution. Finally, overall performance was taken as accuracy divided by simulation time.
Figure 5 shows the results of this performance analysis. The top panel shows how simulation time varied with the error tolerance condition. All methods became slower with decreasing error tolerance, as expected. RK was the slowest method in all conditions here. PS was the fastest method, with better than real-time speed in all conditions in the one-spike experiments. In the ten-spike simulations, PS was between 1.26 (c1) and 3.27 (c15) times slower than the equivalent one-spike simulations, but was still faster than RK and BS in all conditions.
Fig. 5.
HH model results. (Top) Mean simulation time for 1 s benchmark simulation. (Middle) Mean accuracy, with accuracy values taken as the inverse of the mean absolute voltage difference between test and reference solutions. (Bottom) Overall performance (accuracy/time)
The middle panel of Fig. 5 shows mean accuracy values plotted against the error tolerance condition. As with the Izhikevich model results, RK and BS showed convergence towards the reference solution with reducing error tolerance, justifying the choice of reference. As for the Izhikevich model however, the accuracy measures for both methods plateaued, reaching similar peak levels in each case, with the peak located at c11 (BS, one-spike) or c10 (all other simulations). PS accuracy continued to improve beyond this tolerance level, reaching peak values more than two orders of magnitude greater than the alternatives in the one-spike simulations and roughly one order of magnitude greater in the ten-spike experiments.
The bottom panel of Fig. 5 shows overall performance values for each method. PS recorded the best results here in both the one- and ten-spike simulations. Comparing peak performance values for each method, in the one-spike simulations PS performed roughly four orders of magnitude better than BS and five orders of magnitude better than RK. In the ten-spike simulations, PS performed 9 times better than BS, and 558 times better than RK.
The reference PS simulations took around ten percent longer to run than the c15 simulations in both one- and ten-spike simulations. Unlike the Izhikevich model results, double precision accuracy was not obtained due to error tolerance failures. The PS method only failed around singular points in the equations, but the replacement BS steps were unable to achieve zero error tolerance when PS failed.
Discussion
The Parker-Sochacki method is a promising new numerical integration technique. We have presented the method and shown how it can be applied to the Izhikevich and Hodgkin-Huxley neuronal models.
In Section 2, we summarised major milestones in the development of the Parker-Sochacki method and illustrated its application through examples. We demonstrated how to implement adaptive error control using adaptive order processing in PS. We also showed how power series division and power operations can be used within the Parker-Sochacki framework. For terms with powers greater than 4, Euler’s power method can provide significant computational savings over iterated Cauchy products, but it is the division operation which is likely to be of greater utility. Series division is simple to implement since the major calculation can be carried out using a standard Cauchy product function. With this operation, PS can be directly applied to any equation composed only of numbers, variables, positive integer powers, and the four basic arithmetic operations (addition, subtraction, multiplication and division). This is a far broader class of equations than the polynomials considered in previous articles (Parker and Sochacki 1996; Carothers et al. 2005). Where other expressions are present, it may still be possible to apply the method, but additional work will be required to discover and apply suitable variable substitutions.
In Section 3, we applied PS to the Izhikevich neuron model: a simple model capable of rich dynamic behaviour. We developed an efficient PS solution using a single Cauchy product, showed how to calculate exact spike times within larger time steps using the Newton-Raphson method and presented a simple adaptive order algorithm.
Benchmark simulations in Section 5.1 demonstrated that the Parker-Sochaki method is capable of double precision integration accuracy for Izhikevich model neurons in both current injection and recurrent network simulations. Neither the Bulirsch-Stoer nor the Runge-Kutta methods were capable of the same level of accuracy. Furthermore, in Section 5.1.2 it was shown that integration accuracy had a major effect on network behaviour in a recurrent network simulation, with small solution errors leading to divergent behaviour within a relatively short simulation time period.
In light of the typical parameter uncertainties in neuronal modelling, the question of whether double precision integration accuracy is useful in a given setting will be a matter for the individual investigator to consider. However, the relative time cost of applying zero error tolerance in PS simulations is small; reference PS simulations were always faster than any BS simulations on this model and took less than four times as long to run as 4th-order RK simulations with the same global step size. In general, we would make the following suggestions. First, PS should always be run with zero error tolerance. Second, even if PS is not used for the main simulations in a study, it may still be useful as a reference solution in pilot work.
In Section 4, we applied PS to a Hodgkin-Huxley model. We showed how variable substitutions transform the equations into a form suitable for the application of PS. We also successfully applied the series division operation. However, for equations of the form x/(exp(x) − 1), we encountered a Taylor series divergence problem that we were unable to solve through variable substitutions. There are at least three ways to work around such a problem. First, as we did, an alternative numerical integration method can be used for steps where the PS method fails. Second, polynomial, spline, or rational function approximation methods can be used. Cubic interpolating splines are one attractive option here due to the low order and high accuracy offered (de Boor 2001). Alternatively, Floater and Hormann (2007) proposed a family of rational interpolants that have no poles, and arbitrarily high approximation orders. Finally, there exist alternative equation forms for Hodgkin-Huxley type models that avoid the presence of singular points; one promising option being the Extended Hodgkin-Huxley (EHH) model (Borg-Graham 1999) (Section 8.4.2). Here, the voltage-dependent rate constant equations take on the following generic form:
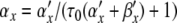 |
66 |
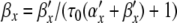 |
67 |
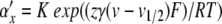 |
68 |
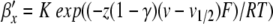 |
69 |
where K is a positive constant, F is Faraday’s constant, R is the gas constant, and T is the temperature in Kelvin. The following method creates a PS solution. First, substitute for the exponential expressions to give α′x = ax, α′x = bx. Next, define a derived variable cx = τ0(ax + bx) + 1. Finally, solve for αx and βx using series division operations on ax/cx and bx/cx, respectively. Furthermore, since K is positive, cx is always positive, avoiding any singularities.
In order to retain the Hodgkin-Huxley model equations used by Brette et al. (2007), benchmark simulations in Section 5.2 used the method-substitution approach. At low error tolerances, this approach appeared to yield greater accuracy than the alternative methods, but was unable to achieve double precision accuracy due to failures in both PS steps close to singular points and the replacement BS steps. Given the lack of singular points in the Extended Hodgkin-Huxley equations, we conjecture that the Maclaurin series would always be convergent for this modelling framework. If simulation testing proves this conjecture to be correct, then the PS method will be able to achieve arbitrary precision and should also run faster than on the standard Hodgkin Huxley model due to an absence of failure and replacement steps.
In all of our simulations, continuous event times were accommodated within a globally clock-driven framework. This modelling approach enables far greater accuracy than traditional clock-driven methods where events are restricted to discrete time points. Event-driven simulation approaches (Mattia and Del Giudice 2000; Delorme and Thorpe 2003; Makino 2003; Brette 2006, 2007), offer comparable event timing precision but are generally restricted to simple neuronal models, while the present approach is far more widely applicable. Morrison et al. (2007) proposed a similar hybrid clock-driven/event-driven approach but, like many standard event-driven techniques, their method was restricted to linear neuron models. In contrast, by using the Parker-Sochacki method, we were able to combine the flexibility of clock-driven simulation methods with the precision of event-driven approaches.
In Appendix A, we present the major routines used in our implementation of the Parker-Sochacki method. We endeavoured to make the code generic, modular and simple to adapt. It should be emphasized that the provided code does not constitute a general neuronal modelling package of the type described by Brette et al. (2007). Rather, it is our hope that the Parker-Sochacki method will be adopted into existing simulation packages as an alternative integration method for highly accurate simulations.
As previously noted, the Parker-Sochacki method is directly applicable to any model with polynomial or rational differential equations. In terms of existing spiking neuron models, the class with polynomial equations includes the leaky (Lapicque 1907; Tuckwell 1988) and quadratic (Ermentrout 1996; Latham et al. 2000) integrate-and-fire models, the resonate-and-fire neuron model (Izhikevich 2001), the Fitzhugh-Nagumo model (Fitzhugh 1961; Nagumo et al. 1962), and the models of spiking and bursting by Hindmarsh and Rose (1982, 1984). The exponential integrate-and-fire model (Fourcaud-Trocmé et al. 2003), including the two-dimensional adaptive version (Brette and Gerstner 2005), can be handled using an exponential variable substitution (see Section 2.4). For Hodgkin-Huxley type models, multiple substitutions will usually be required.
The Hodgkin-Huxley formalism can be viewed as a simple Markov kinetic model (Destexhe et al. 1994). More complex kinetic models have been used to model voltage-gated ion channels (see, for example Vandenberg and Bezanilla 1991; Bezanilla et al. 1994), and it has been suggested that ligand-gated, and second-messenger-gated channels can be modeled along the same lines (Destexhe et al. 1994; Destexhe 2000). Like the HH and EHH models, these general kinetic models usually feature exponential functions that can be handled using the same kind of substitutions.
Compartmental models often use equations similar to single-compartment models for each compartment, plus linear, resistive coupling terms between neighbours. The PS method handles coupling terms in the same way as any other variables; no additional substitutions or manipulations are required. Indeed, PS has already been successfully applied to the n-body problem (Rudmin 1998; Pruett et al. 2003), which features n coupling terms in the equations describing the motion of each body. Thus, the PS method is applicable to a compartmental model provided it is applicable to the equations of individual compartments, with coupling terms.
Calcium modelling introduces calcium ion concentrations as model variables, with equations describing concentration changes (Borg-Graham 1999). Once again, PS handles these new variables in the same way as any others.
In conclusion, the Parker-Sochacki method offers unprecedented integration accuracy in neuronal model simulations, at moderate computational cost, and is applicable in a variety of computational neuroscience settings. It is our hope that this article will help to facilitate its wider adoption.
Acknowledgements
Research supported by The Wellcome Trust. We are grateful to Joseph Rudmin, Stephen Lucas and Jim Sochacki for stimulating discussions and helpful advice.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix A: Parker-Sochacki solution code
The routines below form the core of our implementation of the Parker-Sochacki method. The generic solver routine, , solves a system of differential equations using PS and advances the solution across a single time step. Worker routines and are passed in to and calculate the first and subsequent terms of the PS solution to a specific system. The routine was used with different and functions for both Izhikevich and Hodgkin-Huxley model simulations; the Izhikevich model routines and are included as an example. In order to use to solve a different model, all that is required is to define suitable ‘first’ and ‘iter’ functions specific to the model in question. Also included below are some generic power series operation functions.


Appendix B: Benchmark model neuron parameters
Cellular model parameters for both the Izhikevich and HH models were taken from Brette et al. (2007), and all cells had identical basic parameters. Briefly, the cell area was 20,000 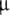 m2, and input resistance was 100 MΩ. Given a specific capacitance of 1
m2, and input resistance was 100 MΩ. Given a specific capacitance of 1 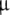 F/cm2, whole cell capacitance was taken as C = 200 pF. Following the published code accompanying Brette et al. (2007), EL was set to − 65 mV; the value of − 60 mV given in the text of the paper was erroneous (Destexhe, personal communication).
F/cm2, whole cell capacitance was taken as C = 200 pF. Following the published code accompanying Brette et al. (2007), EL was set to − 65 mV; the value of − 60 mV given in the text of the paper was erroneous (Destexhe, personal communication).
The HH neuron model was as described in Section 4. In addition to the basic parameters listed above, parameters specific to the HH model were: gL = 10 nS, 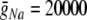 nS,
nS, 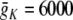 nS, ENa = 50 mV, EK = − 90 mV.
nS, ENa = 50 mV, EK = − 90 mV.
The Izhikevich model neuron parameters were obtained by fitting the model to the HH neuron in Brette et al. (2007). First, 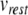 was taken as − 65 mV to match EL. The voltage threshold of − 50 mV was shifted by
was taken as − 65 mV to match EL. The voltage threshold of − 50 mV was shifted by 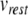 to give
to give 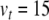 mV. Next,
mV. Next, 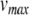 and c were obtained by observing the HH neuron model under constant, supra-threshold current injection. The observed values of 48 mV and −85 mV were shifted relative to
and c were obtained by observing the HH neuron model under constant, supra-threshold current injection. The observed values of 48 mV and −85 mV were shifted relative to 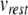 to give
to give 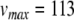 mV and c = − 20 mV. In the same simulations, the rheobase current was found to be around 19 pA. Since the HH neuron from Brette et al. (2007) lacks spike frequency adaptation, ustep was set to zero, and a was set to a value of 0.03 to match the value given by Izhikevich (2007) for a regular spiking cortical neuron.
mV and c = − 20 mV. In the same simulations, the rheobase current was found to be around 19 pA. Since the HH neuron from Brette et al. (2007) lacks spike frequency adaptation, ustep was set to zero, and a was set to a value of 0.03 to match the value given by Izhikevich (2007) for a regular spiking cortical neuron.
Izhikevich (2007) (Ch 5), describes a method for setting b and k given the rheobase current, input resistance, and the resting and threshold potentials. Using this method, values of k = 1.3 and b = − 9.5 were obtained.
Model synapses were conductance-based, and conductances were summed together to form one η and one γ value for each neuron. The conductances decayed exponentially with time constants of 5 ms for η and 10 ms for γ. When fired, excitatory synapses incremented η by 6 nS, while inhibitory synapses incremented γ by 67 nS.
Footnotes
References
- Bezanilla, F., Perozo, E., & Stefani, E. (1994). Gating of shaker k+ channels: Ii. The components of gating currents and a model of channel activation. Biophysical Journal, 66(4), 1011–1021, April. [DOI] [PMC free article] [PubMed]
- Borg-Graham, L. (1999). Interpretations of data and mechanisms for hippocampal pyramidal cell models. In Cerebral cortex (pp. 19–138). New York: Plenum.
- Brette, R. (2006). Exact simulation of integrate-and-fire models with synaptic conductances. Neural Computation, 18(8), 2004–2027. [DOI] [PubMed]
- Brette, R. (2007). Exact simulation of integrate-and-fire models with exponential currents. Neural Computation, 19(10), 2604–2609. [DOI] [PubMed]
- Brette, R., & Gerstner, W. (2005). Adaptive exponential integrate-and-fire model as an effective description of neuronal activity. Journal of Neurophysiology, 94(5), 3637–3642, November. [DOI] [PubMed]
- Brette, R., Rudolph, M., Carnevale, T., Hines, M., Beeman, D., Bower, J. M., et al. (2007). Simulation of networks of spiking neurons: A review of tools and strategies. Journal of Computational Neuroscience, 23(3), 349–398, December. [DOI] [PMC free article] [PubMed]
- Carothers, D. C., Parker, G. E., Sochacki, J. S., & Warne, P. G. (2005). Some properties of solutions to polynomial systems of differential equations. Electronic Journal of Differential Equations, 2005(40), 1–17.
- de Boor, C. (2001). A practical guide to splines. In Applied mathematical sciences, revised edition (Vol. 27). New York: Springer.
- Delorme, A., & Thorpe, S. J. (2003). SpikeNET: An event-driven simulation package for modelling large networks of spiking neurons. Network, 14(4), 613–27. [PubMed]
- Destexhe, A. (2000). Kinetic models of membrane excitability and synaptic interactions. In J. M. Bower & H. Bolouri (Eds.), Computational modeling of genetic and biochemical networks (pp. 225–262). Cambridge: MIT.
- Destexhe, A., Mainen, Z. F., & Sejnowski, T. J. (1994). Synthesis of models for excitable membranes, synaptic transmission and neuromodulation using a common kinetic formalism. Journal of Computational Neuroscience, 1(3), 195–230, August. [DOI] [PubMed]
- Ermentrout, B. (1996). Type i membranes, phase resetting curves, and synchrony. Neural Computation, 8(5), 979–1001, July. [DOI] [PubMed]
- Fitzhugh, R. (1961). Impulses and physiological states in theoretical models of nerve membrane. Biophysical Journal, 1, 445–166. [DOI] [PMC free article] [PubMed]
- Floater, M. S., & Hormann, K. (2007). Barycentric rational interpolation with no poles and high rates of approximation. Numerical Mathematics, 107(2), 315–331. [DOI]
- Fourcaud-Trocmé, N., Hansel, D., van Vreeswijk, C., & Brunel, N. (2003). How spike generation mechanisms determine the neuronal response to fluctuating inputs. Journal of Neuroscience, 23(37), 11628–11640, December. [DOI] [PMC free article] [PubMed]
- Hindmarsh, J. L., & Rose, R. M. (1982). A model of the nerve impulse using two first-order differential equations. Nature, 296(5853), 162–164, March. [DOI] [PubMed]
- Hindmarsh, J. L., & Rose, R. M. (1984). A model of neuronal bursting using three coupled first order differential equations. Proceedings of the Royal Society of London. Series B, Biological Sciences, 221(1222), 87–102, March. [DOI] [PubMed]
- Hodgkin, A. L., & Huxley, A. F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology, 117(4), 500–544, August. [DOI] [PMC free article] [PubMed]
- Izhikevich, E. M. (2001). Resonate-and-fire neurons. Neural networks, 14(6–7), 883–894, July–September. [DOI] [PubMed]
- Izhikevich, E. M. (2003). Simple model of spiking neurons. IEEE Transactions on Neural Networks, 14(6), 1569–1572. [DOI] [PubMed]
- Izhikevich, E. M. (2004). Which model to use for cortical spiking neurons? IEEE Transactions on Neural Networks, 15(5), 1063–1070, September. [DOI] [PubMed]
- Izhikevich, E. M. (2007). Dynamical systems in neuroscience: The geometry of excitability and bursting. Cambridge: MIT.
- Knuth, D. E. (1997). The art of computer programming: Seminumerical algorithms (Vol. 2, 3rd ed.). Boston: Addison-Wesley Longman.
- Lapicque, L. (1907). Recherches quantitatives sur l’excitation électrique des nerfs traitée comme une polarisation. Journal de Physiologie et de Pathologie Générale, 9, 620–35.
- Latham, P. E., Richmond, B. J., Nelson, P. G., & Nirenberg, S. (2000). Intrinsic dynamics in neuronal networks. I. Theory. Journal of Neurophysiology, 83(2), 808–827, February. [DOI] [PubMed]
- Makino, T. (2003). A discrete-event neural network simulator for general neuron models. Neural Computing & Applications, 11, 210–223. [DOI]
- Mattia, M., & Del Giudice, P. (2000). Efficient event-driven simulation of large networks of spiking neurons and dynamical synapses. Neural Computation, 12(10), 2305–2329. [DOI] [PubMed]
- Morrison, A., Straube, S., Plesser, H. E., & Diesmann, M. (2007). Exact subthreshold integration with continuous spike times in discrete-time neural network simulations. Neural Computation, 19(1), 47–79, January. [DOI] [PubMed]
- Nagumo, J., Arimoto, S., & Yoshizawa, S. (1962). An active pulse transmission line simulating nerve axon. Proceedings of the IRE, 50, 2061–2070. [DOI]
- Parker, G. E., & Sochacki, J. S. (1996). Implementing the Picard iteration. Neural, Parallel & Scientific Computations, 4(1), 97–112.
- Parker, G. E., & Sochacki, J. S. (2000). A Picard-Maclaurin theorem for initial value PDE’s. Abstract and Applied Analysis, 5(1), 47–63. [DOI]
- Press, W., Teukolsky, S., Vetterling, W., & Flannery, B. (1992). Numerical recipes in C (2nd ed.). Cambridge: Cambridge University Press.
- Pruett, C. D., Rudmin, J. W., & Lacy, J. M. (2003). An adaptive N-body algorithm of optimal order. Journal of Computational Physics, 187(1), 298–317. [DOI]
- Rudmin, J. W. (1998). Application of the Parker-Sochacki method to celestial mechanics. Technical Report, James Madison University.
- Traub, R. D., & Miles, R. (1991). Neuronal networks of the hippocampus. New York: Cambridge University Press.
- Tuckwell, H. (1988). Introduction to theoretical neurobiology: Linear cable theory and dendritic structure (Vol. 1). Cambridge: Cambridge University Press.
- Vandenberg, C. A., & Bezanilla, F. (1991). A sodium channel gating model based on single channel, macroscopic ionic, and gating currents in the squid giant axon. Biophysical Journal, 60(6), 1511–1533, December. [DOI] [PMC free article] [PubMed]
- Vogels, T. P., & Abbott, L. F. (2005). Signal propagation and logic gating in networks of integrate-and-fire neurons. Journal of Neuroscience, 25(46), 10786–10795, November. [DOI] [PMC free article] [PubMed]