Abstract
Living cells are a fascinating demonstration of nature’s most intricate and well-coordinated micromechanical objects. They crawl, spread, contract, and relax—thus performing a multitude of complex mechanical functions. Alternatively, they also respond to physical and chemical cues that lead to remodeling of the cytoskeleton. To understand this intricate coupling between mechanical properties, mechanical function and force-induced biochemical signaling requires tools that are capable of both controlling and manipulating the cell microenvironment and measuring the resulting mechanical response. In this review, the power of microfluidics as a functional tool for research in cell mechanics is highlighted. In particular, current literature is discussed to show that microfluidics powered by soft lithographic techniques offers the following capabilities that are of significance for understanding the mechanical behavior of cells: (i) Microfluidics enables the creation of in vitro models of physiological environments in which cell mechanics can be probed. (ii) Microfluidics is an excellent means to deliver physical cues that affect cell mechanics, such as cell shape, fluid flow, substrate topography, and stiffness. (iii) Microfluidics can also expose cells to chemical cues, such as growth factors and drugs, which alter their mechanical behavior. Moreover, these chemical cues can be delivered either at the whole cell or subcellular level. (iv) Microfluidic devices offer the possibility of measuring the intrinsic mechanical properties of cells in a high throughput fashion. (v) Finally, microfluidic methods provide exquisite control over drop size, generation, and manipulation. As a result, droplets are being increasingly used to control the physicochemical environment of cells and as biomimetic analogs of living cells. These powerful attributes of microfluidics should further stimulate novel means of investigating the link between physicochemical cues and the biomechanical response of cells. Insights from such studies will have implications in areas such as drug delivery, medicine, tissue engineering, and biomedical diagnostics.
INTRODUCTION
Mechanical processes like pushing, pulling, and squeezing play a remarkably significant role in the biological function of a cell.1, 2 For example, such forces turn out to affect biological processes such as cell growth, division, migration, and death. In addition, mechanical forces at the cellular level impact the function of biological structures ranging from large length scales such as tissues to small length scales such as genes. As a result it is not surprising that when living cells are affected by disease, changes in their mechanical properties also occur. Indeed cell mechanics has been implicated in a variety of diseases, such as cancer,3, 4, 5 malaria,6 sickle cell anemia,7 asthma, and glaucoma.8 For example, Cross et al.5 studied individual cells taken from the tissues of suspected patients with various cancers. From physiology it is suggested that metastatic cancer cells must be more deformable than healthy cells, to be able to invade tissue. Consistent with this notion, individual metastatic cancer cells were indeed found to have lower Young’s moduli measured via atomic force microscope. Also, in suspended state, cancerous and metastatic cells could be distinguished from healthy breast epithelial cells via their resistance to optical stretching.9 Thus there exists an intimate link between the mechanical properties and the diseased state of living cells.
From a mechanics perspective it is well known that intracellular forces are generated and supported via an intracellular framework called the cytoskeleton. This network, containing actin filaments, microtubules, and intermediate filaments, is dynamic in nature, and is regulated via polymerization and depolymerization rates, which in turn are controlled via molecular motors and ATP.10 This coupling between intracellular mechanics and chemistry appears to be universal.11, 12 It allows cells to respond mechanically to external stresses by activating biochemical signaling cascades and also underlies the possibility to restore the mechanical properties of diseased cells by pharmacological interventions. Thus cellular mechanotransduction studies that involve understanding and the control of interactions between mechanical forces and biochemical signaling pathways could lead to novel therapeutic strategies to treat diseases.
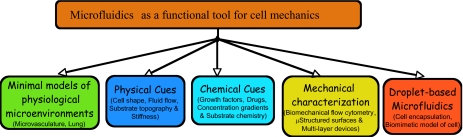
Fundamentally, understanding cell mechanics is a complex problem. This is because cells are not only micromechanical objects that support and generate mechanical forces, but they are also capable of sensing their microenvironment and adapting to it. To understand this intricate coupling requires tools that are capable of both manipulating the cellular microenvironment in a controlled manner and also quantifying the resulting mechanical response. We believe this is precisely where tools such as microfluidics and microfabrication present an immense opportunity for studies in cell mechanics. As summarized in Fig. 1, these tools are not only capable of creating and manipulating the physicochemical environment of cells but also measuring the intrinsic mechanical properties of cells. The goal of this review is to highlight this power of microfluidics and its significance for fundamental and applied studies of cell mechanics. The importance of other micro- and nanotechnological tools for investigating cell mechanics has been recently reviewed by Le Duc and co-workers.13 Recent advances in the basic understanding of cytoskeleton remodeling14 and the broad suite of techniques2, 15 available for characterizing the mechanical properties of the cytoskeleton have been reported elsewhere. In addition, soft lithographic procedures to fabricate microfluidic channels, structured elastomeric substrates, and multilayer devices have been reviewed elsewhere.16, 17
Figure 1.
Various attributes of microfluidics that are useful for fundamental and applied investigations in cell mechanics.
MICROFLUIDIC DEVICES MIMIC IN VIVO CELLULAR MICROENVIRONMENT
The last decade has witnessed revolutionary advances in the manipulation of fluids at small scales.18 This growth has been driven by the potential that microfluidic devices can perform fast, reliable, and high throughput biochemical analysis using only minute quantities of samples and reagents. In addition, microfluidics has become such a powerful tool that its application has been extended from analyzing simple systems such as proteins and DNA to more complicated biological systems such as living cells. This concept of cells on chips19 is appealing from a cell mechanics point of view, because microfluidic systems can be built that mimic the in vivo environment of cells, allowing mechanical behavior of cells to be studied under almost physiological conditions, with minimal consumption of reagents.
One general consideration that needs to be assessed during cell culture in chips is the degree of cell viability in a given microfabricated device.20 Apoptosis assays, especially on adherent cells, are employed as a sensitive test to ensure that the microfluidic device and the culture conditions used do not negatively influence the cellular behavior.21 Several studies22, 23 have shown normal cell viability in polydimethylsiloxane (PDMS) chips for as long as a few weeks. Besides PDMS, cell culture has been performed in devices made with other materials such as silicon dioxide and glass. Even in these devices, good cell viability has been demonstrated.21 However, devices fabricated with different materials require different strategies to ensure delivery of gases (oxygen and carbon dioxide). In glass chips, this has been achieved by gas dissolution in the liquid, whereas in PDMS devices, gas permeation through the walls suffices. In one study,24 the thickness of the PDMS was varied between 100 microns and several millimeters, without noticeable effects on cell viability. Also, nongaseous nutrients and secretion products need to be transported to∕away from the cell. Since diffusion is slow, liquid flow is often needed to ensure this.24, 25 However, careful design of the channel geometry is needed to ensure undesirable shear-induced activation of gene expression. The amount of shear stress that can be endured without significant changes in cell-state depends on the cell type.26, 27 While this implies that different (maximum) shear stresses apply for different types of cells, it should be noted that the chip design is generally flexible—changing the channel dimensions is a straightforward way to change the flow stress. Apart from flow fields, cell viability under the application of external fields that are often incorporated into microfluidic devices for cell manipulation has also been addressed. For example, the sorting of cells using dielectrophoresis28 or optical forces29 does not appear to perturb cell physiology appreciably. Under well-controlled conditions, also ultrasound has been applied to cells inside microfluidic chips, without significant loss of viability.30
Given that many studies have shown good viability of living cells in microfluidic devices, there is ample scope to use microtechnology to design and mimic in vivo cellular microenvironments. An excellent case, for example, is the recent growth in using microfluidics to develop minimal models of the microvasculature. A number of studies31, 32, 33, 34 have begun to use microfluidic devices, instead of parallel plate chambers, to grow endothelial cells and expose them to shear. These microfluidic channels of 10–100 μm in size are better mimics of the capillaries in the microvasculature because of the matching of the length scales of channels and cells. Such microfluidic channels have also been integrated with membrane valves and pumps to achieve an automated in vitro microvascular system for long term culturing and shearing of endothelial cells.34 In addition, shear stresses of 1–10 Pa are quite easily achieved in microfluidic devices mimicking the hemodynamic shear stresses experienced by endothelial cells. Apart from using microfluidic devices for microvasculature studies, a recent development has been the application of two-phase microfluidic systems to mimic the physiology and pathophysiology associated with pulmonary airways. Huh et al.35 were able to demonstrate mechanical injury of airway epithelial cells due to rupture of liquid plugs in microfluidic channels.
Microfluidic devices are also being increasingly used as in vitro model systems for investigating the role of blood cell mechanics in hematological diseases.36 Shelby et al.37 used narrow microfluidic channels to investigate the deformability of red blood cells infected with the malarial parasite Plasmodium falciparum. Healthy red blood cells were able to pass through these narrow constrictions readily, compared to infected cells, indicating that infected cells were mechanically more rigid than the healthy cells. Higgins et al.38 studied sickle cell vasoocclusion in a microfluidic device. The microfluidic format enabled them to vary a number of parameters (geometric confinement, flow rates, oxygen concentration) and identify the conditions for the onset of vasoocclusion. More recently, Fletcher and co-workers39 used a microfluidic device consisting of successively bifurcating channels to characterize the single-cell transit times of red blood cells and neutrophils. The transit time distributions of the cells infected with sepsis and leukostasis were found to be significantly different from that of the corresponding healthy cells.
MICROFLUIDICS AS A MEANS TO DELIVER PHYSICAL CUES TO CELLS
Microfluidics and microfabrication are versatile means of manipulating physical cues that affect cell mechanical behavior such as fluid flow, substrate topography and stiffness, and cell shape. For example, cells can be easily exposed to laminar shear and extentional flows in microfluidic devices.40 More complex flows including pulsatile17, 41 and chaotic flows42, 43 can also be generated in microfluidic devices. The capability of producing such a broad range of flow types in microchannels opens up new possibilities for studying flow-induced cytoskeleton remodeling,11 particularly for cells that are sensitive to fluid forces such as endothelial cells.
Soft lithographic techniques allow varying the topography of substrate with feature size ranging from microns to nanometers.44 Remarkably, cells can sense substrate topography as well as stiffness.45, 46 A recent study has shown that the elastic modulus of cells grown on softer substrates is lower than those grown on harder substrates and that cells tend to tune their stiffness to the stiffness of the substrate.47 Cancer cells have also been identified via their growth on soft agar gels.4 In certain stem cells it has been demonstrated that their development into highly specialized cells can be steered via the stiffness of the substrate.48 As a result, microfluidic devices should find increasing application in such studies, as the substrate stiffness of these devices can be easily tuned. For example, the stiffness of substrates fabricated using soft lithography can be varied from ∼1 kPa to 100 kPa.45, 49 Microfluidic strategies also allow cells to be grown in 3D microstructures that could be used to study anchorage dependent cells.33, 50, 51
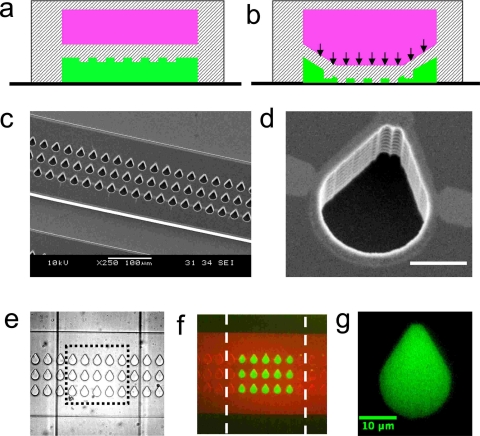
Apart from the use of microfluidics to control the physical microenvironment of the cell, it also allows control over the shape of individual cells. Patterning techniques such as microcontact printing52 are well equipped to control the shape of single cells, including introducing anisotropy in shape. The basic principle exploits the affinity of adherent cells to regions where the extracellular matrix is present. Although patterning techniques work well for cells on open substrates, coupling them to microfluidic channels requires additional steps including cumbersome alignment. The advantage of coupling microfluidic networks and patterning techniques is that the channels provide a conduit for delivery of either physical or chemical cues in a precise manner. Such functionality could be useful in endothelial cell mechanics to address the link between cell morphology and flow-induced cytoskeleton remodeling. In this regard, we recently developed a multi-layer device that has the capability to pattern cells inside microchannels53 (see Fig. 2). These devices were fabricated in a manner similar to membrane valves, except that topographical structures were introduced on the roof of the fluid channel. The working principle of this device is based on using the deformation of the structured elastomeric membrane (SEM) to mask a certain area on the microchannel surface and passivate the unmasked area with a blocking agent. Subsequently extracellular matrix proteins can be introduced that preferentially adsorb in the previously protected area. Thus, depending on the shape of the feature on the SEM, adhesive islands of various shapes can be obtained. Figure 3 shows that this technique can indeed provide good control over the shape of a cell on a microchannel surface.
Figure 2.
A multilayer microfluidic device capable of patterning cells inside microchannels (a) Cross-sectional view of the device showing the fluid channel in green and control channel in pink. Sandwiched in between the two channels is the structured elastomeric membrane. In the top view (not shown) the fluid and control channels are oriented perpendicular to each other. (b) Actuation of the control channel by pressure deforms the membrane, allowing it to touch the floor. (c) Scanning electron micrograph of the mold showing the fluid channel with recessed features. (d) High resolution scanning electron micrograph of the feature that has a tear drop shape. Scale bar corresponds to 10 μm. (e) Optical micrograph of the PDMS-based multilayer device. The fluid channel is oriented horizontally and the control channel is oriented vertically. Both the channels are 200 μm in width. The features are also shown. (f) Selective patterning of proteins on the microchannel surface. Fluorescently labeled bovine serum albumin protein and immunostained fibronectin are shown in red and green, respectively. (g) High resolution image of fibronectin patterned on the microchannel surface. Additional details regarding the fabrication of devices and experimental procedures on protein patterning are reported in Ref. 53.
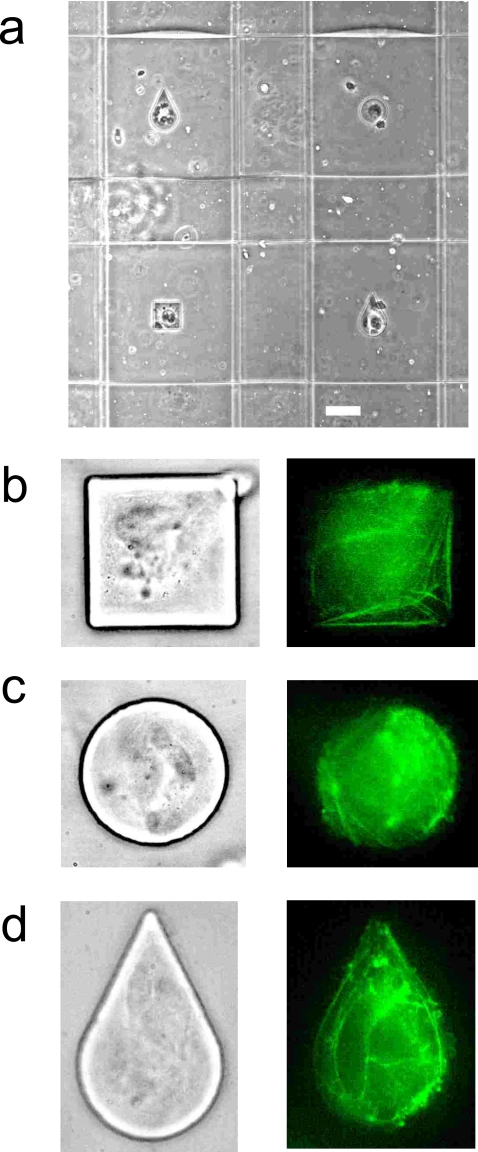
Figure 3.
Patterning of endothelial cells on microchannel surfaces by the multilayered device with structured elastomeric membrane. (a) Phase contrast image showing the device. Scale bar is 50 μm. [(b), (c), and (d)] High-resolution phase contrast (left) and fluorescent (right) images of the endothelial cells showing high fidelity shape control. Fluorescence is due to staining of actin network with FITC-phalloidin. In some cases multiple cells adhered on the same fibronectin patterned island.
MICROFLUIDICS AS A MEANS TO EXPOSE CELLS TO CHEMICAL CUES
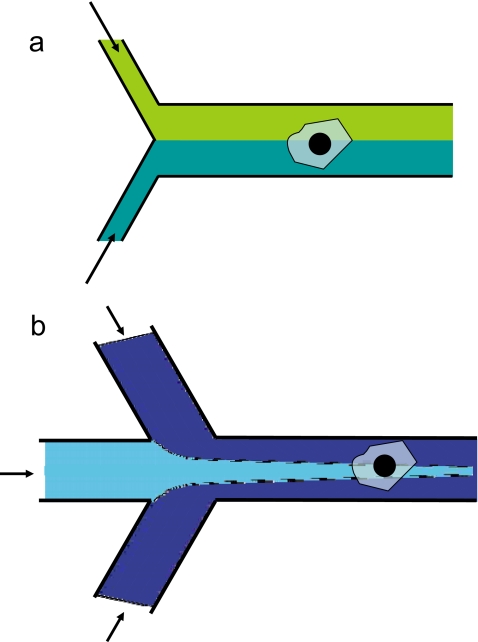
Microfluidics is capable of delivering chemical cues such as growth factors and drugs to living cells that in turn affect their mechanical behavior. The laminar nature of microfluidic flows provides unique possibilities for stimulating cells chemically. Coflowing streams can be used to deliver chemical cues locally to individual cells [see Fig. 4a]. Takayama et al.54 were the first to apply this principle and demonstrate selective labeling of mitochondria with different fluorescent dyes in endothelial cells. The laminar interface in such flows can also be controlled with a high degree of spatiotemoral resolution.55 Local chemical stimuli have also been delivered using microfluidic flow-focusing as shown in Fig. 4b.56 Gradients in concentration of chemical cues including extracellular matrix proteins on a substrate can also be easily set up using microfluidic laminar flows. Motivated by studies on chemotaxis, several groups have reported a variety of microfluidic networks that are capable of generating simple to complex chemical gradients.57
Figure 4.
Microfluidic delivery of chemical cues locally to single cells using (a) co-flowing laminar flows and (b) flow-focusing streams. The arrows show direction of fluid flow.
MICROFLUIDIC TOOLS TO CHARACTERIZE THE MECHANICAL PROPERTIES OF CELLS
So far we have discussed how microfluidics allows regulating the physicochemical environment of cells. We now discuss microfluidic strategies that have been developed to characterize cellular mechanical properties. In particular, biomechanical flow cytometry is well suited for analyzing suspended cells and microstructured elastomeric surfaces and multi-layer devices are suitable for characterizing the mechanical behavior of adherent cells. Below we discuss these tools in detail.
Biomechanical flow cytometry: Recently, several groups9, 39, 58, 59, 60 have demonstrated microfluidic strategies to screen suspended cells mechanically in microchannels. Akin to conventional flow cytometry where cells are selectively detected based on fluorescence, these recent methods detect cells mechanically in microfluidic devices by exploiting characteristics that are linked to their individual mechanical properties. Such approaches therefore fall under the category of biomechanical flow cytometry. (Recently Fletcher and co-workers collectively referred to these approaches as biophysical flow cytometry; however, we suggest it is more appropriate to categorize these approaches as biomechanical flow cytometry since they are based on cell mechanics.) Typically mechanical property measurements obtained from biomechanical flow cytometry are qualitative compared to more precise techniques such as atomic force microscopy and micropipette aspiration. However, their advantage lies in throughput (see Table 1 for throughput reported by various approaches) compared to these precise techniques, which analyze a few cells per day. Hence biomechanical flow cytometric approaches are suitable for rapid biomechanical diagnosis of diseased state of cells.
Table 1.
A summary of the throughput generated by current microfluidic-based biomechanical flow cytometric devices.
In one of its simplest manifestation, biomechanical flow cytometry involves pumping suspended cells through narrow microfluidic channels and subsequently recording the changes in cell size due to shear-induced deformation.61 Alternative approaches involve measuring the transit time for single cells to pass through a narrow channel. The transit times can be recorded using optoelectronic62 or resistive pulse detection63 methods. Instead of using a single pore, recently, Fletcher and co-workers39 performed many parallel measurements using a microfluidic binary tree network that consists of successive bifurcations of a large channel into smaller channels. In this study, cell size and transit times were measured using automated image processing methods. This allowed them to delineate the dependence of transit times on cell size and cell deformability and thus establish a link between transit time distributions, cell deformability, and their diseased state.
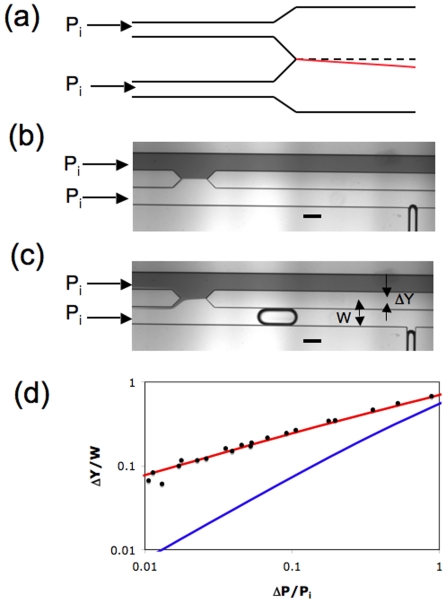
Instead of measuring transit time due to cell passage through a pore, Abkarian et al.58 reported a novel technique called microfluidic manometer to measure the excess pressure drop due to passage of a cell in a confined channel. The microfluidic manometer is a fluidic analog of a voltage comparator.64 It typically consists of two identical channels—the reference and test channels—that are connected downstream to form the comparator region (see Fig. 5). To measure the excess pressure drop due to the confined cell, fluid flow is generated in both the channels with the cell being present only in the test channel. When equal driving pressures are imposed at the channel inlets, any increase in the hydrodynamic resistance due to the presence of the cell in the test channel yields a corresponding displacement of the fluid-fluid interface in the comparator region. In practice, prior to the introduction of the cell into the test channel, a calibration curve is generated between known excess pressure drop (ΔP) and measured interface displacement (ΔY). This relation together with the measured interface displacement directly yields the excess pressure drop induced by the traversing cell. Using this approach Abkarian et al.58 were able to demonstrate pressure drop variation due to drug-induced changes in the cytoskeleton mechanical properties.
Figure 5.
(a) Schematic diagram of the comparator as reported by Abkarian et al. (Ref. 58). The top and bottom channels are reference and test channels, respectively. When equal input pressures (Pi) are applied, the fluid-fluid interface position lies on the dashed symmetry line. However, when a cell is passing through the test channel, the interface moves and is shown by the red line. (b) The modified comparator design as reported by Vanapalli et al. (Ref. 65). Due to the application of equal input pressures, the comparator is balanced. Scale bar is 200 μm. (c) The interface gets displaced due to hydrodynamic resistance of a single confined moving droplet. Scale bar is 200 μm. (d) For the same excess pressure drop, our comparator design (red line) yields higher interface displacement compared to the design of Abkarian et al. (Ref. 58) (blue line). The lines are derived from numerical calculations and the symbols are experimentally measured data points in which known ΔP was imposed and ΔY was measured. Additional details regarding the comparator design and microfluidic drop experiments are reported in Refs. 65, 66.
A key determinant of the performance of the manometer for measuring cellular-scale hydrodynamics is the calibration curve as it determines the sensitivity of the measurement. In Abkarian et al.58 the interface displacement was measured from the symmetry line of the comparator and also their geometry involved a long comparator section that could smear the interface due to diffusive mixing downstream. We recently modified this previous comparator design by introducing a split in the downstream section of the comparator as shown in Fig. 5b. The interface displacement due to a soft object (droplet) was measured from the vertex of the angular split as shown in Fig. 5c. Figure 5d shows interface displacement as a function of excess pressure drop, obtained via numerical calculations of the flow field in the comparator. Also shown is the curve for the comparator geometry used by Abkarian et al.58 The results demonstrate that for the smallest excess pressure drops, the interface displacement is about an order of magnitude higher in our comparator geometry compared to that of Abkarian et al. We also investigated the effect of the aspect ratio (width∕height) of the geometry and we found that the sensitivity increases with increase in the aspect ratio of the channel.65 We recently demonstrated the applicability of this improved comparator design by measuring the excess pressure drop due to the flow of a single confined droplet in a microchannel.66
Apart from microfluidic devices that use shear flows to assess cellular mechanical properties, other field-based approaches have also been developed to determine cellular mechanical properties. In one technique, called the optical stretcher, optical forces are used to induce surface stresses on the cell and subsequently determine the cell deformability.67 This approach has been successfully coupled to a microfluidic format to screen healthy and metastasized cancer cells.9 Recently, Bao et al.59 reported electroporative-induced cell swelling in a microfluidic device as a marker for cellular biomechanical properties. Using this approach, these authors were able to demonstrate that cancer cells showed higher degree of swelling compared to healthy cells.
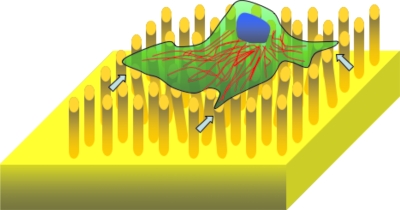
Microstructured elastomeric surfaces: So far we have discussed microfluidic devices capable of screening suspended cells. However, the majority of the cells in the human body are adherent cells that prefer to stick to substrates. The process of cell adhesion to a substrate involves binding to the extracellular matrix, spreading, and subsequent generation of traction forces that are mediated by the intracellular contractile apparatus. Thus, there is significant generation of mechanical forces during cell adhesion. To characterize these traction forces microfabricated elastomeric substrates that typically have microtopography in the form of pillars or posts have been developed.45, 68, 69 The basic principle for extracting the traction forces relies on measuring the deflection of the posts, when a cell exerts forces on the substrate (see Fig. 6). In the linear regime, the deflection of the post (δ) is proportional to the exerted force, F=3EIδ∕L3, where E is the Young’s modulus of the elastomer (typically PDMS), I is the moment of inertia of the (usually cylinder shaped) post, and L is the length of the post. Thus, the structured elastomeric substrates provide a direct measure of traction forces with high spatial and temporal resolution. Recently Chen and co-workers70 introduced elastomeric substrates with magnetic posts to actively manipulate cellular traction forces locally.
Figure 6.
Schematic of a microstructured elastomeric surface to measure cellular traction forces. The arrows show some deformed pillars.
Using this approach, Balaban et al.45 and Tan et al.69 showed that cells exert a constant stress (of ∼5 nN∕μm2) at focal adhesions irrespective of the area and the magnitude of force corresponding to each focal adhesion site. By controlling the morphology of the cell and therefore the spread area, Tan et al.69 further showed that the average force exerted on each post is proportional to the area of cell spreading. Compared to these two studies, du Roure et al.68 employed a higher density of elastomeric pillars and were able to demonstrate that the adhesion forces at the edge of a monolayer are higher than those measured for a single cell. In most of these studies, elastomeric substrates with pillars that are typically 2–3 μm in diameter, 6–10 μm in length, and spaced 4–9 μm apart have been used. The availability of microstructured elastomeric substrates capable of both measuring and manipulating cellular adhesion forces should open up new possibilities to understand the link between external and internal forces that regulate cell mechanics.
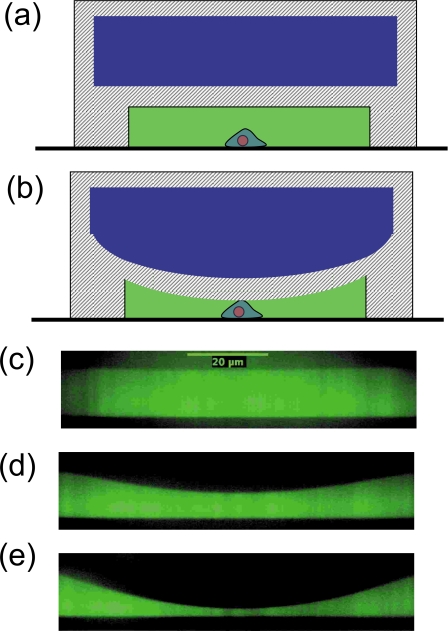
Multi-layer devices: Microfluidic devices with deformable membranes have also been recently demonstrated to be capable of measuring cellular mechanical properties. These multi-layer devices made using soft lithography typically consist of an elastomeric membrane that is sandwiched between two fluid channels that are oriented orthogonal to each other.17 The basic principle underlying the measurement of cellular mechanical properties using this technique is illustrated in Fig. 7. In the approach devised by Solomon and co-workers,49 stress is imposed on the cell by pressurizing the control channel which deforms the membrane. The resulting strain in the membrane is measured by visualizing the membrane deflection using confocal microscopy. Structural mechanics calculations based on finite element methods are then used to extract the relative contributions of the membrane and the cell to the measured deformation. Using this approach, the authors were able to measure the Young’s modulus of bacterial biofilms to be ∼1 kPa. Additional design calculations revealed that this technique is capable of characterizing soft objects with elastic moduli in the range of 102–105 Pa. Since the elastic modulus of mammalian cells is typically reported to be ∼1 kPa, this technique is potentially useful for high throughput mechanical characterization of mammalian cells as well. In an alternative approach, Kim et al.71 used much stronger PDMS membranes compared to Solomon and co-workers to essentially compress the cell and measure the resulting deformation (or bulge). Such cell deformability measurements were shown to be a biomechanical marker to distinguish breast cancer cells and healthy cells.
Figure 7.
[(a) and (b)] Schematic of the principle of operation of the multilayer microfluidic device for cell mechanical characterization. The control channel is pressurized to impose deformation on the cell. [(c), (d), and (e)] Confocal cross-sectional images of the fluid channel showing the deformation of the membrane with increasing control channel pressure (0, 0.2, and 0.4 bar). The width of the fluid and control channels are 120 and 100 μm, respectively. The depth of both channels is 20 μm and the thickness of the membrane is 11 μm. Buffer solution containing fluorescein was introduced into the fluid channel for visualization by confocal microscopy.
DROPLET BASED MICROFLUIDICS
Besides the continuous flow methods discussed so far, also droplet-based microfluidic systems72, 73 can be used for creating and manipulating physicochemical environments for cells inside microfluidic chips. Having individual cells inside microscopic aqueous droplets (surrounded by air or oil phase) offers additional possibilities for diagnosis or treatment. This could, for example, be the controlled addition of a drug, or a mechanical test. One distinct advantage of having a droplet environment is that the volume, and with it the exchange of material, can be kept under control. For example, drugs can be kept inside as long as needed for incubation. Additional control over intradroplet concentrations is possible by letting a cell-droplet coalesce with a cell-free droplet having well-defined amounts of chemicals (nutrients, drugs) and volume. Using relatively large droplets enables transport of cells without significant stress due to the flow of its surrounding liquid. Just as in continuous microfluidic systems, cell viability in aqueous droplets is an important consideration. Judicious choice of oil phase and surfactants is required in order to ensure cell viability. Fluorocarbon oils combined with fluorosurfactants have been shown to exhibit good biocompatibility and also induce less swelling of PDMS-based microfluidic devices.74
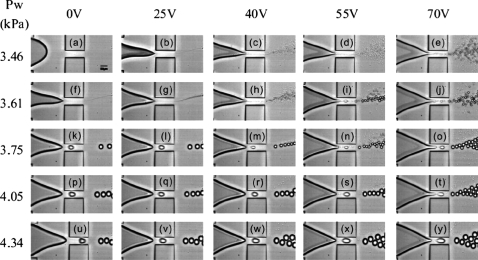
These possibilities have recently come within reach, due to tremendous developments in both droplet generation and manipulation.72, 75, 76 Droplet generation in miniaturized devices has been typically achieved using either passive (T-junction and flow-focusing geometries) or active (electric, electrowetting, and acoustic) methods. There exists plenty of scope to combine both passive and active methods to achieve enhanced flexibility and finer control over drop generation and size. In this context, we recently developed an electrowetting-based microfluidic flow-focusing device77, 78 that offers continuous and much finer digital control over drop size than purely hydrodynamics-based devices (see Fig. 8).
Figure 8.
Images showing droplet generation from an electrowetting-controlled microfluidic flow-focusing device. The continuous phase is driven at a constant flow rate (capillary number=0.072) by a syringe pump and the pressure (Pw) at the aqueous inlet is adjusted using a hydrostatic head. The voltage and Pw have been varied to generate this diagram. Reproduced from Ref. 77.
With respect to single cell encapsulation in droplets, different methods have seen the light in the past few years, with impressive improvements in reliability, high throughput, and downsizing.79 In principle, each of these methods could be combined with a downstream microfluidics module for further cell processing. For applications in biomedical diagnostics that favor portability, digital microfluidic platforms driven by electrowetting76 are well suited. This could involve an array of individually addressable electrodes with a size and pitch tweaked to the size of the droplets. Electrowetting could then be used to achieve merging and mixing (with nutrient or reagent), transport, and holding (at a detector site or an on-chip reservoir) of each individual cell-droplet. The first implementation of digital microfluidics for cell-based assays has been recently realized,80 but considering its potential, it is expected that many will follow. Moreover, with electrowetting based manipulation of cell-laden droplets, unwanted interactions of cells and electric fields are negligible, since there is practically no field inside the droplet at low frequencies of the voltage.
Another interesting application of droplets is as in vitro models for living cells. Confined fluid volumes have been used to interrogate enzyme kinetics, mimicking the confined microenvironment of cells.81 Also confined cytoskeletal networks have been generated using droplets demonstrating the important role that confinement plays on the mechanics of cytoskeletal networks.82 Thus, significant potential exists for using droplets to unravel the biochemical and biophysical machinery driving the mechanical behavior of living cells.
CONCLUSIONS
Living cells are complex and dynamic systems. Understanding the coupling between the sheer biological complexity and the cellular mechanical behavior requires tools capable of probing and manipulating living cells. In this review, we have highlighted the importance of microfluidics as an enabling tool for fundamental investigations in cell mechanical behavior. While the current studies primarily make use of relatively simple microfluidic channels, the design flexibility of microfluidics and the degree of control of both chemical and physical parameters will allow for analyzing the cells’ response in unparalleled detail under conditions that will reflect more and more aspects of the complexity of life. Physical models—theoretical and numerical—will have to be developed to model the cell mechanical behavior and to interpret quantitatively the information that becomes available from such experiments. For example, there is a need for quantitative modeling to extract membrane and cytoskeleton mechanical properties from micofluidic manometer measurements. In turn, numerical models can be used to optimize the design of dedicated microfluidic devices for extracting specific properties. Given the easy optical access, the full spectrum of optical-microscopy-based techniques for analyzing intracellular processes such as signaling pathways is readily combined with microfluidics and thereby provides access to the regulatory network behind the cell’s mechanical response. Simultaneously efforts need to be pursued to integrate established rheometric methods such as atomic force microscopy, particle-tracking microrheology, and magnetic twisting cytometry into microfluidic devices in order to extract more specific mechanical information. Insights from such combined studies may lead to therapeutic strategies for the control and prevention of diseases and the development of miniaturized devices for biomedical diagnostics.
ACKNOWLEDGMENTS
We acknowledge the support from the Cell Stress Program at the University of Twente. We are grateful to Dirk van den Ende for performing the numerical calculations reported in Fig. 5 and Michael J. Solomon from the University of Michigan for providing the preprint on the multilayer microfluidic device for mechanical characterization of soft matter. S. A. V. thanks the Texas Tech University start-up funds for supporting this research.
References
- Zhu C., Bao G., and Wang N., Annu. Rev. Biomed. Eng. 10.1146/annurev.bioeng.2.1.189 2, 189 (2000); [DOI] [PubMed] [Google Scholar]; Bao G. and Suresh S., Nature Mater. 10.1038/nmat1001 2, 715 (2003). [DOI] [PubMed] [Google Scholar]
- Kasas S. and Dietler G., Pfluegers Arch. Eur. J. Physiol. 10.1007/s00424-008-0448-y 456, 13 (2008). [DOI] [PubMed] [Google Scholar]
- Suresh S., Acta Biomater. 3, 413 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E., Janmey E. P., and Wang Y. L., Science 10.1126/science.1116995 310, 1139 (2005). [DOI] [PubMed] [Google Scholar]
- Cross S. E., Jin Y. S., Rao J., and Gimzewski J. K., Nat. Nanotechnol. 10.1038/nnano.2007.388 2, 780 (2007). [DOI] [PubMed] [Google Scholar]
- Lee G. Y. H. and Lim C. T., Trends Biotechnol. 25, 111 (2007). [DOI] [PubMed] [Google Scholar]
- Nash G. B., Johnson C. S., and Meiselman H. J., Blood 63, 73 (1984). [PubMed] [Google Scholar]
- Ingber D. E., Ann. Med. 35, 564 (2003). [DOI] [PubMed] [Google Scholar]
- Guck J., Schinkinger S., Lincoln B., Wottawah F., Ebert S., Romeyke M., Lenz D., Erickson H. M., Ananthakrishnan R., Mitchell D., Kas J., Ulvick S., and Bilby C., Biophys. J. 10.1529/biophysj.104.045476 88, 3689 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. and Borisy G. G., Cell 10.1016/S0092-8674(03)00120-X 112, 453 (2003); [DOI] [PubMed] [Google Scholar]; Bausch A. R. and Kroy K., Nat. Phys. 2, 231 (2006). [Google Scholar]
- Davies P. F., Physiol. Rev. 75, 519 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A. S., Annu. Rev. Physiol. 54, 135 (1992); [DOI] [PubMed] [Google Scholar]; Gillespie P. G. and Walker R. G., Nature (London) 10.1038/35093011 413, 194 (2001); [DOI] [PubMed] [Google Scholar]; Hamill O. P. and Martinac B., Physiol. Rev. 81, 685 (2001); [DOI] [PubMed] [Google Scholar]; Watson P. A., FASEB J. 5, 2013 (1991); [DOI] [PubMed] [Google Scholar]; Wang N., Butler J. P., and Ingber D. E., Science 10.1126/science.7684161 260, 1124 (1993). [DOI] [PubMed] [Google Scholar]
- Kumar S. and LeDuc P. R., Exp. Mech. 10.1007/s11340-007-9063-7 (2007); [DOI]; LeDuc P. R. and Bellin R. M., Ann. Biomed. Eng. 10.1007/s10439-005-9008-1 34, 102 (2006). [DOI] [PubMed] [Google Scholar]
- Bursac P., Lenormand G., Fabry B., Oliver M., Weitz D. A., Viasnoff V., Butler J. P., and Fredberg J. J., Nature Mater. 10.1038/nmat1404 4, 557 (2005); [DOI] [PubMed] [Google Scholar]; Hoffman B. D., Massiera G., Van Citters K. M., and Crocker J. C., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0510348103 103, 10259 (2006); [DOI] [PMC free article] [PubMed] [Google Scholar]; Kasza K. E., Rowat A. C., Liu J. Y., Angelini T. E., Brangwynne C. P., Koenderink G. H., and Weitz D. A., Curr. Opin. Cell Biol. 10.1016/j.ceb.2006.12.002 19, 101 (2007); [DOI] [PubMed] [Google Scholar]; Trepat X., Deng L. H., An S. S., Navajas D., Tschumperlin D. J., Gerthoffer W. T., Butler J. P., and Fredberg J. J., Nature (London) 10.1038/nature05824 447, 592 (2007); [DOI] [PMC free article] [PubMed] [Google Scholar]; Ingber D. E., J. Cell. Sci. 10.1242/jcs.00360 116, 1397 (2003); [DOI] [PubMed] [Google Scholar]; Sultan C., Stamenovic D., and Ingber D. E., Ann. Biomed. Eng. 10.1023/B:ABME.0000019171.26711.37 32, 520 (2004). [DOI] [PubMed] [Google Scholar]
- Van Vliet K. J., Bao G., and Suresh S., Acta Mater. 10.1016/j.actamat.2003.09.001 51, 5881 (2003); [DOI] [Google Scholar]; Lele T. P., Sero J. E., Matthews B. D., Kumar S., Xia S., Montoya-Zavala M., Polte T., Overby D., Wang N., and Ingber D. E., Methods Cell Biol. 83, 443 (2007). [DOI] [PubMed] [Google Scholar]
- Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 10.1021/ac980656z 70, 4974 (1998); [DOI] [PubMed] [Google Scholar]; Xia Y. N. and Whitesides G. M., Annu. Rev. Mater. Sci. 10.1146/annurev.matsci.28.1.153 28, 153 (1998). [DOI] [Google Scholar]
- Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 10.1126/science.288.5463.113 288, 113 (2000). [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Nature (London) 10.1038/nature05058 442, 368 (2006); [DOI] [PubMed] [Google Scholar]; Squires T. M. and Quake S. R., Rev. Mod. Phys. 10.1103/RevModPhys.77.977 77, 977 (2005); [DOI] [Google Scholar]; Stone H. A., Stroock A. D., and Ajdari A., Annu. Rev. Fluid Mech. 10.1146/annurev.fluid.36.050802.122124 36, 381 (2004). [DOI] [Google Scholar]
- El-Ali J., Sorger P. K., and Jensen K. F., Nature (London) 10.1038/nature05063 442, 403 (2006); [DOI] [PubMed] [Google Scholar]; Andersson H. and van den Berg A., Sens. Actuators B 10.1016/S0925-4005(03)00266-1 92, 315 (2003). [DOI] [Google Scholar]
- Kim L., Toh Y. C., Voldman J., Yu H., Lab Chip 7, 681 (2007). [DOI] [PubMed] [Google Scholar]
- Wolbers F., ter Braak P., Le Gac S., Luttge R., Andersson H., Vermes I., and van den Berg A., Electrophoresis 10.1002/elps.200600203 27, 5073 (2006). [DOI] [PubMed] [Google Scholar]
- Davidsson R., Boketoft A., Bristulf J., Kotarsky K., Olde B., Owman C., Bengtsson M., Laurell T., and Emneus J., Anal. Chem. 76, 4715 (2004). [DOI] [PubMed] [Google Scholar]
- Blau A. W. and Ziegler C. M., J. Biochem. Biophys. Methods 10.1016/S0165-022X(01)00163-4 50, 15 (2001); [DOI] [PubMed] [Google Scholar]; Lee J. N., Jiang X., Ryan D., and Whitesides G. M., Langmuir 20, 11684 (2004); [DOI] [PubMed] [Google Scholar]; Prokop A., Prokop Z., Schaffer D., Kozlov E., Wikswo J., Cliffel D., and Baudenbacher F., Biomed. Microdevices 6, 325 (2004); [DOI] [PubMed] [Google Scholar]; Tourovskaia A., Figueroa-Masot X., and Folch A., Lab Chip 10.1039/b405719h 5, 14 (2005); [DOI] [PubMed] [Google Scholar]; Gomez-Sjoberg R., Leyrat A. A., Pirone D. M., Chen C. S., and Quake S. R., Anal. Chem. 79, 8557 (2007); [DOI] [PubMed] [Google Scholar]; Futai N., Gu W., Song J. W., and Takayama S., Lab Chip 10.1039/b510901a 6, 149 (2006). [DOI] [PubMed] [Google Scholar]
- Komen J., Wolbers F., Franke H. R., Andersson H., Vermes I., and van den Berg A., Biomed. Microdevices 10, 727 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. M., Meyvantsson I., Shkel I. A., and Beebe D. J., Lab Chip 5, 1089 (2005); [DOI] [PubMed] [Google Scholar]; Hung P. J., Lee P. J., Sabounchi P., Lin R., and Lee L. P., Biotechnol. Bioeng. 10.1002/bit.20289 89, 1 (2005). [DOI] [PubMed] [Google Scholar]
- Li Y. S., Haga J. H., and Chien S., J. Biomech. 10.1016/j.jbiomech.2004.09.030 38, 1949 (2005). [DOI] [PubMed] [Google Scholar]
- Healy Z. R., Lee N. H., Gao X. Q., Goldring M. B., Talalay P., Kensler T. W., and Konstantopoulos K., Proc. Natl. Acad. Sci. U.S.A. 102, 14010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voldman J., Gray M., Toner M., and Schmidt M., Anal. Chem. 10.1021/ac0256235 74, 3984 (2002). [DOI] [PubMed] [Google Scholar]
- Wang M. M., Tu E., Raymond D. E., Yang J. M., Zhang H. C., Hagen N., Dees B., Mercer E. M., Forster A. H., Kariv I., Marchand P. J., and Butler W. F., Nat. Biotechnol. 10.1038/nbt1050 23, 83 (2005). [DOI] [PubMed] [Google Scholar]
- Hultstrom J., Manneberg O., Dopf K., Hertz H. M., Brismar H., and Wiklund M., Ultrasound Med. Biol. 10.1016/j.ultrasmedbio.2006.07.024 33, 145 (2007); [DOI] [PubMed] [Google Scholar]; Li H., Friend J. R., and Yeo L. Y., Biomaterials 10.1016/j.biomaterials.2007.06.005 28, 4098 (2007). [DOI] [PubMed] [Google Scholar]
- Gray B. L., Lieu D. K., Collins S. D., Smith R. L., and Barakat A. I., Biomed. Microdevices 4, 9 (2002). [Google Scholar]
- Frame M. D. and Sarelius I. H., Microcirculation (Philadelphia) 7, 419 (2000); [PubMed] [Google Scholar]; Cinamon G. and Alon R., J. Immunol. Methods 273, 53 (2003); [DOI] [PubMed] [Google Scholar]; Frame M. D. S., Chapman G. B., Makino Y., and Sarelius I. H., Biorheology 35, 245 (1998); [DOI] [PubMed] [Google Scholar]; Schaff U. Y., Xing M. M. Q., Lin K. K., Pan N., Jeon N. L., and Simon S. I., Lab Chip 7, 448 (2007); [DOI] [PubMed] [Google Scholar]; Tanaka Y., Kikukawa Y., Sato K., Sugh Y., and Kitamori T., Anal. Sci. 23, 261 (2007); [DOI] [PubMed] [Google Scholar]; Young E. W. K., Wheeler A. R., and Simmons C. A., Lab Chip 7, 1759 (2007); [DOI] [PubMed] [Google Scholar]; Ku C. J., Oblak T. D., and Spence D. M., Anal. Chem. 80, 7543 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu K., Pitchimani R., Dang D., Bayer K., Harrington T., and Pappas D., Langmuir 24, 5955 (2008); [DOI] [PubMed] [Google Scholar]; Shamloo A., Ma N., Poo M. M., Sohn L. L., and Heilshorn S. C., Lab Chip 8, 1292 (2008). [DOI] [PubMed] [Google Scholar]
- Borenstein J. T., Terai H., King K. R., Weinberg E. J., Kaazempur-Mofrad M. R., and Vacanti J. P., Biomed. Microdevices 4, 167 (2002). [DOI] [PubMed] [Google Scholar]
- Song J. W., Gu W., Futai N., Warner K. A., Nor J. E., and Takayama S., Anal. Chem. 10.1021/ac050131o 77, 3993 (2005). [DOI] [PubMed] [Google Scholar]
- Huh D., Fujioka H., Tung Y. C., Futai N., Paine R., Grotberg J. B., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0610868104 104, 18886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antia M., Herricks T., and Rathod P. K., Cell. Microbiol. 10, 1968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby J. P., White J., Ganesan K., Rathod P. K., and Chiu D. T., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2433968100 100, 14618 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. M., Eddington D. T., Bhatia S. N., and Mahadevan L., Proc. Natl. Acad. Sci. U.S.A. 104, 20496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth M. J., Lam W. A., and Fletcher D. A., Lab Chip 10.1039/b802931h 8, 1062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. Y., Chu L. Y., Chueh B. H., Shen M. W., Hazarika B., Phadke N., and Takayama S., Analyst (Cambridge, U.K.) 129, 1026 (2004); [DOI] [PubMed] [Google Scholar]; Atencia J. and Beebe D. J., Lab Chip 10.1039/b514070f 6, 567 (2006); [DOI] [PubMed] [Google Scholar]; Hudson S. D., Phelan F. R., Handler M. D., Cabral J. T., Migler K. B., and Amis E. J., Appl. Phys. Lett. 10.1063/1.1767594 85, 335 (2004); [DOI] [Google Scholar]; Lee J. S., Dylla-Spears R., Teclemariam N. P., and Muller S. J., Appl. Phys. Lett. 10.1063/1.2472528 90, 074103 (2007). [DOI] [Google Scholar]
- Yobas L., Tang K. C., Yong S. E., and Ong E. K. Z., Lab Chip 8, 660 (2008); [DOI] [PubMed] [Google Scholar]; Gu W., Zhu X. Y., Futai N., Cho B. S., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0404353101 101, 15861 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroock A. D., Dertinger S. K. W., Ajdari A., Mezic I., Stone H. A., and Whitesides G. M., Science 10.1126/science.1066238 295, 647 (2002). [DOI] [PubMed] [Google Scholar]
- Groisman A. and Steinberg V., Nature (London) 10.1038/35073524 410, 905 (2001). [DOI] [PubMed] [Google Scholar]
- Zhao X. M., Xia Y. N., and Whitesides G. M., J. Mater. Chem. 10.1039/a700145b 7, 1069 (1997). [DOI] [Google Scholar]
- Balaban N. Q., Schwarz U. S., Riveline D., Goichberg P., Tzur G., Sabanay I., Mahalu D., Safran S., Bershadsky A., Addadi L., and Geiger B., Nat. Cell Biol. 10.1038/35074532 3, 466 (2001). [DOI] [PubMed] [Google Scholar]
- Bischofs I. B. and Schwarz U. S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1233544100 100, 9274 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J., Levental I., Sengupta K., Georges P. C., and Janmey P. A., Biophys. J. 10.1529/biophysj.106.101386 93, 4453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L., and Discher D. E., Cell 10.1016/j.cell.2006.06.044 126, 677 (2006). [DOI] [PubMed] [Google Scholar]
- Hohne D. N., Younger J. G., and Solomon M. J., “Flexible microfluidic device for mechanical property characterization of soft viscoelastic solids such as bacterial biofilms,” Langmuir (submitted). [DOI] [PMC free article] [PubMed]
- Griffith L. G. and Swartz M. A., Nat. Rev. Mol. Cell Biol. 7, 211 (2006). [DOI] [PubMed] [Google Scholar]
- Khetani S. R. and Bhatia S. N., Nat. Biotechnol. 10.1038/nbt1361 26, 120 (2008). [DOI] [PubMed] [Google Scholar]
- Kane R. S., Takayama S., Ostuni E., Ingber D. E., and Whitesides G. M., Biomaterials 10.1016/S0142-9612(99)00165-9 20, 2363 (1999). [DOI] [PubMed] [Google Scholar]
- Vanapalli S. A., Wijnperle D., van den Berg A., Mugele F., and Duits M. H. G., “Programmable structured elastomeric membranes as a multifunctional microfluidic tool,” Lab Chip (submitted). [DOI] [PubMed]
- Takayama S., Ostuni E., LeDuc P. R., Naruse K., Ingber D. E., and Whitesides G. M., Nature (London) 10.1038/35082637 411, 1016 (2001). [DOI] [PubMed] [Google Scholar]
- Kuczenski B., LeDuc P. R., and Messner W. C., Lab Chip 10.1039/b617065j 7, 647 (2007). [DOI] [PubMed] [Google Scholar]
- Wang F., Wang H., Wang J., Wang H. Y., Rummel P. L., Garimella S. V., and Lu C., Biotechnol. Bioeng. 10.1002/bit.21737 100, 150 (2008). [DOI] [PubMed] [Google Scholar]
- Dertinger S. K. W., Chiu D. T., Jeon N. L., and Whitesides G. M., Anal. Chem. 10.1021/ac001132d 73, 1240 (2001); [DOI] [Google Scholar]; Irimia D., Geba D. A., and Toner M., Anal. Chem. 10.1021/ac0518710 78, 3472 (2006); [DOI] [PMC free article] [PubMed] [Google Scholar]; Campbell K. and Groisman A., Lab Chip 10.1039/b610011b 7, 264 (2007). [DOI] [PubMed] [Google Scholar]
- Abkarian M., Faivre M., and Stone H. A., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0507171102 103, 538 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao N., Zhan Y. H., and Lu C., Anal. Chem. 80, 7714 (2008). [DOI] [PubMed] [Google Scholar]
- Drochon A., Med. Eng. Phys. 27, 157 (2005); [DOI] [PubMed] [Google Scholar]; Lee W. G., Bang H., Yun H., Lee J., Park J., Kim J. K., Chung S., Cho S., Chung C., Han D. C., and Chang J. K., Lab Chip 10.1039/b614912j 7, 516 (2007). [DOI] [PubMed] [Google Scholar]
- Korin N., Bransky A., and Dinnar U., J. Biomech. 40, 2088 (2007); [DOI] [PubMed] [Google Scholar]; Gifford S. C., Frank M. G., Derganc J., Gabel C., Austin R. H., Yoshida T., and Bitensky M. W., Biophys. J. 84, 623 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesewetter H., Dauer U., Teitel P., Schmidschonbein H., and Trapp R., Biorheology 19, 737 (1982). [DOI] [PubMed] [Google Scholar]
- Frank R. S. and Hochmuth R. M., J. Biomech. 109, 103 (1987); [DOI] [PubMed] [Google Scholar]; Frank R. S. and Tsai M. A., J. Biomech. 112, 277 (1990). [DOI] [PubMed] [Google Scholar]
- Groisman A., Enzelberger M., and Quake S. R., Science 10.1126/science.1083694 300, 955 (2003). [DOI] [PubMed] [Google Scholar]
- Vanapalli S. A., van den Ende D., Duits M. H. G., and Mugele F., Appl. Phys. Lett. 10.1063/1.2713800 90, 114109 (2007). [DOI] [Google Scholar]
- Vanapalli S. A., Banpurkar A. G., van den Ende D., Duits M. H. G., and Mugele F., “Hydrodynamic resistance of single confined moving drops in rectangular microchannels” Lab Chip 10.1039/B815002H. [DOI] [PubMed]
- Guck J., Ananthakrishnan R., Mahmood H., Moon T. J., Cunningham C. C., and Kas J., Biophys. J. 81, 767 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Roure O., Saez A., Buguin A., Austin R. H., Chavrier P., Siberzan P., and Ladoux B., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0408482102 102, 2390 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0235407100 100, 1484 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniadecki N., Anguelouch A., Yang M. T., Lamb C. M., Liu Z., Kirschner S. B., Liu Y., Reich D. H., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0611613104 104, 14553 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Park S. J., and Park J. K., Ann. Phys. 133, 1432 (2008). [Google Scholar]
- Teh S. Y., Lin R., Hung L. H., and Lee A. P., Lab Chip 10.1039/b715524g 8, 198 (2008); [DOI] [PubMed] [Google Scholar]; Christopher G. F. and Anna S. L., J. Phys. D 10.1088/0022-3727/40/19/R01 40, R319 (2007). [DOI] [Google Scholar]
- Song H., Chen D. L., and Ismagilov R. F., Angew. Chem., Int. Ed. 10.1002/anie.200601554 45, 7336 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach S. L., Song H., and Ismagilov R. F., Anal. Chem. 77, 785 (2005); [DOI] [PMC free article] [PubMed] [Google Scholar]; Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angile F. E., Schmitz C. H., Koester S., Duan H., Humphry K. J., Scanga R. A., Johnson J. S., Pisignano D., and Weitz D. A., Lab Chip 10.1039/b806706f 8, 1632 (2008). [DOI] [PubMed] [Google Scholar]
- Mugele F. and Baret J. C., J. Phys.: Condens. Matter 10.1088/0953-8984/17/28/R01 17, R705 (2005). [DOI] [Google Scholar]
- Fair R. B., Microfluid. Nanofluid. 10.1007/s10404-007-0161-8 3, 245 (2007). [DOI] [Google Scholar]
- Gu H., Malloggi F., Vanapalli S. A., and Mugele F., Appl. Phys. Lett. 10.1063/1.3013567 93, 183507 (2008). [DOI] [Google Scholar]
- Malloggi F., Vanapalli S. A., Gu H., van den Ende D., and Mugele F., J. Phys.: Condens. Matter 10.1088/0953-8984/19/46/462101 19, 462101 (2007). [DOI] [Google Scholar]
- Edd J. F., Di Carlo D., Humphry K. J., Koster S., Irimia D., Weitz D. A., and Toner M., Lab Chip 10.1039/b805456h 8, 1262 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]; Chabert M. and Viovy J. L., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0708321105 105, 3191 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]; He M. Y., Edgar J. S., Jeffries G. D. M., Lorenz R. M., Shelby J. P., and Chiu D. T., Anal. Chem. 10.1021/ac0480850 77, 1539–1544 (2005). [DOI] [PubMed] [Google Scholar]
- Barbuloviv-Nad I., Yang H., Park P. S., and Wheeler A. R., Lab Chip 10.1039/b717759c 8, 519 (2008). [DOI] [PubMed] [Google Scholar]
- Rondelez Y., Tresset G., Tabata K. V., Arata H., Fujita H., Takeuchi S., and Noji H., Nat. Biotechnol. 10.1038/nbt1072 23, 361 (2005); [DOI] [PubMed] [Google Scholar]; Jung S. Y., Liu Y., and Collier C. P., Langmuir 24, 4439 (2008); [DOI] [PubMed] [Google Scholar]; Song H. and Ismagilov R. F., J. Am. Chem. Soc. 10.1021/ja0354566 125, 14613 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens M. M. A. E., Tharmann R., Kroy K., and Bausch A. R., Nat. Phys. 2, 186 (2006). [Google Scholar]