Abstract
Hepatitis B virus (HBV) infection is a global health problem that causes a wide spectrum of liver disease, including acute or fulminant hepatitis, inactive carrier state, chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). The pathogenesis of hepatocyte damage associated with HBV is mainly through immune-mediated mechanisms. On the basis of the virus and host interactions, the natural history of HBV carriers who are infected in early life can be divided into four dynamic phases. The frequency, extent, and severity of hepatitis flares or acute exacerbation in the second immune clearance and/or fourth reactivation phase predict liver disease progression in HBV carriers. In the past decade, hepatitis B viral factors including serum HBV DNA level, genotype, and naturally occurring mutants predictive of clinical outcomes have been identified. The higher the serum HBV DNA level after the immune clearance phase, the higher the incidence of adverse outcomes over time. In addition, high viral load, genotype C, basal core promoter mutation, and pre-S deletion correlate with increased risk of cirrhosis and HCC development. As to the treatment of chronic hepatitis B, patients with high HBV DNA level and genotype C or D infection are shown to have a worse response to interferon therapy. In conclusion, serum HBV DNA level, genotype, and naturally occurring mutants are identified to influence liver disease progression and therapy of chronic hepatitis B. More investigations are needed to clarify the molecular mechanisms of the viral factors involved in the pathogenesis of each stage of liver disease and the response to antiviral treatments.
Keywords: Chronic hepatitis B, HBV DNA, Genotype, Mutant, HBeAg seroconversion, Hepatocellular carcinoma, Therapy
Introduction
Hepatitis B virus (HBV) is an important public health problem and the major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) worldwide [1]. HBV is the smallest human DNA virus, with a genome of 3,200 base pairs, and belongs to the family Hepadnaviridae [2]. The partially double-stranded circular DNA encodes four overlapping open reading frames including surface (S), core (C), polymerase (P), and X genes [3] (Fig. 1). Owing to the error rate of the viral reverse transcriptase, the HBV genome evolves and the estimated rate of nucleotide substitution is around 1.4 to 3.2 × 10−5 per site per year [4]. This unique replication strategy accounts for the majority of point mutations and deletions or insertions observed in the HBV genome. The long-time evolution of HBV therefore leads to the occurrence of various genotypes, subgenotypes, mutants, recombinants, and even quasispecies [5]. In this article, recent advances in the impact of hepatitis B viral factors including serum HBV DNA level, genotype, subgenotype, and naturally occurring mutants on the natural course and therapy of chronic hepatitis B will be reviewed and discussed.
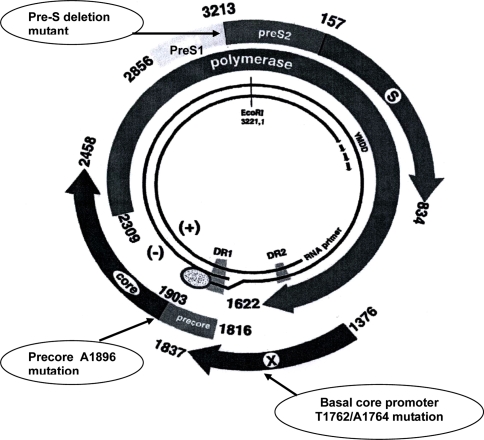
Fig. 1.
The partially double-strand circular genome of hepatitis B virus: S for the surface gene, C for the core gene, P for the polymerase gene, and X for the X gene. Naturally occurring mutants including mutations in precore/basal core promoter and deletion mutation in pre-S gene have been reported to be associated with progressive liver disease including development of cirrhosis and hepatocellular carcinoma
Natural course of chronic hepatitis B
Hepatitis B virus infection is prevalent in Asia, Africa, Southern Europe, and South America, where the prevalence of HBsAg in the general population ranges from 2 to 20% [1]. HBV causes both acute and chronic infection in humans. Acute infection may result in classical acute hepatitis or fulminant hepatitis. In chronic infection, HBV replication persists throughout the course of chronic infection, and host immune response plays a pivotal role in HBV-related liver injury and control of HBV replication [5].
The clinical course of chronic HBV infection is different between Asian and Western patients. Asian HBV carriers mostly acquire the virus in the perinatal period or early childhood [6]. On the basis of the virus and host interactions, the natural history of perinatally acquired HBV infection can be divided into four dynamic phases [7, 8] (Table 1). The first immune tolerance phase, typically represented in the chronically infected children or young adults, is associated with high serum HBV DNA level (>108 copies/ml), limited immunological reactivity to HBV, extensive intrahepatic replication of HBV, and positivity of HBeAg. The liver disease activities are low in this phase despite active HBV replication [9]. A recent report from Hong Kong indicated that adult HBV carriers who remain in the immune tolerance phase have minimal disease progression [10]. After 2–3 decades of persistent infection, chronic hepatitis B enters the second immune clearance phase. During this phase, the previously symptomless carriers may start to have bouts of symptoms and signs suggestive of acute hepatitis flare [11, 12]. With intense immune-mediated cytotoxic response toward infected hepatocytes, the liver cells suffer continuous or repeated bouts of damage, documented by elevated serum alanine aminotransferase (ALT) levels. After these immune attacks, the serum HBV DNA level will be suppressed and these patients may undergo HBeAg seroconversion with the loss of HBeAg and subsequent gain of anti-HBe. However, a certain proportion of HBV carriers may only experience transient and mild elevation of serum ALT levels before HBeAg seroconversion [13]. Several factors have been reported to be associated with seroconversion of HBeAg (Table 2). In the third low replication phase, active replication of HBV ceases; however, HBsAg is continuously expressed from liver cells that contain the integrated HBV genome. Because of the absence of active HBV replication in the liver, the liver cells are spared from immune attacks and patients are in the inactive HBV carrier status. Thus this phase is characterized by absence of HBeAg, presence of anti-HBe, persistently normal ALT level, and low or undetectable serum HBV DNA (<2000 IU/ml). In a minority of patients (0.1–0.8% per year for Asian HBV carriers), after a relatively short duration of follow-up, seroclearance of HBsAg may occur [8]. In contrast, a recent observation indicated an appreciably higher rate of HBsAg seroclearance during a long-term follow-up [14]. HBV carriers in this phase usually confer a favorable prognosis [15]; however, a significant proportion of Asian carriers still develop HCC from a background of cirrhosis [16]. HBV carriers negative for HBeAg were usually thought to have nonreplicative HBV infection, and their serum ALT levels were normal or nearly normal. In the early 1980s, it became apparent that HBV could replicate in the absence of HBeAg, thus the fourth HBV reactivation phase was designated. Reactivation of HBV may occur spontaneously [17] or as a result of immunosuppression [18]. The annual rate of spontaneous reactivation of HBV following HBeAg seroconversion was 2.2–3.3% in asymptomatic HBsAg carriers or patients with chronic hepatitis B [13, 19], while the annual rate of spontaneous reactivation in inactive HBsAg carriers was 1.5%, with a cumulative incidence of 20% after 20 years of follow up [17]. Of particular note is that reactivation of HBV in inactive carriers correlated significantly with patient age on study entry [17], suggesting a favorable outcome of earlier HBeAg seroconversion. In addition, reactivation of HBV was significantly more frequent in male gender [13] and genotype C patients [20]. In this phase, detectable HBV DNA level (>2,000 IU/ml), elevated serum ALT level, and progressive liver disease may develop.
Table 1.
Characteristics of four dynamic phases of chronic hepatitis B virus infection
| Immune tolerance (minimally active) | Immune clearance (HBeAg-positive CHB) | Low replication (inactive carrier state) | Reactivation (HBeAg-negative CHB) | |
|---|---|---|---|---|
| HBeAg | Positive | Positive | Negative | Negative |
| Precore/core promoter | Wild-type | Most wild-type | Most mutants | Most mutants |
| HBV DNA Level (copies/ml)a | Very High (>108) | High (>105) | Low (<104) | Moderate (>104) |
| ALT level | Normal | Elevated | Normal | Elevated |
| Liver histology | Normal or minimal change | Chronic inflammation and fibrosis | Normal or minimal change | Chronic inflammation and fibrosis |
| Candidate for approved therapy | No | Yes | No | Yes |
Abbreviations: CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; ALT, alanine aminotransferase
aFive copies = 1 IU
Table 2.
Factors associated with a higher spontaneous HBeAg seroconversion rate
| Advancing age: |
| <5 years, 0.8% per year |
| 5–15 years, 3% per year |
| >15 years, 8–15% per year |
| Age of infection (older > younger) |
| Mode of transmission (horizontal > vertical) |
| Serum ALT level (higher > lower) |
| Serum AFP level (higher > lower) |
| Immune status (competence > suppression) |
| Ethnicity (Non-Asian > Asian) |
| HLA haplotype (HLA-B61 and -DQB1*0503 > others) |
| Genotype (B > C; D > A) |
Abbreviations: ALT, alanine aminotransferase; AFP, α-fetoprotein.
In contrast, Western HBV carriers usually acquire the virus at older ages through horizontal transmission and do not have the prolonged immune tolerance phase as Asian carriers. They thus enter the immune clearance phase shortly after the establishment of persistent infection [21]. This difference in age of HBV infection may explain, at least in part, the different clinical course and treatment response to immunomodulatory agents between Asian and Western HBV carriers [22].
Factors associated with liver disease progression
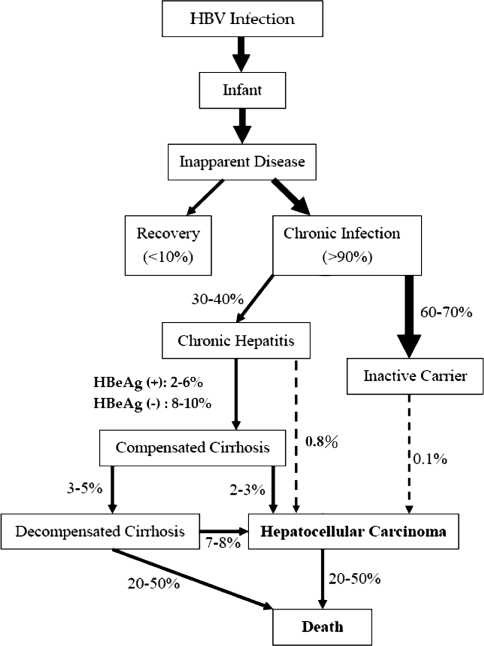
In a population study, the relative risk of HCC in Taiwanese patients with persistent HBV infection was estimated to be 200 times higher than in noninfected individuals [23]. The lifetime risk of HBV carriers to develop cirrhosis, liver failure, or HCC may be as high as 15–40% [24]. The annual incidence of cirrhosis has been estimated to be 2–6% for HBeAg-positive and 8–10% for HBeAg-negative chronic hepatitis B patients [19, 25] (Fig. 2). The annual incidence of HCC has been estimated to be less than 1% for chronic hepatitis patients and 2–3% for those with cirrhosis [25, 26].
Fig. 2.
Annual rates of liver disease progression in hepatitis B virus carriers
Host and environmental factors
The identification of risk factors for the development of end-stage liver disease, including cirrhosis and HCC, in HBV carriers is important for developing appropriate treatment. The risk factors associated with the development of HCC include chronic infection with either HBV or HCV, the presence of cirrhosis, carcinogen exposure (especially aflatoxin), alcohol abuse, genetic factors, male gender, cigarette smoking, and advanced age [27, 28]. These factors have been found to act synergistically to induce cirrhosis and HCC. Among these risk factors, chronic hepatitis viral infections, particularly those with cirrhosis and HBsAg-positive family members of patients with HBV-related HCC, show the strongest association with the development of HCC in Asian countries (Table 3) [29–31].
Table 3.
Risk factors associated with the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis B
| Virus | Host | Environment |
|---|---|---|
| Persistently high HBV replication | Male gender | Concurrent HCV, HDV or HIV infection Alcohol drinking |
| Genotype (C > B; D > A) | Advanced age or longer duration of infection | Cigarette smoking |
| Specific HBV mutants (core promoter mutant, pre-S deletion) X gene transactivation |
Family history of HCC Ethnicity (Asian, African > Caucasian) Genetic alteration Repeated hepatitis flare |
Aflatoxin exposure Diabetes mellitus Obesity Hepatic steatosis |
HBV factors
Serum HBV DNA level
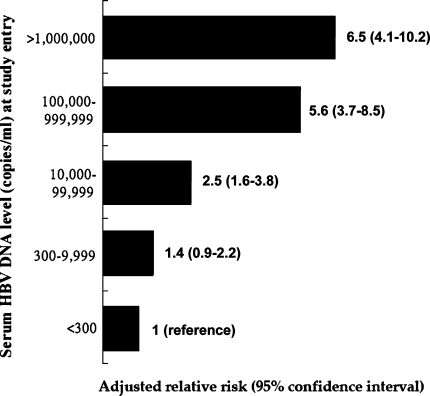
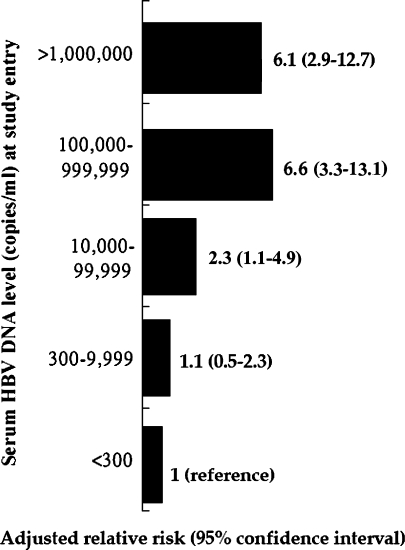
The impact of baseline viral load at enrollment on the risk of cirrhosis and HCC development over time has been increasingly recognized in HBV carriers aged between 30 and 65 years. In a population-based prospective cohort including 7 townships in Taiwan, 3582 untreated HBV carriers were enrolled (REVEAL-HBV study). Of them, 85% were HBeAg-negative and were followed for a mean duration of 11 years [32, 33]. According to this important study, the higher the serum HBV DNA level at early 1990s, the higher the risk of cirrhosis and HCC at early 2000s (Figs. 3 and 4). The cumulative incidence of cirrhosis increased with serum HBV DNA level and ranged from 4.5 to 36.2% for patients with a hepatitis B viral load of less than 60 IU/ml and 2 × 105 IU/ml or more, respectively (P < 0.001). After adjusting for HBeAg status and serum ALT level among other variables, hepatitis B viral load was the strongest predictor of progression to cirrhosis. The relative risk started to increase at an entry HBV DNA level of 2 × 103 IU/ml (relative risk 2.5; 95% CI, 1.6–3.8). Those with HBV-DNA levels of 2 × 105 IU/ml or more had the greatest risk 6.5 (relative risk 6.5; 95% CI, 4.1–10.2; P < 0.001). Of particular note, in HBeAg-negative patients with normal serum ALT levels at entry, the risk of cirrhosis was also increased significantly as serum HBV DNA level increased [32]. The relationship between serum HBV DNA level and risk of HCC was also evaluated in several prospective clinical and population-based studies. In the same prospective cohort in Taiwan, the risk factors associated with the development of HCC were assessed in 3653 HBV carriers aged between 30 and 65 years [33]. A significant biological gradient of HCC risk by serum HBV DNA level from 60 IU/ml (undetectable) to 2 × 105 IU/ml or greater was observed. The dose-response relationship was most prominent for participants who were seronegative for HBeAg with normal serum ALT levels and no cirrhosis at study entry. Participants with persistent elevation of serum HBV DNA level during follow-up had the highest HCC risk. These results were consistent with another prospective cohort study in 2,763 adult HBV carriers with 11 years of follow-up from Haimen city in China that assessed the relationship between HBV viral load at entry and mortality [34]. Compared to HBV carriers without detectable viremia, the relative risk for HCC mortality in those with low viral load (≤2 × 104 IU/ml) was 1.7 (95% CI, 0.5–5.7) and 11.2 (95% CI, 3.6–35.0) in those with high viral load (≥2 × 104/ml). Overall, the ample evidence from these studies strongly indicate that the best predictor of adverse outcomes (cirrhosis, HCC, and death from liver disease) in adult HBV carriers is the serum HBV DNA level at enrollment, independent of HBeAg status, baseline serum ALT level, and other risk factors. However, whether there exists a “safe” serum HBV DNA level for nonprogressive liver disease in the real world remains to be determined. In a recent report, the serum samples of the subjects from the REVEAL-HBV study were reanalyzed by using a more sensitive molecular method with the detection limit of 20 IU/ml. The results showed that the hazard ratio of HCC development significantly increased with elevation of serum HBV DNA level after adjustment for other factors [35]. These data suggest that even a relatively low serum HBV DNA level (<2,000 IU/ml) increases HCC risk when compared to an extremely low HBV DNA level (<20 IU/ml).
Fig. 3.
Adjusted relative risks of cirrhosis in 3,582 hepatitis B virus carriers stratified by baseline serum HBV DNA levels in a population-based prospective cohort study [32]
Fig. 4.
Adjusted relative risks for hepatocellular carcinoma in 3,653 hepatitis B virus carriers stratified by baseline serum HBV DNA levels in a population-based prospective cohort study [33]
Owing to the heterogeneity of HCC populations, risk factors may differ between old HCC patients and young HCC patients. Our previous case–control study showed that young (≤40 years old) HCC patients had lower serum HBV DNA levels than old HCC patients (log10HBV DNA: 4.2 vs. 4.8, P = 0.056). In addition, high serum HBV DNA levels were associated with the development of HCC in old patients (odds ratio [OR], 1.584; 95% confidence interval [CI], 1.075–2.333; P = 0.02) rather than in young patients (OR, 0.848; 95% CI, 0.645–1.116; P = 0.239) [36]. Thus, viral factors in association with the development of HCC in young and old patients await more investigation.
Even in patients already with HCC, serum HBV DNA level appears to impact their outcomes. Recently, Yeo et al. reported that high hepatitis B viral load in unresectable HCC patients prior to systemic chemotherapy was associated with a high incidence of hepatitis flare due to HBV reactivation and thus had an adverse effect on these patients’ survival [37]. Therefore, prophylactic antiviral therapy may be needed to reduce this disastrous event and improve survival for HBV-related HCC patients undergoing chemotherapy.
Genotype and subgenotype
According to the heterogeneity of the virus sequence, at least eight HBV genotypes (A–H) are defined by divergence in the entire HBV genomic sequence >8% [38]. Epidemiologic studies have shown that each genotype has its distinct geographic and ethnic distribution [38, 39]. Genotypes A and D occur frequently in Africa, Europe, and India, while genotypes B and C are prevalent in Asia. Genotype E is restricted to West Africa, and F is found in Central and South America. Genotype G has been reported in France, Germany, and the United States. Lastly, the eighth genotype H was described in Central America. Interestingly, it is noted that genotypes B and C are prevalent in highly endemic areas, such as Asian countries, where perinatal or vertical transmission plays an important role in spreading the virus and the same genotype may be conserved in the same population, whereas genotypes A, D, E, F, and G are frequently found in areas where horizontal transmission is the main mode of transmission.
Extensive phylogenetic analyses have shown that HBV genotypes can be further subdivided into subgenotypes by at least 4% difference in entire genome sequence. Except for genotype E and G, all genotypes have subgenotypes [40]. Epidemiologic data show that in genotypes A, B, and C, respective subgenotypes A1 (Aa)/A2 (Ae), B1 (Bj)/B2 (Ba), and C1 (Cs)/C2 (Ce) differ widely in many virologic aspects. These subgenotypes also display distinct geographic distribution as genotypes do [41]. For example, in Asian countries, subgenotype B1 dominates in Japan and B2 in China and Vietnam. Subgenotype C1 is common in southern China and Southeast Asia and C2 in Taiwan, Japan, Korea, and northern China [42].
Contrary to HCV genotyping, HBV genotyping remained an investigational tool with limited application to the study of the natural history and treatment of HBV infection until a few years ago. Nevertheless, ample evidence has recognized that HBV genotypes influence the natural course of liver disease in HBV carriers, especially in Asian countries where genotypes B and C prevail. Several recent reviews have summarized current knowledge on this issue [26, 38, 39, 41–43]. In addition, owing to the unique distribution of HBV genotypes in Asian and Western countries, the clinical significance and virologic characteristics of HBV genotype could be reliably compared only between genotypes B and C (Table 4) or genotypes A and D (Table 5). In general, genotype B has been shown to be associated with less progressive liver disease than genotype C, and genotype D has been shown to have a less favorable prognosis than genotype A. The clinical significance of genotypes E to H remains to be examined. Nevertheless, it should be emphasized that all HBV genotypes could lead to end-stage liver disease, including cirrhosis and HCC.
Table 4.
Clinical and virologic differences between hepatitis B virus genotypes B and C
| Genotype B | Genotype C | |
|---|---|---|
| Clinical differences | ||
| Positivity of HBeAg | Lower | Higher |
| Seroconversion of HBeAg | Earlier | Later |
| Immunoclearance phase | Shorter | Longer |
| Th1 immune response | Higher | Lower |
| Seroclearance of HBsAg | More | Less |
| Reactivation after HBeAg seroconversion | Less | More |
| Histologic activity | Lower | Higher |
| Clinical outcome (cirrhosis and hepatocellular carcinoma) | Better | Worse |
| Recurrence after liver transplantation | Lower | Higher |
| Virologic differences | ||
| Serum HBV DNA level | Lower | Higher |
| Frequency of precore A1896 mutation | Higher | Lower |
| Frequency of basal core promoter T1762/A1764 mutation | Lower | Higher |
Table 5.
Clinical and virologic differences between hepatitis B virus genotypes A and D
| Genotype A | Genotype D | |
|---|---|---|
| Clinical differences | ||
| Acute hepatitis | Less | More |
| Chronic hepatitis | More | Less |
| Reactivation after HBeAg seroconversion | Less | More |
| Histologic activity | Lower | Higher |
| Clinical outcome (cirrhosis and hepatocellular carcinoma) | Better | Worse |
| Recurrence after liver transplantation | Lower | Higher |
| Virologic differences | ||
| Frequency of precore A1896 mutation | Lower | Higher |
| Frequency of basal core promoter T1762/A1764 mutation | Higher | Lower |
Hepatitis B e antigen seroconversion has been considered an important event in the natural course of perinatally acquired chronic HBV infection, the earlier the seroconversion of HBeAg, the better the clinical outcome of HBV carriers [13]. To this end, the cut-off value for age of HBeAg seroconversion has been suggested; patients with HBeAg seroconversion above 40 years of age were shown to have a substantially high rate of progression to cirrhosis than those below 40 years of age [44].
Previous longitudinal studies showed that genotype B patients had an earlier and more frequent HBeAg seroconversion than genotype C patients [20, 45, 46], suggesting HBV genotype B may be more immunogenic and susceptible to host immunity. This speculation is strengthened by a recent immunologic study that HBV genotype B induced a greater Th1 and lesser Th2 response during the immune clearance phase than genotype C [47].
Most previous retrospective or case–control studies indicated that genotype C patients have more severe liver disease, including cirrhosis and HCC, than do genotype B patients [48–50]. Our recent 14-year prospective study on 4,841 Taiwanese men who were HBV carriers also demonstrated that HBV genotype C was associated with an increased risk of HCC compared with other HBV genotypes (adjusted OR = 5.11, 95% CI = 3.20–8.18) [51]. A community-based prospective cohort study further showed that HBV genotype C was associated with the risk of HCC development [52]. These findings indicated that genotype C was correlated with a higher risk of developing HCC. Of particular note, we also found genotype B was significantly more common in patients with HCC aged less than 50 years compared with age-matched asymptomatic carriers in Taiwan (80% vs. 52%; P = 0.03). This predominance was more remarkable in younger patients with HCC, being 90% in those aged less than 35 years, and most were noncirrhotic [48]. Similar findings were observed in a recent report from China, in which all HCC patients younger than 35 years were infected with genotype B [53]. These data thus suggested that certain genotype B strains might be associated with the development of HCC in young HBV carriers. Studies from Japan, Hong Kong, and China already confirmed the findings that HBV genotype C was associated with the development of HCC [49, 50, 54, 55]. Recent data from the REVEAL-HBV study showed that the hazard ratio of cirrhosis after adjustment for age, gender, smoking, alcohol use, and HBV DNA level was 1.9 for genotype C compared to genotype B, suggesting genotype C was an independent risk factor for cirrhosis development in HBV carriers [56].
Hepatitis B virus genotypes also influence the clinicopathological features of patients with resectable HCC. Among our 193 patients with resectable HBV-related HCC, genotype B patients were less associated with cirrhosis compared with genotype C patients (33% vs. 51%, P = 0.01). Pathologically, genotype B patients had a higher rate of solitary tumor (94% vs. 86%, P = 0.048) and more satellite nodules (22% vs. 12%, P = 0.05) than genotype C patients [57]. These characteristics may contribute to the recurrence patterns and prognosis of HBV-related HCC patients with genotype B or C infection [58].
Although the influence of HBV genotypes on disease progression and clinical outcome has been increasingly recognized, the virologic and molecular mechanisms involved remain largely unknown. Sugiyama et al. recently reported the intra- and extracellular expression of HBV DNA and antigen [59]. The intracellular expression of HBV DNA and HBcAg were higher for genotypes B and C than genotypes A and D. The extracellular expression of HBV DNA and HBeAg were also higher for genotypes B and C than genotypes A and D (Table 6). The intracellular accumulation of HBV DNA and antigens may play a role in inducing liver cell damage. In addition, the highest replication capacity of genotype C may explain the association of genotype C with more severe histological liver damage than other genotypes. On the other hand, a strong extracellular virion secretion may endow a high infectious capacity to blood from individuals infected with this genotype. These data suggest that virologic differences indeed exist among HBV genotypes that may influence their clinical outcomes and epidemiological characteristics. Nevertheless, whether immunopathogenesis differs between various HBV genotypes needs further studies. A timely relevant study showed that the frequency and IFN-α-producing capacity of peripheral blood plasmacytoid dendritic cells (pDCs) were dramatically reduced in chronic hepatitis B patients at the immunoactive phase, and genotype C patients harbored an even lower reduction in IFN-α production than genotype B patients [60]. This observation may correlate with different outcomes of immunomodulatory treatment and the progression of liver disease in HBV carriers infected with different genotypes.
Table 6.
Influence of hepatitis B virus genotypes (A to D) on intra- and extracellular expression of viral DNA and antigens [59]
| Expression | Genotype |
|---|---|
| Intracellular | |
| HBV DNA | C > B > D > A |
| HBcAg | B > C > D > A |
| Extracellular | |
| HBV DNA | B > C = D > A |
| HBeAg | B > C > D > A |
| HBsAg | A > B > C > D |
Although the clinical significance of HBV genotype has become recognized, limited studies have examined the clinical relevance of HBV subgenotypes. HBV subgenotype A1 appears to be associated with low serum HBV DNA levels as well as a low prevalence of serum HBeAg and is implicated in the high incidence of HBV-related HCC in Africa [61]; whereas subgenotype A2 has a higher rate of sustained remission after HBeAg seroconversion and a lower rate of liver-related death than other genotypes during long-term follow-up [62, 63]. Sugauchi et al. found that positivity of HBeAg is significantly more frequent in carriers of subgenotype B1 than B2 [64]. An additional study analyzed the distribution of HBV subgenotypes in 296 HBV-related HCC patients collected from all over Japan [65]. They found HBV subgenotype B2 in 4.4%, B1 in 7.4%, and genotype C in 86.5%. Interestingly, in the Tohoku district and Okinawa, subgenotypes B2 and B1 and genotype C were found in 6.7, 40.0, and 48.9%, respectively, compared to 4.0, 1.6, and 93.2% in the other districts in Japan. In addition, subgenotype B1 was more frequently found in those older than 65 years while subgenotype B2 was found in all age groups. These data suggest that HBV subgenotype B1 may run a more indolent course than subgenotype B2 (Table 7).
Table 7.
Virologic and clinical differences between HBV subgenotypes B1 and B2
| B1 | B2 | |
|---|---|---|
| Positivity of HBeAg | Lower | Higher |
| Precore stop codon mutation | More | Less |
| Basal core promoter mutation | Less | More |
| Risk of hepatocellular carcinoma | Lower | Higher |
Abbreviations: B1, HBV genotype B without recombination with genotype C (G-1838); B2, HBV genotype B and C recombinant (A-1838).
Hepatitis B virus genotype C has been divided into subgenotypes C1 to C4. In Hong Kong, 80% of HBV genotype C patients belonged to subgenotype C1, and the remaining 20% belonged to subgenotype C2 [66]. When subgenotypes C1 and C2 were compared, subgenotype C1 was associated with a higher tendency to develop basal core promoter (BCP) mutations (80% vs. 50%; P = 0.14), a higher prevalence of C at nucleotide 1,858 (C-1858) (95% vs. 0%; P < 0.001), and a lower prevalence of precore stop codon mutations (5% vs. 50%; P = 0.002). It is thus proposed that subgenotypes C1 and C2 have different epidemiological distributions and virologic characteristics. To test this speculation, we studied the distribution of HCV subgenotypes in 242 Taiwanese HBV carriers with various stages of liver disease, and found that HBV subgenotype C2 was the predominant subgenotype in Taiwan. In addition, there was no significant difference in the distribution of the HBV genotype C subgenotypes among patients with different stages of liver disease, suggesting subgenotypes of genotype C may have minimal impact on liver disease progression of chronic hepatitis B in Taiwan [67]. Similarly, a cross-sectional study of 211 patients with various stages of liver disease in China showed the distribution of HBV genotype C was greater among cirrhosis and HCC patients, while genotype B was common in chronic hepatitis patients. In addition, no significant differences in clinical features were found between patients with HBV subgenotypes B2, C1, and C2 [68]. Taken together, further elaborated studies are needed to examine the clinical impact of each HBV subgenotype on the pathogenesis and progression of liver diseases.
Naturally occurring mutants
Owing to the spontaneous error rate of viral reverse transcription, naturally occurring HBV mutants arising during the course of a patient’s infection under the pressure of host immunity or specific therapy [69]. These HBV mutants could display alteration of epitopes important in the host immune recognition, enhanced virulence with increased levels of HBV replication, resistance to antiviral therapies, or facilitated cell attachment/penetration and thus have implications at both the clinical and epidemiological levels. Several HBV mutant strains including mutations in precore, core promoter, and deletion mutation in pre-S/S genes have been reported to be associated with the pathogenesis of fulminant or progressive liver disease, including cirrhosis and HCC [70] (Fig. 1). HBV precore nucleotide 1,896 mutation from guanine (G) to adenine (A) as well as changes of two nucleotides, an adenine (A) to thymine (T) transversion at nucleotide 1,762 together with a guanine (G) to adenine (A) transition at nucleotide 1,764 within the BCP, lead to a proportion of HBeAg-negative patients who continue to have moderate levels of HBV replication and active liver disease [71–73]. Although these mutants can also be found in asymptomatic hepatitis B carriers, the BCP T1762/A1764 mutation has been shown to increase the risk of liver disease progression and HCC development for both genotypes B and C infection [74–78].
In HBV-related acute fulminant hepatitis, universally specific genomic mutational pattern of HBV has not yet been defined. Our previous study indicated that genomic variability of HBV contributes little to the development of fulminant and subfulminant hepatitis in Taiwanese HBV carriers. Although precore A1896 stop codon mutation was frequently found in these patients, it was also commonly encountered in control patients [79, 80]. These findings suggest host factors rather than viral factors are linked with these catastrophic events.
As to the progression of liver disease, in a cohort study of 250 genotype B- or C-infected HBV carriers with different stages of liver disease, we found that genotype C patients had a higher prevalence of BCP T1762/A1764 mutation than genotype B (OR, 5.18; 95% CI, 2.59–10.37; P < 0.001). The likelihood of T1762/A1764 mutation paralleled the progression of liver disease, from 3% in inactive carriers to 64% in HCC patients (OR, 20.04; 95% CI, 7.25–55.41; P < 0.001). Patients with BCP T1762/A1764 mutation were significantly associated with the development of HCC than those without (OR, 10.60; 95% CI, 4.92–22.86; P < 0.001), and the risk was observed for both genotypes B and C [81]. These findings are confirmed by a long-term follow-up study involving 400 HBV carriers in the United States and a case–control study from Hong Kong, in which the presence of BCP T1762/A1764 mutation were independent predictors for the risk of HCC development [49, 82]. In the Philippines, a cross-sectional study of 100 HBV carriers with various stages of liver disease revealed that 51 HBV genotype A1, 22 genotype B and 27 genotype C strains, and genotypes B and C were more prevalent than genotype A in cirrhosis and HCC patients (P < 0.02). In addition, the prevalence of BCP T1762/A1764 mutant was higher in HCC patients with genotypes B and C. Multivariate analysis indicated that age and core promoter mutation were risk factors for HCC development [83]. Taking these lines of evidence together, BCP T1762/A1764 mutation seems to play an important role in the pathogenesis of liver disease progression and serve as the strongest viral factor associated with HCC risk in HBV carriers.
Other naturally occurring mutations or deletions in the pre-S gene of HBV genome frequently occur in chronic HBV infection [84, 85]. The deletion over pre-S gene may affect the expression of middle and small surface proteins, resulting in intracellular accumulation of large surface protein [86], and contribute to more progressive liver cell damage and finally hepatocarcinogenesis [87, 88]. In our recent case–control study, pre-S deletion mutant of HBV were determined in 202 asymptomatic carriers and 64 HCC patients with chronic HBV genotype B or C infection. The presence of pre-S deletion mutant was independently associated with the development of HCC (OR, 3.72; 95% CI, 1.44–9.65; P = 0.007) [77]. Our further mapping study of pre-S region revealed that all the deletion regions encompassed T- and B-cell epitopes, and most of them lost one or more functional sites, including polymerized human serum albumin-binding site and nucleocapsid-binding site. These findings lend support to the biologic significance of emerging HBV pre-S deletion mutants, which may contribute to more progressive liver cell damage and finally hepatocarcinogenesis.
Potential interactions between known HBV factors
In light of these emerging data, HBV genotypes, mutant stains and HBV DNA level are closely associated with the long-term outcomes of HBV-related chronic liver disease. In our earlier study, we already found that genotype C has a higher frequency of BCP T1762/A1764 mutation than genotype B, that is, 50% vs. 6% [89]. However, it is unclear whether a specific combination of these factors is associated with the development of a more severe liver disease. In a prospective study, Yu et al. provide strong evidence that the risk of HCC was increased approximately 5-fold among men infected with HBV genotype C compared with genotype B. They also found HBV viral load was higher with HBV genotype C than with HBV genotype B, and men who had both HBV genotype C and a very high HBV viral load had a 26-fold higher risk of HCC than those with other genotypes and low or undetectable viral loads [51]. These observations suggest additive associations of viral load and HBV genotype C with HCC risk. We recently investigated the independent and interactive effects of each known viral factor on the development of HCC. Compared with patients with low HBV load and the BCP A1762/G1764 wild-type strain, the adjusted odds ratio of developing HCC was more than 30-fold in patients with an HBV load ≥20,000 IU/ml and the BCP T1762/A1764 mutant, irrespective of the presence of the precore A1896 stop codon mutation and viral genotype [75]. Similar to that in cirrhotic HCC, BCP T1762/A1764 mutation and viral load ≥20,000 IU/ml were independently associated with the risk of noncirrhotic HCC [76]. We also addressed the interactions among pre-S deletion, precore mutation, and BCP T1762/A1764 mutation in various stages of chronic HBV infection [77]. The results revealed that the presence of pre-S deletion and BCP T1762/A1764 mutation was significantly associated with the development of progressive liver diseases. In addition, a combination of mutations rather than a single mutation was associated with the development of progressive liver diseases, especially in combination with pre-S deletion. Similarly, Yuen et al. from Hong Kong examined the risks for HCC with respect to HBV genotypes, specific viral mutants, serum HBV DNA levels, and cirrhosis in a case–control study, and multivariate analysis showed that core promoter mutant, T1653, HBV DNA ≥2,000 IU/ml, and cirrhosis were independent factors for HCC. In addition, the risks remarkably increased in HBV carriers with these factors in combination [90]. In Japan, an age-, sex-, and HBeAg status-matched cross-sectional control study was conducted to determine HCC-associated mutations of the HBV genome in the entire X, core promoter, and precore/core regions among 80 patients infected with HBV subgenotype C2 with HCC and 80 without HCC. They found that the prevalence of the T1653 mutation in the box alpha region and V1753 and T1762/A1764 mutations in the BCP region was significantly higher in the HCC group than in the non-HCC group. Further multivariate analysis showed that the presence of T1653, V1753, and a low platelet count were independent predictive factors for HCC in patients with HBV subgenotype C2 [91]. In China, a cross-sectional study indicated that V1753 and BCP T1762/A1764 mutations seem to be associated with HCC development, especially in patients with HBV subgenotype C1 [53]. These findings altogether suggest that in addition to HBV DNA level, accumulation of complex viral mutants with precore mutation and BCP T1762/A1764, T1653, and pre-S deletion mutations may affect the long-term outcomes of chronic HBV infection.
With variables that possibly are risk factors for HCC development, a risk function nomogram for predicting HCC in HBV carriers was developed by using noninvasive clinical and virologic information collected from the REVEAL-HBV study [92]. Chen et al. found that this predictive model had good characteristics, and the combination of serum HBV DNA level ≥2,000 IU/ml and HBV genotype C had the greatest HCC risk. However, this nomogram needs further validation and, once validated, will simplify the communication of individual HCC risk using noninvasive variables.
Current therapy of chronic hepatitis B
At the time of this writing, no antiviral treatment has been shown to cure chronic HBV infection. Therefore, the treatment goals are to control hepatitis activity, obtain hepatitis B e antigen (HBeAg) seroconversion or sustained suppression of HBV replication, and improve necroinflammatory activity and fibrosis of the liver [93, 94]. Currently, six drugs have been approved for the treatment of chronic hepatitis B: conventional interferon (IFN) alfa, lamivudine, adefovir dipivoxil, entecavir, pegylated interferon alfa-2a, and telbivudine [94]. The latest Asian-Pacific Association for the Study of the Liver (APASL) consensus recommendations suggest that IFN-alfa, pegylated IFN alfa-2a, lamivudine, and adefovir dipivoxil can be used as first-line therapies for chronic hepatitis B [93]. The APASL working party also raised unresolved issues for further studies, including the role of combination therapy or other immunomodulatory agents. In addition, the updated treatment algorithm for the management of chronic hepatitis B virus infection developed by a US expert panel and the 2007 update of the American Association of the Study of Liver Disease (AASLD) guidelines for chronic hepatitis B recommend that treatment may be initiated with any of the six approved agents, but pegylated interferon, adefovir dipivoxil, or entecavir are preferred for HBeAg-positive or HBeAg-negative chronic hepatitis B patients [94, 95].
HBV factors and therapy of chronic hepatitis
Serum HBV DNA level
As reviewed earlier, persistent elevation of serum HBV DNA level is known to be associated with liver disease progression in chronic hepatitis B patients. Therefore, this would suggest that the primary aim of hepatitis B treatment is to reduce and maintain serum HBV DNA at the lowest possible level. Profound and sustained viral suppression may also lead to other aims of therapy, such as biochemical normalization, HBeAg seroconversion, histologic improvement, and reduced drug resistance [93–95].
For the treatment of chronic hepatitis B with standard interferon, several host and viral factors including baseline serum ALT levels, baseline HBV DNA, degree of histological activity, age at acquisition, and sex have been found to be associated with a better virologic response in HBeAg-positive patients [93, 96]. Among them, HBV DNA <200 pg/ml (∼5 × 107 copies/ml) is more frequently associated with a virologic response [96, 97]. As to pegylated interferon, multivariate analysis of HBeAg-positive patients treated with 48 weeks of pegylated interferon alfa-2a indicated that a low HBV DNA level (<109 copies/ml) predicted a high HBeAg seroconversion rate [98]. Among patients with low HBV DNA levels, HBeAg seroconversion was observed in more than 50%. For the treatment of HBeAg-negative patients, multivariate analysis of a phase III study using pegylated interferon alfa-2a has found that a low baseline HBV DNA level could be predictive of a sustained virologic response when evaluated at 24 weeks of therapy [99]. During treatment, patients who became HBV DNA undetectable after 12 weeks of pegylated interferon alfa-2a therapy have been shown to have a better-sustained virologic response [100].
In contrast, baseline serum HBV DNA level has not been shown to predict the response to nucleoside or nucleotide analogues [94]. However, a higher serum HBV DNA level may be associated with a higher cumulative incidence of drug resistance to lamivudine and time to HBV DNA breakthrough [101]. Further studies are awaited to examine whether various baseline HBV DNA levels affect HBeAg seroconversion rates of patients treated with oral antiviral agents, as is observed with interferon treatment.
During treatment with nucleoside or nucleotide analogues, early virologic response has been shown to be an important factor in predicting sustained response and reducing drug resistance. In a study to explore risk factors associated with HBV DNA breakthrough during prolonged lamivudine treatment, Yuen et al. found that patients with HBV DNA levels of >200 IU/ml after 6 months of lamivudine therapy had a 63% chance of subsequently developing drug-resistant mutants [102]. Recently, of HBeAg-positive patients treated with telbivudine who achieved undetectable serum HBV DNA (<60 IU/ml) at 6 months of therapy, 49% achieved HBeAg seroconversion, 85% achieved ALT normalization, 86% achieved sustained viral suppression, and 98% without resistance at year 2. Similarly, of HBeAg-negative patients, 83% achieved ALT normalization, 88% achieved sustained viral suppression, and 98% without resistance at year 2 [103]. Taking these data together, on-treatment HBV DNA monitoring strategies to identify early viral suppression to an undetectable level would be predictive of better therapeutic outcomes and a greater likelihood of detecting drug resistance.
Genotypes
The role of the HBV genotype in predicting response to both immunomodulatory treatment and nucleoside analogue has gained considerable attention in recent years. Owing to the unique distribution of HBV genotypes in Eastern and Western countries, the therapeutic implications of HBV genotype could only be compared between genotypes B and C or genotypes A and D. For further details, please see reviews [43, 104].
In HBeAg-positive chronic hepatitis B patients treated with standard IFN for 4–6 months, the response rate as defined by normalization of serum ALT level and HBeAg seroconversion was better in patients with genotypes A and B than in those with genotypes C and D [105–108]. However, these studies, on the association of HBV genotype with response to standard interferon were limited by small sample sizes and retrospective study design. Our recent randomized study on 119 HBeAg-positive patients also showed that genotype B patients have a better sustained HBeAg seroconversion rate to 32 weeks of standard IFN-based therapy than genotype C patients (44% vs. 22%, P = 0.02) [109]. The relationship between viral genotype and response to pegylated interferon is similar to that observed with standard interferon. Cooksley et al. showed that the response rate of using 24-week pegylated IFN alfa-2a or standard IFN alfa-2a was higher in genotype B than genotype C (33% vs. 21%; 25% vs. 6%; respectively) in a phase 2 study [110]. In a multicenter phase 3 study using a relatively low dose of pegylated interferon alfa-2b for 48 weeks, the overall response rate differed according to HBV genotype: genotype A, 47%; genotype B, 44%; genotype C, 28%; and genotype D, 25% [111]. Subsequent follow-up showed that the loss of HBsAg differed according to HBV genotype: 14% for genotype A, 9% for genotype B, 3% for genotype C, and 2% for genotype D (A vs. D: P = 0.006) [112]. In another multicenter phase 3 trial using a standard dose of pegylated IFN-α 2a for 48 weeks, genotype A patients had the highest HBeAg seroconversion rate (50%) but the difference between genotype B and C patients was not significant (30% vs. 31%) [113]. In a recent randomized controlled trial from China, Zhao et al. assessed the efficacy of low-dose, 24-week IFN or pegylated IFN treatment as well as factors predicting sustained response in patients with HBeAg-positive chronic hepatitis B [114]. They consistently found that HBV genotype B and younger age were independent factors associated with sustained response, and suggested low-dose IFN regimen might be cost-effective for the treatment of younger genotype B patients. In addition, Hou et al. prospectively investigated the relationship between HBV genetic characteristics and the outcome of short (16 weeks) or prolonged (32 weeks) treatment with standard IFN in 103 European HBeAg-positive patients and found that HBV genotype A responded earlier to IFN than other genotypes, which was associated with its molecular characteristics [115]. Collectively, the optimal duration of interferon treatment for chronic hepatitis B may vary among different HBV genotypes. For example, HBV genotype B is somewhat like HCV genotype 2 or 3, which may need only 24 weeks of low-dose pegylated IFN. In contrast, genotype C acts like HCV genotype 1, which may require a longer duration of high-dose pegylated IFN [116]. Nevertheless, whether the discrepancies between these pegylated interferon studies is attributable to the differences in dose, duration or antiviral potency of particular interferons, patient characteristics or a combination of both need to be clarified in clinical trials stratified by different HBV genotypes and treatment regimens. Furthermore, we failed to identify an IFN sensitivity-determining region (ISDR) within the genome of HBV genotype B2 [117], suggesting host factors and virus–host interactions may be more important in determining the response to IFN treatment.
For the treatment of HBeAg-negative CHB patients with pegylated IFN-α 2a, Bonino et al. investigated the effect of pretreatment factors on posttreatment responses. They found that baseline ALT and HBV DNA levels, age, gender, and HBV genotype significantly influenced combined response at 24 weeks posttreatment. Patients with genotype B or C had a higher chance of response than genotype D infected patients (P < 0.001), and the latter responding better to the combination than to peginterferon alpha-2a monotherapy (P = 0.015) [99]. Although thymosin alfa-1 has not been widely approved for the treatment of chronic hepatitis B, a recent study indicated that genotype B was associated with a higher response rate to this immunomodulatory therapy than genotype C [118].
In patients treated with lamivudine, previous studies from Taiwan and Hong Kong indicated that HBV genotype had no impact on the therapeutic response to lamivudine [119–121]. In Spain, Buti et al. suggested that the outcome after lamivudine treatment was also comparable between genotypes A and D [122]. These data imply that HBV genotype may have no substantial impact on the response to lamivudine treatment. However, Chien et al. reported that the sustained response rate to lamivudine was much higher in patients with genotype B than those with genotype C [123]. As to the emergence of drug resistance to lamivudine after prolonged therapy, we found that the development of lamivudine resistance was similar between genotypes B and C [119]. Buti et al. suggested that the emergence of lamivudine resistance were also comparable between genotypes A and D [122]. Nevertheless, studies from Germany and China indicated that there exist different YMDD mutational patterns among different lamivudine-resistant HBV genotypes [124, 125]. For example, the frequency of rtM204I mutants is higher in genotypes C and D, whereas that of rtM204V is higher in genotypes B and A.
As to nucleoside or nucleotide analogues other than lamivudine, several studies have shown that virologic response to treatment with adefovir or entecavir is not affected by viral genotype [126–128]. However, patients with adefovir resistance were more likely to be infected with genotype D [129]. In a phase 3 multicenter trial of latest approved telbivudine, reduction in serum HBV DNA level for genotypes B and C patients at year 2 was 5.9 vs. 5.8 log copies/ml, respectively, for HBeAg-positive patients and 4.7 vs. 5.0 log copies/ml, respectively, for HBeAg-negative patients; the difference was not statistically significant [103]. In summary, these lines of evidence imply that HBV genotype appears to have no substantial impact on the response to nucleoside or nucleotide analogue treatment.
In summary, HBV genotype correlates well with the response to standard IFN but not nucleoside/nucleotide-based therapy (Tables 8 and 9). The association between HBV genotype and response to pegylated IFN remains controversial. Alternatively, future genotype-based stratification trials should be considered to clarify the impact of HBV genotype.
Table 8.
Therapeutic differences for HBeAg-positive chronic hepatitis B between genotypes B and C
| HBV genotype | |||
|---|---|---|---|
| B | C | ||
| Standard IFN/thymosin | Better | Worse | |
| Pegylated IFN | Controversial | ||
| Lamivudine | |||
| Response | No difference | ||
| Relapse | Lower | Higher | |
| Resistance | No difference | ||
| Mutational pattern | More rtM204V | More rtM204I | |
| Adefovir dipivoxil | No difference | ||
| Entecavir | No difference | ||
| Telbivudine | No difference | ||
Table 9.
Therapeutic differences for HBeAg-positive chronic hepatitis B between genotypes A and D
| HBV genotype | |||
|---|---|---|---|
| A | D | ||
| Standard IFN | Better | Worse | |
| Pegylated IFN | Better | Worse | |
| Lamivudine | |||
| Response | No difference | ||
| Relapse | No data | ||
| Resistance | No difference | ||
| Mutational pattern | More rtM204V | More rtM204I | |
| Adefovir dipivoxil | No difference | ||
| Entecavir | No difference | ||
| Telbivudine | No difference | ||
Naturally occurring mutants
It has been reported that HBeAg-negative chronic hepatitis B is associated with a high relapse rate and a poor sustained response rate of only 10% to standard IFN therapy [130]. Although precore A1896 stop codon mutant and BCP T1762/A1764 mutant are known to abolish or reduce the production of HBeAg, leading to the development of HBeAg-negative chronic hepatitis B [131, 132], the influence of these two common HBV mutants on the response to currently available anti-HBV agents remains largely unknown. Earlier studies showed that precore A1896 stop codon mutant might be associated with the poor response to IFN therapy in patients with HBeAg-negative chronic hepatitis B [130]; however, the results seem controversial [133]. Recently, Erhardt et al. performed sequence analysis of several HBV subgenomic regions in 96 HBeAg-positive and HBeAg-negative patients treated with standard interferon [134]. The overall sustained response (SR) rate to IFN was 30% with no significant difference between HBeAg-positive and HBeAg-negative patients. IFN responsiveness correlated with the number of mutations in the complete BCP, especially the nucleotide region 1,753–1,766 and mutations at nucleotide 1,762 and 1,764. Sustained response to IFN was associated with a high number of mutations in the BCP and the nucleotide region 1,753–1,766 as well as mutations at nucleotide 1,764 in HBeAg-positive patients. In contrast, SR to IFN correlated with a low number of mutations in the BCP and nucleotide region 1,753–1,766 and a wild-type sequence at nucleotide 1,764 in HBeAg-negative patients. In addition, IFN response did not correlate with the occurrence of the precore A1896 mutation. These data suggest that mutations located within the BCP of HBV genome may determine a response to IFN therapy. However, whether similar phenomenon could be observed in the era of pegylated interferon awaits further studies.
For the treatment of HBeAg-negative patients with lamivudine, Lok et al. reported that core promoter mutation was associated with the selection of lamivudine-resistant mutants in addition to the duration of treatment. They also found that some patients with lamivudine-resistant mutations had reversion of the precore A1896 stop codon mutation with reappearance of HBeAg [135]. Our recent study on HBeAg-positive patients further showed that lamivudine therapy may result in the rapid development of BCP T1762/A1764 mutation of the HBV genome, but this mutation may reverse to wild type gradually after cessation of therapy. In addition, there was no significant change of precore sequences before and during lamivudine therapy [136]. A recent report from Australia indicated that the presence of precore A1896 stop codon mutant was an independent predictor of the early development of lamivudine resistance, while genotype did not influence treatment outcome or time to onset of resistance [137]. Since relevant data are limited, further studies should focus on the contribution of viral factors other than genotype to the treatment outcome of existing antiviral agents.
Non-naturally occurring mutations in the polymerase gene play an important role in developing resistance to nucleoside or nucleotide analogues [138], but do not appear to have an adverse effect on the outcome of IFN therapy.
Conclusions
Several hepatitis B viral factors such as serum HBV DNA level, genotype, and naturally occurring mutants have already been identified to influence liver disease progression and therapy of chronic hepatitis B. On the basis of these emerging data, it is recommended that HBV carriers be routinely genotyped to help identify those who may be at a higher risk of liver disease progression and who are most appropriate for interferon treatment. In the future, more investigations are needed to clarify the molecular and virologic mechanisms of these viral factors involved in the pathogenesis of each stage of liver disease and response to antiviral treatments.
Abbreviations
- ALT
Alanine aminotransferase
- BCP
Basal core promoter
- HBeAg
Hepatitis B e antigen
- HCC
Hepatocellular carcinoma
- HBV
Hepatitis B virus
- ULN
Upper limit of normal
References
- 1.Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis 2002;2:395–403. [DOI] [PubMed]
- 2.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem 1987;56:651–93. [DOI] [PubMed]
- 3.Hunt CM, McGill JM, Allen MI, Condreay LD. Clinical relevance of hepatitis B viral mutations. Hepatology 2000;31:1037–44. [DOI] [PubMed]
- 4.Okamoto H, Imai M, Kametani M, Nakamura T, Mayumi M. Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through materno-fetal transmission. Jpn J Exp Med 1987;57:231–6. [PubMed]
- 5.Lau JY, Wright TL. Molecular virology and pathogenesis of hepatitis B. Lancet 1993;342:1335–40. [DOI] [PubMed]
- 6.Stevens CE, Beasley RP, Tsui J, Lee WC. Vertical transmission of hepatitis B antigen in Taiwan. N Engl J Med 1975;292:771–4. [DOI] [PubMed]
- 7.Chen DS. Natural history of chronic hepatitis B virus infection: new light on an old story. J Gastroenterol Hepatol 1993;8:470–5. [DOI] [PubMed]
- 8.Kao JH, Chen DS. The natural history of hepatitis B virus infection. In: Lai CL, Locarnini S, editors. Hepatitis B virus. London: International Medical Press Ltd.; 2002. p. 161–72.
- 9.Chu CM, Liaw YF. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology 1987;92:220–5. [DOI] [PubMed]
- 10.Hui CK, Leung N, Yuen ST, Zhang HY, Leung KW, Lu L, et al., for the Hong Kong Liver Fibrosis Study Group. Natural history and disease progression in Chinese chronic hepatitis B patients in immune-tolerant phase. Hepatology 2007;46:395–401. [DOI] [PubMed]
- 11.Liaw YF, Chu CM, Su IJ, Huang MJ, Lin DY, Chang-Chien CS. Clinical and histological events preceding hepatitis B e antigen seroconversion in chronic type B hepatitis. Gastroenterology 1983;84:216–9. [PubMed]
- 12.Liaw YF, Yang SS, Chen TJ, Chu CM. Acute exacerbation in hepatitis B e antigen positive chronic type B hepatitis. A clinicopathological study. J Hepatol 1985;1:227–33. [DOI] [PubMed]
- 13.Chu CM, Hung SJ, Lin J, Tai DI, Liaw YF. Natural history of hepatitis B e antigen to antibody seroconversion in patients with normal serum aminotransferase levels. Am J Med 2004;116:829–34. [DOI] [PubMed]
- 14.Chu CM, Liaw YF. HBsAg seroclearance in asymptomatic carriers of high endemic areas: appreciably high rates during a long-term follow-up. Hepatology 2007;45:1187–92. [DOI] [PubMed]
- 15.Chen YC, Sheen IS, Chu CM, Liaw YF. Prognosis following spontaneous HBsAg seroclearance in chronic hepatitis B patients with or without concurrent infection. Gastroenterology 2002;123:1084–9. [DOI] [PubMed]
- 16.Huo TI, Wu JC, Lee PC, Chau GY, Lui WY, Tsay SH, et al. Sero-clearance of hepatitis B surface antigen in chronic carriers does not necessarily imply a good prognosis. Hepatology 1998;28:231–6. [DOI] [PubMed]
- 17.Chu CM, Liaw YF. Spontaneous relapse of hepatitis in inactive HBsAg carriers. Hepatol Int 2007;1:311–5. [DOI] [PMC free article] [PubMed]
- 18.Lok AS, Liang RH, Chiu EK, Wong KL, Chan TK, Todd D. Reactivation of hepatitis B virus replication in patients receiving cytotoxic therapy: report of a prospective study. Gastroenterology 1991;100:182–8. [DOI] [PubMed]
- 19.Hsu YS, Chien RN, Yeh CT, Sheen IS, Chiou HY, Chu CM, et al. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 2002;35:1522–7. [DOI] [PubMed]
- 20.Chu CM, Liaw YF. Genotype C hepatitis B virus infection is associated with a higher risk of reactivation of hepatitis B, progression to cirrhosis than genotype B: a longitudinal study of hepatitis B e antigen-positive patients with normal aminotransferase levels at baseline. J Hepatol 2005;43:411–7. [DOI] [PubMed]
- 21.Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen-negative chronic hepatitis B. Hepatology 2001;34:617–24. [DOI] [PubMed]
- 22.Yuen MF. Revisiting the natural history of chronic hepatitis B: impact of new concepts on clinical management. J Gastroenterol Hepatol 2007;22:973–6. [DOI] [PubMed]
- 23.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet 1981;2:1129–33. [DOI] [PubMed]
- 24.Lok AS. Chronic hepatitis B. N Engl J Med 2002;346:1682–3. [DOI] [PubMed]
- 25.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 2004;127 Suppl 1:S35–S50. [DOI] [PubMed]
- 26.Kao JH. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology 2003;46:400–7. [DOI] [PubMed]
- 27.Colombo M. Hepatocellular carcinoma. J Hepatol 1992;15:225–36. [DOI] [PubMed]
- 28.Kao JH, Chen DS. Changing disease burden of hepatocellular carcinoma in the Far East and Southeast Asia. Liver Int 2005:25:696–703. [DOI] [PubMed]
- 29.Chen DS. From hepatitis to hepatoma: lessons from type B viral hepatitis. Science 1993;262:369–70. [DOI] [PubMed]
- 30.Kao JH, Chen DS. Overview of hepatitis B and C viruses. In: Goedert JJ, editor. Infectious causes of cancer: targets for intervention. Tottowa: Humana Press Inc.; 2000. p. 313–30.
- 31.Kao JH, Chen DS. Hepatocellular carcinoma in Taiwan: progress in the past decade 1988–1997. In: Leong ASY, Liew CT, Lau JWY, Johnson PJ, editors. Hepatocellular carcinoma: contemporary diagnosis, investigation and management. London: Arnold; 1999. p. 235–50.
- 32.Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. Predicting liver cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 2006;130:678–86. [DOI] [PubMed]
- 33.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006;295:65–73. [DOI] [PubMed]
- 34.Chen G, Lin W, Shen F, Iloeje UH, London WT, Evans AA. Past HBV viral load as predictor of mortality and morbidity from HCC and chronic liver disease in a prospective study. Am J Gastroenterol 2006;101:1797–803. [DOI] [PubMed]
- 35.Chen CJ, Iloeje UH, Yang HI. Serum hepatitis B virus DNA as a predictor of the development of cirrhosis and hepatocellular carcinoma. Curr Hepat Rep 2007;6:9–16.
- 36.Tsai FC, Liu CJ, Chen CL, Chen PJ, Lai MY, Kao JH, et al. Lower serum viral loads in young patients with hepatitis-B-virus-related hepatocellular carcinoma. J Viral Hepat 2007;14:153–60. [DOI] [PubMed]
- 37.Yeo W, Mo FK, Chan SL, Leung NW, Hui P, Lam WY, et al. Hepatitis B viral load predicts survival of HCC patients undergoing systemic chemotherapy. Hepatology 2007;45:1382–9. [DOI] [PubMed]
- 38.Kao JH. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J Gastroenterol Hepatol 2002;17:643–50. [DOI] [PubMed]
- 39.Kao JH, Chen DS. HBV genotypes: epidemiology and implications regarding natural history. Curr Hepat Rep 2006;5:5–13.
- 40.Kato H, Fujiwara K, Gish RG, Sakugawa H, Yoshizawa H, Sugauchi F, et al. Classifying genotype F of hepatitis B virus into F1 and F2 subtypes. World J Gastroenterol 2005;11:6295–304. [DOI] [PMC free article] [PubMed]
- 41.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol 2007;13:14–21. [DOI] [PMC free article] [PubMed]
- 42.Liu CJ, Kao JH. Clinical implications of hepatitis B virus genotype. Hepatol Rev 2006;3:33–40.
- 43.Liu CJ, Kao JH, Chen DS. Therapeutic implications of hepatitis B virus genotypes. Liver Int 2005;25:1097–107. [DOI] [PubMed]
- 44.Chu CM, Liaw YF. Chronic hepatitis B virus infection acquired in childhood: special emphasis on prognostic and therapeutic implication of delayed HBeAg seroconversion. J Viral Hepat 2007;14:147–52. [DOI] [PubMed]
- 45.Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 2002;122:1756–62. [DOI] [PubMed]
- 46.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol 2004;72:363–9. [DOI] [PubMed]
- 47.Yuen MF, Wong DK, Zheng BJ, Chan CC, Yuen JC, Wong BC, et al. Difference in T helper responses during hepatitis flares in hepatitis B e antigen (HBeAg)-positive patients with genotypes B and C: implication for early HBeAg seroconversion. J Viral Hepat 2007;14:269–75. [DOI] [PubMed]
- 48.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 2000;118:554–9. [DOI] [PubMed]
- 49.Yuen MF, Tanaka Y, Mizokami M, Yuen JC, Wong DK, Yuan HJ, et al. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis 2004;25:1593–8. [DOI] [PubMed]
- 50.Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut 2004;53:1494–8. [DOI] [PMC free article] [PubMed]
- 51.Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst 2005;97:265–72. [DOI] [PubMed]
- 52.Yang HI, Chen PJ, Yeh SH, et al. Risk of hepatocellular carcinoma associated with genotypes and mutants of hepatitis B virus: a community-based prospective cohort study. J Hepatol 2006;44 Suppl 2:27A. 16290909
- 53.Yuan J, Zhou B, Tanaka Y, Kurbanov F, Orito E, Gong Z, et al. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J Clin Virol 2007;39:87–93. [DOI] [PubMed]
- 54.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, et al. A case–control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 2001;33:218–23. [DOI] [PubMed]
- 55.Ding X, Mizokami M, Yao G, Xu B, Orito E, Ueda R, et al. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shanghai, China. Intervirology 2001;44:43–7. [DOI] [PubMed]
- 56.Chen YL, Chen PJ, Yang HI, Iloeje UH, Su J, et al. Risk of liver cirrhosis associated with genotype and mutants of hepatitis B virus. EASL 2007; Abstract no. 222.
- 57.Lin CL, Chen JD, Liu CJ, Lee PH, Chen PJ, Lai MY, et al. Clinicopathological differences between hepatitis B viral genotype B- and C-related resectable hepatocellular carcinoma. J Viral Hepat 2007;14:64–9. [DOI] [PubMed]
- 58.Chen JD, Liu CJ, Lee PH, Chen PJ, Lai MY, Kao JH, et al. Hepatitis B genotypes correlate with tumor recurrence after curative resection of hepatocellular carcinoma. Clin Gastroenterol Hepatol 2004;2:64–71. [DOI] [PubMed]
- 59.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 2006;44:915–24. [DOI] [PubMed]
- 60.Wang K, Fan X, Fan Y, Wang B, Han L, Hou Y. Study on the function of circulating plasmacytoid dendritic cells in the immunoactive phase of patients with chronic genotype B and C HBV infection. J Viral Hepat 2007;14:276–82. [DOI] [PubMed]
- 61.Kramvis A, Kew MC, Bukofzer S. Hepatitis B virus precore mutants in serum and liver of Southern African Blacks with hepatocellular carcinoma. J Hepatol 1998;28:132–41. [DOI] [PubMed]
- 62.Tanaka Y, Hasegawa I, Kato T, et al. A case–control study for differences among hepatitis B virus infections of genotypes A (subtypes Aa and Ae) and D. Hepatology 2004;40:747–55. [DOI] [PubMed]
- 63.Sugauchi F, Kumada H, Acharya SA, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol 2004;85:811–20. [DOI] [PubMed]
- 64.Sugauchi F, Orito E, Ichida T, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol 2002;76:5985–92. [DOI] [PMC free article] [PubMed]
- 65.Orito E, Sugauchi F, Tanaka Y, Ichida T, Sata M, Tanaka E, et al. Differences of hepatocellular carcinoma patients with hepatitis B virus genotypes of Ba, Bj or C in Japan. Intervirology 2005;48:239–45. [DOI] [PubMed]
- 66.Chan HL, Tse CH, Ng EY, Ng EY, Au TC, Yuen L, et al. Epidemiological and virological characteristics of 2 subgroups of hepatitis B virus genotype C. J Infect Dis 2005;191:2022–32. [DOI] [PubMed]
- 67.Tseng TC, Liu CJ, Chen PJ, Lai MY, Lin CL, Kao JH, et al. Subgenotypes of hepatitis B virus genotype C do not correlate with disease progression of chronic hepatitis B in Taiwan. Liver Int 2007;7:983–8. [DOI] [PubMed]
- 68.Wang Z, Tanaka Y, Huang Y, Kurbanov F, Chen J, Zeng G, et al. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol 2007;45:1491–6. [DOI] [PMC free article] [PubMed]
- 69.Gunther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res 1999;52:125–37. [DOI] [PubMed]
- 70.Hunt CM, McGill JM, Allen MI, Condreay LD. Clinical relevance of hepatitis B viral mutations. Hepatology 2000;31:1037–44. [DOI] [PubMed]
- 71.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet 1989;2:588–91. [DOI] [PubMed]
- 72.Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, et al. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol 1990;64:1298–303. [DOI] [PMC free article] [PubMed]
- 73.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, et al. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol 1994;68:8102–10. [DOI] [PMC free article] [PubMed]
- 74.Lin CL, Liao LY, Wang CS, Chen PJ, Lai MY, Chen DS, et al. Basal core promoter mutant of hepatitis B virus and progression of liver disease in hepatitis B e antigen-negative chronic hepatitis B. Liver Int 2005;25:564–70. [DOI] [PubMed]
- 75.Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, et al. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis 2006;193:1258–65. [DOI] [PubMed]
- 76.Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, et al. Role of hepatitis B virus precore/core promoter mutations and serum viral load on noncirrhotic hepatocellular carcinoma: a case–control study. J Infect Dis 2006;194:594–9. [DOI] [PubMed]
- 77.Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology 2006;130:1153–68. [DOI] [PubMed]
- 78.Kuang SY, Jackson PE, Wang JB, Lu PX, Munoz A, Qian GS, et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA 2004;101:3575–80. [DOI] [PMC free article] [PubMed]
- 79.Liu CJ, Kao JH, Lai MY, Chen PJ, Chen DS. Precore/core promoter mutations and genotypes of hepatitis B virus in chronic hepatitis B patients with fulminant or subfulminant hepatitis. J Med Virol 2004;72:545–50. [DOI] [PubMed]
- 80.Chu CM, Yeh CT, Chiu CT, Sheen IS, Liaw YF. Precore mutant of hepatitis B virus prevails in acute and chronic infections in an area in which hepatitis B is endemic. J Clin Microbiol 1996;34:1815–8. [DOI] [PMC free article] [PubMed]
- 81.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 2003;124:327–34. [DOI] [PubMed]
- 82.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Precore/basal core promoter mutants and hepatitis B viral DNA levels as predictors for liver deaths and hepatocellular carcinoma. World J Gastroenterol 2006;12:6620–6. [DOI] [PMC free article] [PubMed]
- 83.Sakamoto T, Tanaka Y, Orito E, Co J, Clavio J, Sugauchi F, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines. J Gen Virol 2006;87:1873–82. [DOI] [PubMed]
- 84.Gerken G, Kremsdorf D, Capel F, Petit MA, Dauguet C, Manns MP, et al. Hepatitis B defective virus with rearrangements in the preS gene during chronic HBV infection. Virology 1991;183:555–65. [DOI] [PubMed]
- 85.Fernholz D, Stemler M, Brunetto M, Bonino F, Will H. Replicating and virion secreting hepatitis B mutant virus unable to produce preS2 protein. J Hepatol 1991;13:S102–4. [DOI] [PubMed]
- 86.Xu Z, Yen TSB. Intracellular retension of surface protein by a hepatitis B virus mutant that releases virion particles. J Virol 1996;70:133–40. [DOI] [PMC free article] [PubMed]
- 87.Fan YF, Lu CC, Chen WC, Yao WJ, Wang HC, Chang TT, et al. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology 2001;33:277–86. [DOI] [PubMed]
- 88.Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, et al. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol 2003;70:537–44. [DOI] [PubMed]
- 89.Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 2000;33:998–1002. [DOI] [PubMed]
- 90.Yuen MF, Tanaka Y, Shinkai N, Poon RT, But DY, Fong DY, et al. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut 2007 (in press). [DOI] [PubMed]
- 91.Shinkai N, Tanaka Y, Ito K, Mukaide M, Hasegawa I, Asahina Y, et al. Influence of hepatitis B virus X, core promoter mutations on hepatocellular carcinoma among patients with subgenotype C2. J Clin Microbiol 2007 (in press). [DOI] [PMC free article] [PubMed]
- 92.Chen CJ, Yang HI, Iloeje UH, Su J, Jen CL, You SL, et al. A risk function nomogram for predicting HCC in patients with chronic hepatitis B: the REVEAL-HBV study. EASL 2007 Abstract no. 475.
- 93.Liaw YF, Leung N, Guan R, Lau GK, Merican I, McCaughan G, et al. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2005 update. Liver Int 2005;25:472–89. [DOI] [PubMed]
- 94.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology 2007;45:507–39. [DOI] [PubMed]
- 95.Keeffe EB, Dieterich DT, Han SH, Jacobson IM, Martin P, Schiff ER, et al. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States: an update. Clin Gastroenterol Hepatol 2006;4:936–62. [DOI] [PubMed]
- 96.Kao JH. Appropriate use of interferon for treatment of chronic hepatitis B. Hepatol Res 2007;37 Suppl 1:S47–54. [DOI] [PubMed]
- 97.Perrillo RP. Therapy of hepatitis B—viral suppression or eradication? Hepatology 2006;43 2 Suppl 1:S182–93. [DOI] [PubMed]
- 98.Cooksley G, Lau GKK, Liaw Y-F, Marcellin P, Chow WC, Thongsawat S, et al. Effect of genotype and other baseline factors on response to PEGInterferon-2a (40 kDa) (Pegasys®) in HBeAg-positive chronic hepatitis B: results from a large, randomized study [Abstract]. J Hepatol 2005;42 Suppl 2:S30.
- 99.Bonino F, Marcellin P, Lau GK, Hadziyannis S, Jin R, Piratvisuth T, et al. Predicting response to peginterferon alpha-2a, lamivudine and the two combined for HBeAg-negative chronic hepatitis B. Gut 2007;56:699–705. [DOI] [PMC free article] [PubMed]
- 100.Farci P, Marcellin P, Lu Z-M, Diago M, Lai M-Y, Gurel S, et al. On-treatment predictors of sustained biochemical and virological response in patients with HBeAg-negative chronic hepatitis B (CHB) treated with PEGInterferon-2a (40 kDa) (Pegasys®) [abstract]. J Hepatol 2005;42 Suppl 2:S175.
- 101.Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis 2003;36:687–96. [DOI] [PubMed]
- 102.Yuen MF, Sablon E, Hui CK, Yuan HJ, Decraemer H, Lai CL. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology 2001;34:785–91. [DOI] [PubMed]
- 103.Ruiz-Sancho A, Sheldon J, Soriano V. Telbivudine: a new option for the treatment of chronic hepatitis B. Expert Opin Biol Ther 2007;7:751–61. [DOI] [PubMed]
- 104.Liu CJ, Kao JH. Hepatitis B genotype: what should the clinician know? Curr Hepat Rep 2007;6:17–23.
- 105.Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 2000;33:998–1002. [DOI] [PubMed]
- 106.Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology 2002;36:1425–30. [DOI] [PubMed]
- 107.Hou J, Schilling R, Janssen HLA. Molecular characteristics of hepatitis B virus genotype A confer a higher response to interferon treatment. J Hepatol 2001;34 Suppl 1:15.
- 108.Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, et al. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut 2005;54:1009–13. [DOI] [PMC free article] [PubMed]
- 109.Liu CJ, Lai MY, Chao YC, Liao LY, Yang SS, Hsiao TJ, et al. Interferon alpha-2b with and without ribavirin in the treatment of hepatitis B e antigen-positive chronic hepatitis B: a randomized study. Hepatology 2006;43:742–9. [DOI] [PubMed]
- 110.Cooksley WG, Piratvisuth T, Lee SD, Mahachai V, Chao YC, Tanwandee T, et al. Peginterferon alpha-2a (40 kDa): an advance in the treatment of hepatitis B e antigen-positive chronic hepatitis B. J Viral Hepat 2003;10:298–305. [DOI] [PubMed]
- 111.Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, et al., for the HBV 99-01 Study Group, Rotterdam foundation for liver research. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 2005;365:123–9. [DOI] [PubMed]
- 112.Flink HJ, van Zonneveld M, Hansen BE, de Man RA, Schalm SW, Janssen HL, for the HBV 99-01 Study Group. Treatment with Peg-interferon alpha-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am J Gastroenterol 2006;101:297–303. [DOI] [PubMed]
- 113.Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, et al. Peginterferon alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med 2005;352:2682–95. [DOI] [PubMed]
- 114.Zhao H, Kurbanov F, Wan MB, Yin YK, Niu JQ, Hou JL, et al. Genotype B and younger patient age associated with better response to low-dose therapy: a trial with pegylated/nonpegylated interferon-alpha-2b for hepatitis B e antigen-positive patients with chronic hepatitis B in China. Clin Infect Dis 2007;44:541–8. [DOI] [PubMed]
- 115.Hou JL, Schilling R, Janssen HLA, Hansen BE, Heijtink R, Sablon E, et al. Genetic characteristics of hepatitis B virus genotypes as a factor for interferon-induced HBeAg clearance. J Med Virol 2007;79:1055–63. [DOI] [PubMed]
- 116.Lin CL, Kao JH. To genotype or not to genotype: toward an optimal tailoring of treatment of chronic hepatitis B. Clin Infect Dis 2007;44:1665–6. [DOI] [PubMed]
- 117.Liu CJ, Chen PJ, Lai MY, Chen TC, Wang HY, Huang WL, et al. Lack of interferon sensitivity-determining region in the genome of hepatitis B virus genotype Ba. Antivir Ther 2004;9:895–903. [PubMed]
- 118.Chien RN, Lin CY, Yeh CT, Liaw YF. Hepatitis B virus genotype B is associated with better response to thymosin alpha1 therapy than genotype C. J Viral Hepat 2006;13:845–50. [DOI] [PubMed]
- 119.Kao JH, Liu CJ, Chen DS. Hepatitis B viral genotypes and lamivudine resistance. J Hepatol 2002;36:303–4. [DOI] [PubMed]
- 120.Yuen MF, Wong DK, Sablon E, Yuan HJ, Sum SM, Hui CK, et al. Hepatitis B virus genotypes B and C do not affect the antiviral response to lamivudine. Antivir Ther 2003;8:531–4. [PubMed]
- 121.Chan HL, Wong ML, Hui AY, Chim AM, Tse AM, Hung LC, et al. Hepatitis B virus genotype has no impact on hepatitis B e antigen seroconversion after lamivudine treatment. World J Gastroenterol 2003;9:2695–7. [DOI] [PMC free article] [PubMed]
- 122.Buti M, Cotrina M, Valdes A, Jardi R, Rodriguez-Frias F, Esteban R. Is hepatitis B virus subtype testing useful in predicting virological response and resistance to lamivudine? J Hepatol 2002;36:445–6. [DOI] [PubMed]
- 123.Chien RN, Yeh CT, Tsai SL, Chu CM, Liaw YF. Determinants for sustained HBeAg response to lamivudine therapy. Hepatology 2003;38:1267–73. [DOI] [PubMed]
- 124.Zollner B, Petersen J, Puchhammer-Stockl E, Kletzmayr J, Sterneck M, Fischer L, et al. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology 2004;39:42–50. [DOI] [PubMed]
- 125.Pan XP, Li LJ, Du WB, Li MW, Cao HC, Sheng JF. Differences of YMDD mutational patterns, precore/core promoter mutations, serum HBV DNA levels in lamivudine-resistant hepatitis B genotypes B and C. J Viral Hepatitis 2007 (in press). [DOI] [PubMed]
- 126.Westland C, Delaney W IV, Yang H, Chen SS, Marcellin P, Hadziyannis S, et al. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil. Gastroenterology 2003;125:107–16. [DOI] [PubMed]
- 127.Lurie Y, Manns MP, Gish RG, Chang TT, Yurdaydin C, Lai CL, et al. The efficacy of entecavir is similar regardless of disease-related baseline subgroups in treatment of nucleoside-naive, HBeAg(+) and HBeAg(−) patients with chronic hepatitis B. J Hepatol 2005;42 Suppl 2:184.
- 128.Rivkin A. Entecavir: a new nucleoside analogue for the treatment of chronic hepatitis B. Drugs Today (Barc) 2007;43:201–20. [DOI] [PubMed]
- 129.Fung SK, Chae HB, Fontana RJ, Conjeevaram H, Marrero J, Oberhelman K, et al. Virologic response and resistance to adefovir in patients with chronic hepatitis B. J Hepatol 2006;44:283–90. [DOI] [PubMed]
- 130.Brunetto MR, Oliveri F, Colombatto P, Capalbo M, Barbera C, Bonino F. Treatment of chronic anti-HBe-positive hepatitis B with interferon-alpha. J Hepatol 1995;22:42–4. [PubMed]
- 131.Lin CL, Liao LY, Liu CJ, Chen PJ, Lai MY, Kao JH, et al. Hepatitis B genotypes and precore/basal core promoter mutants in HBeAg-negative chronic hepatitis B. J Gastroenterol 2002;37:283–7. [DOI] [PubMed]
- 132.Liu CJ, Chen PJ, Lai MY, Lin FY, Wang T, Kao JH, et al. Viral factors correlate with hepatitis B e antigen seroconversion in patients with chronic hepatitis B. Liver Int 2006;26:949–55. [DOI] [PubMed]
- 133.Lampertico P, Del Ninno E, Manzin A, Donato MF, Rumi MG, Lunghi G, et al. A randomized, controlled trial of a 24-month course of interferon alfa 2b in patients with chronic hepatitis B who had hepatitis B virus DNA without hepatitis B e antigen in serum. Hepatology 1997;26:1621–5. [DOI] [PubMed]
- 134.Erhardt A, Reineke U, Blondin D, Gerlich WH, Adams O, Heintges T, et al. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology 2000;31:716–25. [DOI] [PubMed]
- 135.Lok AS-F, Hussain M, Cursano C, MargottiI M, Gramenzi A, Grazi GL, et al. Evolution of hepatitis B virus polymerase gene mutations in hepatitis B e antigen negative patients receiving lamivudine therapy. Hepatology 2000;32:1145–53. [DOI] [PubMed]
- 136.Lin CL, Liao LY, Wang CS, Chen PJ, Lai MY, Chen DS, et al. Evolution of hepatitis B virus precore/basal core promoter gene in HBeAg-positive chronic hepatitis B patients receiving lamivudine therapy. Liver Int 2004;24:9–15. [DOI] [PubMed]
- 137.Thompson AJ, Ayres A, Yuen L, Bartholomeusz A, Bowden DS, Iser DM, et al. Lamivudine resistance in patients with chronic hepatitis B: role of clinical and virological factors. J Gastroenterol Hepatol 2007;22:1078–85. [DOI] [PubMed]
- 138.Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, et al. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 2007;46:254–65. [DOI] [PubMed]