Abstract
Aberrant hypermethylation of gene promoter regions is one of the mechanisms for inactivation of tumour suppressor genes in many human cancers including breast carcinoma. In the current study, we aimed to assess by MSP, the methylation pattern of two cancer-related genes involved in DNA repair: hMLH1 (mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) and BRCA1 (breast cancer 1, early onset) in 78 primary breast cancers from Tunisian patients. The methylation frequencies were 24.36% for hMLH1 and 46% for BRCA1. BRCA1 methylation correlated with age at diagnosis (P = .015) and 5-years disease free survival (P = .016) while hMLH1 methylation was more frequent in larger tumors (P = .002) and in presence of distant metastasis (P = .004). Furthermore, methylation of hMLH1 significantly correlated with high level of P53 expression (P = .006) and with overall survival (P = .015) suggesting that silencing of hMLH1 through aberrant promoter methylation could be used as a poor prognosis indicator in breast cancer.
1. Introduction
Breast cancer is the first cause of cancer mortality among women worldwide and the overall lifetime risk for developing invasive breast cancer is estimated to 10% [1]. In Tunisia, breast cancer is the second most frequent cancer among females and in the south area the incidence is of 30/100000 [2]. Tunisian breast cancer is characterized by a particular aggressive profile compared with Western countries as the incidence is more prevalent in young (<35 years) than older patients [3]. Breast cancer results of abnormal genetic as well as epigenetic changes [4]. Promoter-CpG islands hypermethylation, associated with global hypomethylation, are common molecular defects in cancer cells [5].
CpG islands, frequently located at the 5′-end regulatory regions of genes, are subject to epigenetic modifications including DNA methylation and histone modification that are known to play an important role in regulating gene expression [5]. In normal cells, the majority of promoter CpG islands are protected from this epigenetic event and thus they are unmethylated. Conversely, in cancer cells, several promoter CpG islands are hypermethylated and form a closed repressive chromatin configuration that affects the transcription initiation of the corresponding genes [6–8].
In breast cancer, more than 40 genes have been shown to be inactivated by methylation including those involved in DNA repair, cell-cycle regulation, tumor suppression, cell adhesion and cell signalling [9–12]. The identification of these methylated genes has significantly contributed to elucidate the molecular pathways which are altered in breast carcinoma and provided potential targets for molecular detection.
Recent studies suggested that methylation profiles of cancers depend on tumor type and ethnicity [13, 14]. In this context, the methylation status of breast cancer patients from Western population is well documented whereas that of Tunisian breast cancer is still not well studied except a recent work on the relationship between SV40 status and promoter hypermethylation in patients from the central region of Tunisia [15].
Therefore, our study was initiated with the aim to assess the methylation profile of two genes involved in DNA repair: hMLH1 (mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) and BRCA1 (breast cancer 1, early onset). We next explored for associations between gene methylation and clinicopathological features, hormone receptors, and patients survival.
BRCA1 and hMLH1 have been shown to be methylated in tumors leading to the downregulation of gene expression [16–20]. The hMLH1 gene is a member of the DNA mismatch repair system which corrects DNA base-pairing errors in newly replicated DNA [21]. Deficiencies in this system result in mutation rates 100-fold higher than those observed in normal cells [22, 23]. Besides its involvement in DNA repair, BRCA1 is a tumor suppressor gene located on 17q12-21. Loss of the wild-type allele (LOH) is required for tumorigenesis in germ line mutation carriers. Hypermethylation of the BRCA1 promoter has been shown to be one of the mechanisms leading to functional inactivation of BRCA1 [24].
2. Material and Methods
2.1. Patients Characteristics
A total of 78 fresh-frozen primary breast cancer tissues were collected from the Anatomo-Pathology; CHU Habib Bourguiba of Sfax in Tunisia with Institutional Review Board approval. The age at diagnosis ranged from 28 to 73 years, with a mean of 50.6. All cases were graded according to the modified Scarff- Bloom-Richardson system [25]. The clinical stage of the disease was determined according to TNM classification of the International Union Against Cancer [26]. Clinical-pathological data (age, histological grade, tumor size, lymph node status, and distant metastasis) are summarized in Table 1.
Table 1.
Summary of primer sequences, annealing temperature, product size and number of cycle used in MSP assay. M: methylated DNA, U: unmethylated DNA, F: forward, R: Reverse.
| Gene | Sequence (5′-3′) | Tm (°C) | Size (bp) | PCR cycles | |
|---|---|---|---|---|---|
| hMLH1 (U) | F | TTTTGATGTAGATGTTTTATTAGGGTTGT | 60 | 114 | 30 |
| R | ACCACCTCATCATAACTACCCACA | ||||
| hMLH1 (M) | F | ACGTAGACGTTTTATTAGGGTCGC | 58 | 110 | 30 |
| R | CCTCATCGTAACTACCCGCG | ||||
| BRCA1I (U) | F | GGTTAATTTAGAGTTTTGAGAGATG | 60 | 75 | 30 |
| R | TCAACAAACTCACACCACACAATCA | ||||
| BRCA1 (M) | F | GGTTAATTTAGAGTTTCGAGAGACG | 60 | 86 | 30 |
| R | TCAACGAACTCACGCCGCGCAATCG |
2.2. DNA Extraction
Genomic DNA was extracted from fresh-frozen specimens using the standard protocol with Proteinase K in the presence of SDS at 37°C overnight, followed by phenol/chloroform extraction as described previously [27]. Briefly tissues sections were incubated in 100 μL TE (10 mM Tris-Hcl, pH 8.0, 1 mM EDTA) containing 40 μg proteinase K at 55°C for 4 hours. DNA was extracted with a mixture of phenol-chloroform-isoamyl alcohol (25 : 24 : 1) and precipitated with ethanol at −20°C. The quantity of DNA was checked by spectrophotometer and stored at −20°C for further use.
2.3. Methylation-Specific-PCR (MSP)
DNA samples (1 to 2 μg) were modified with sodium bisulfite that converts the unmethylated cytosines but not the methylated ones to uracil using the Methyl Detector Kit as recommended by Active Motif (Belgium). The bisulfite-treated DNA was amplified using specific primers for methylated and unmethylated alleles as described by Herman et al. [28]. Sequences of the primers, annealing temperature, and product size are listed in Table 1.
PCR was carried out in a total volume of 25 μL containing 2 μM of each primer pair, 200 μM dNTP, 1.5 mM MgCl2, 1X PCR buffer, and 1 U of Taq DNA polymerase (Fermentas). Each PCR reaction underwent initial denaturation at 95°C for 10 minutes, and 35 cycles of the following profile: 30 seconds at 94°C, 30 seconds at optimal T° (Table 1), and 30 seconds at 72°C followed by a final 10 minutes extension at 72°C. Fully methylated DNA (Active Motif, Belgium) and lymphocytes DNA from healthy individual were used as positive controls for the methylated and unmethylated reactions, respectively. Blank control without DNA was included in each PCR assay. After amplification, products were electrophoresed using 2% agarose gels, stained with ethidium bromide and visualized under UV illumination.
2.4. Immunohistochemistry
Immunostaining of Estrogen (ER) and Progesteron (PR) receptors was performed for all specimens, while HER/neu and p53 protein expression was investigated in 64 and 39 specimens, respectively. Four micrometers sections attached on silanized slides were de-waxed in xylene, rehydrated in graded ethanol, and covered with 10 mM citrate buffer (pH 6). Slides were incubated for 30 minutes with primary monoclonal antibodies against: ER (Dako, clone 1D5, 1 : 25), PR (Dako, clone PgR636, 1 : 50), HER2/neu (Dako, clone 124, 1 : 100), and p53 (clone DO-7, Dako Cytomation 1 : 50) followed by incubation with biotin-labelled secondary antibodies. The streptavidin-peroxidase complex was visualized using diaminobenzidine as a chromogenic substrate. All slides were evaluated without knowledge of the clinical outcome. Sections were considered to be positive if more than 5% of tumor cells were stained. For each run of staining, positive control slides were prepared from breast carcinoma known to be positive for the proteins under study. The HER2/neu and p53 immunostaining were scored from 0 to 3 according to the criteria set by Dako. The staining was scored as negative (0) when no membrane staining was observed for HER2/neu, and no nuclear staining for p53, or when staining was observed in less than 10% of the tumor cells, weak positive (1+) if weak focal staining was seen in more than 10% of the tumor cells, intermediate (2+), if weak to moderate, complete staining was seen in more than 10% of the tumor cells and strongly positive (3+) if intense and complete staining in more than 10% of the tumor cells. In the final analysis, scores 0, 1, and 2 were considered as negative, only score 3 was retained as positive in HER2/neu overexpression cases. However, scores 0 and 1 were negative and score 2 and 3 were considered as positive for p53.
2.5. Data Analysis and Statistics
The association between aberrant hypermethylation and clinicopathological parameters was checked by the χ2 test. All variables (age, stage, histological grade, lymph node involvement, distal metastasis, hormone receptor status, and CpG island hypermethylation) were also subjected to multivariate analysis using Logistic regression. Survival curves were constructed according to the method of Kaplan Meier. For differences between curves, the P value was calculated using the log rank test. All the statistical analysis was performed using the software SPSS for Windows version 13.0. P < .05 was considered as statistically significant.
3. Results and Discussion
3.1. Methylation Profiles of BRCA1 and hMLH1 in Tumor Tissues
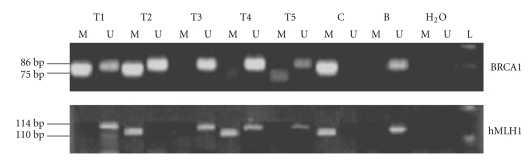
Aberrant methylation of the promoter region is considered as one of the major mechanisms for silencing of cancer-related genes, resulting in downregulation of gene expression. It has been demonstrated that CpG islands hypermethylation is implicated in the loss of expression of critical tumor suppressor and growth regulatory genes leading to cancer development and progression [5–8]. Analysis of promoter methylation of BRCA1 and hMLH1 was carried out in 78 and 74 invasive primary breast tissues, respectively. Figure 1 shows representative examples of MSP results. The frequency of promoter hypermethylation was 44.8% (35 out of 78) for BRCA1 and 24.3% (17 out of 74) for hMLH1. Among the 78 malignant tissues, more than a half exhibited at least one methylated gene (62%). In other hand, for some samples, we observed both the methylated and unmethylated alleles, this can be probably explained by the presence of infiltrating lymphocytes and/or nonmalignant epithelial cells in the primary tumors.
Figure 1.
Electrophoresis of PCR products spanning the BRCA1 and hMLH1 promoters from bisulfite-treated DNA in breast cancer tissues. Each lane contains products generated from separate PCR reactions using primers specific for unmethylated (U) or methylated (M) DNA template. Fully methylated DNA from Active Motif and Blood from healthy individual used as a positive control (C) for methylated and unmethylated DNA (B). The lanes marked (H2O) indicates water only without the addition of DNA and (L) 100-bp ladder as molecular weight markers (Fermentas).
When we reviewed the literature, the methylation frequency ranged from 8 to 43.5% for hMLH1 and from 11 to 60% for BRCA1 [20, 29–32]. In our series, the frequencies of BRCA1 and hMLH1 methylation were in the middle of the range of previous studies. These differences may be explained by several factors: (i) unmethylated DNA from the normal cells infiltrating the tumor might attenuate the methylation levels and (ii) number of CpG within the region of interest. To avoid misinterpretation of our results, we included in each assay, both methylated and unmethylated DNA as controls.
On the other hand, several investigations have attempted to identify the racial/ethnic differences in promoter hypermethylation. Bae et al. evaluated promoter methylation status in American and Korean breast cancers patients and they found no significant interethnic differences [13]. More recently, it was reported that there are many similarities of promoter hypermethylation profiles between Korean and Caucasian women, but also dissimilarities that characterize tumors of one ethnicity from the other [14].
3.2. Relationship between Methylation Status and Clinicopathological Characteristics
The relationship between promoter hypermethylation of hMLH1 and BRCA1 genes and clinicopathological parameters was summarized in Table 2. Statistical analysis showed that BRCA1 hypermethylation was associated with age at diagnosis (P = .015, Table 2) since 72% of patients ≤45 years displayed the methylated pattern. This observation suggests that loss of BRCA1 expression through aberrant promoter methylation occurred more frequently in young women with breast carcinoma which is known to have a particular more aggressive feature. Despite age, no other association was found with patients' characteristics and BRCA1 methylation status. Nevertheless, by multivariate analysis, we found a significant association of BRCA1 promoter methylation with clinical stage, tumor size and 5-year survival (P = .022, .037 and .017, resp., Table 3).
Table 2.
Association between gene promoter methylation and clinicopathological features ER and p53 expression in breast cancer.
| BRCA1 | hMLH1 | ||||||
|---|---|---|---|---|---|---|---|
| M (%) | U (%) | P(b) | M (%) | U (%) | P(b) | ||
| N | 78 | 35 (44.8) | 43 (55.1) | 17 (21.7) | 57 (73) | ||
| Age | |||||||
| ≤45 | 30 | 19 (63.3) | 11 (36.7) | 6 (21.4) | 22 (78.6) | ||
| >45 | 48 | 16 (33.3) | 32 (66.7) | 11 (23.9) | 35 (76.1) | ||
| .01 | .8 | ||||||
| Tumor size | |||||||
| T ≤ 30 mm | 38 | 18 (47.4) | 20 (52.6) | 3 (8.1) | 34 (91.9) | ||
| T > 30 mm | 40 | 17 (42.5) | 23 (57.5) | 14 (37.8) | 23 (62.2) | ||
| .66 | .002 | ||||||
| (a)Metastasis | |||||||
| No | 37 | 18 (48.6) | 19 (51.4) | 1 (2.8) | 35 (97.2) | ||
| Yes | 14 | 8 (37.1) | 6 (42.9) | 4 (30.8) | 9 (69.2) | ||
| .58 | .004 | ||||||
| (a)DFS | |||||||
| <5 years | 22 | 7 (31.8) | 15 (68.2) | 3 (14.3) | 18 (87.5) | ||
| >5 years | 12 | 9 (75) | 3 (25) | 1 (8.3) | 11 (91.7) | ||
| .015 | .6 | ||||||
| (a)Response to radiotherapy | |||||||
| Radiosensitive | 37 | 18 (48.6) | 19 (51.4) | 1 (2.8) | 35 (97.2) | ||
| Radioresistant | 14 | 8 (57.1) | 6 (42.9) | 4 (30.8) | 9 (69.2) | ||
| .58 | .004 | ||||||
| ER | |||||||
| Positive | 19 | 9 (47.4) | 10 (52.6) | 7 (36.8) | 12 (63.2) | ||
| Moderate | 28 | 11 (39.3) | 17 (60.7) | 2 (7.7) | 24 (92.3) | ||
| Negative | 31 | 15 (48.4) | 16 (51.6) | 8 (27.6) | 21 (72.4) | ||
| .75 | .054 | ||||||
| (a)P53 | |||||||
| Positive | 16 | 9 (56.3) | 7 (43.8) | ||||
| Negative | 23 | 10 (43.5) | 13 (56.5) | 5 (68.8) | 11 (31.3) | ||
| 0 (0) | 21 (100) | ||||||
| .44 | .006 | ||||||
DFS: Disease Free Survival, ER: Estrogen Receptor.
(a)Data for DFS, response to radiotherapy, metastasis, and p53 status were not available for all specimens.
(b)Comparisons of DFS and P53 status was made by Fisher's exact test and all other comparisons were by the 2 test.
Table 3.
Multivariate analysis of BRCA1 methylation status and clinical features.
| Clinical features | BRCA1 Methylation | ||
|---|---|---|---|
| P | OR | 95%CI | |
| Age | .11 | 0.17 | 0.02–1.5 |
| TNM | .218 | 0.173 | 0.01–2.81 |
| Clinical stage | .022 | 53.8 | 1.7–1640 |
| Metastasis | .891 | 0.8 | 0.03–16.6 |
| Tumor size | .037 | 0.05 | 0.004–0.84 |
| 5-y survival | .017 | 20.7 | 1.7–251.5 |
The most consistent associations were seen between hMLH1 promoter hypermethylation and tumor size and distant metastasis (P = .002 and .004, resp., Table 2). This result leads us to suggest that hypermethylation of hMLH1 gene promoter seemed to confer advantage for tumors cells invasion, and it may be used as a marker of advanced breast cancer. In this context, it was reported that hypermethylation of hMLH1 was significantly associated with advanced stage and lymphatic metastasis [19, 29]. Our finding is consistent with those studies since a trend of association was found between methylated hMLH1 profile and clinical stage of breast cancer. In fact, 14.3% of hMLH1 methylated tumors are of stage T III, IV versus 4.8% of early stage although the difference was not statistically significant (P = .276). Furthermore, we detected a significant correlation between response to treatment and hMLH1 methylation by univariate (P = .004, Table 2).
3.3. Relationship between Methylation of Individual Genes and ER, PR, HER2/neu, and P53 Status
The ER, PR, and HER2/neu are important prognostic biomarkers and therapeutic targets in primary breast cancer. ER-negative tumors appear to be more malignant, resulting in a poorer prognosis than ER-positive tumors [33, 34]. Expression of HER2/neu, ER, and PR proteins are considered as predictive marker for response to hormone therapy in breast cancer. High to moderate level of ER expression was detected in 60.3% of tumors while 39.7% of cases were ER− (Table 1). Univariate analysis, revealed that ER status was weakly associated with hMLH1 methylation (P = .054, Table 1). No other significant association between the methylation profile of hMLH1 or BRCA1 and PR, HER2/neu status was noted in our series. In breast cancer, methylation has already been connected to hormone regulation, but the correlation is not clear yet. Campan et al., reviewed the DNA methylation profiles of breast, endometrial, ovarian, and proximal colon cancers but did not find evidence for global hormone-specific DNA methylation alterations [35]. On the other hand, Widschwendter et al. reported significant differences in hormone receptor status between clusters of DNA methylation profiles [36]. Recently, it was demonstrated that epigenetic differences between ER-positive and ER-negative breast tumors arise early in cancer development and persist during cancer progression [37].
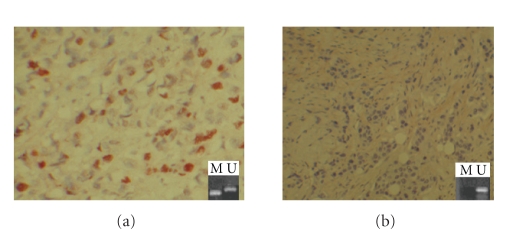
In the current study, p53 expression was assessed by IHC in 39 specimens. Nuclear staining was seen in 41% (16 out of 39) of cases. Figure 2 shows an example of specimen with p53 positive (a) and negative (b) nuclear staining. Interestingly, we found that all tumors displaying unmethylated hMLH1 profile were negative for p53 (P = .006, Table 2, Figure 2). This observation suggests a positive association between methylated hMLH1 and functional p53. Furthermore, p53 expression was significantly connected to hormone receptor status. Indeed, high level of p53 expression was associated with double negative (ER−/PR−, P = .001 and ER−/HER2−, P = .011, Table 4) and triple negative (ER−/PR−/HER2−, P = .01, Table 4) tumors. Our data was consistent with previously reported results showing that triple negative breast cancers correlated with high p53 expression level [37]. It was well established that triple negative breast cancer has aggressive clinical features and reduced survival [38]. In the current work, we showed that triple negative breast cancer correlated with 5-year survival, (P = .002; OR = 0.083, 95%CI = 0.015–0.462) while no statistically significant association was found with overall survival and triple negative tumors in our series. Recently, Rhee et al., reported that most of the relapses in triple negative breast cancer occur within the first 3 years, in contrast to tumors expressing ER, PR, and HER2/neu [37]. Altogether, these findings reflect the aggressiveness of triple negative breast cancer as previously reported [38].
Figure 2.
Representative result of p53 immunohistochemical staining and hMLH1 promoter methylation. (a) Breast tumor specimen with p53 nuclear positivity associated with both hMLH1 methylated and unmethylated DNA. (b) Breast tumor specimen negative for p53 correlated with unmethylated hMLH1 profile.
Table 4.
Correlation between p53 expression level and double negative (ER−/PR−, ER−/HER2− ) and triple negative (ER−/PR−/HER2−) breast cancer.
| N | ER−/PR− | ER−/HER2− | ER−/PR−/HER2− | ||||
|---|---|---|---|---|---|---|---|
| Yes | No | Yes | No | Yes | No | ||
| P53 | |||||||
| Positive | 16 | 10 (62.5%) | 6 (37.5%) | 8 (50%) | 8 (50%) | 7 (43.8%) | 9 (56.3%) |
| Negative | 23 | 3 (13%) | 20 (87%) | 3 (13%) | 20 (87%) | 2 (8.7%) | 21 (91.3%) |
| P (t-test) | .001 | .011 | .01 | ||||
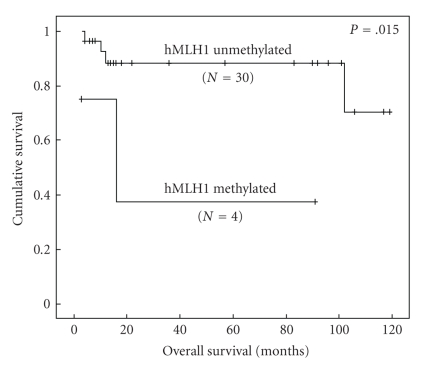
3.4. Relationship between Methylation of hMLH1 and Overall Survival
The survival rate was available for 34 patients and the follow-up time ranged from 3 to 119 months. The Kaplan-Meier method was used to correlate hMLH1 and BRCA1 methylation status with overall survival. As shown on Figure 3, patients with unmethylated hMLH1 gene promoter have a significant prolonged survival rate compared to those with methylated profile (P log rank = .015). To the best of our knowledge, this is the first report about the involvement of hMLH1 hypermethylation with patients' outcome suggesting that it could be an important prognostic factor of breast cancer. However this finding should be confirmed on larger series. In the current study, BRCA1 methylation and survival association was of borderline significance (P log-rank = .065). Recently, it was reported in a large cohort of 851 patients that BRCA1 methylation was associated with breast cancer-specific mortality [39].
Figure 3.
Kaplan-Meier survival curve correlating overall survival with hMLH1 methylated and unmethylated profile.
4. Conclusion
In summary, we examined BRCA1 and hMLH1 promoter methylation status and explored the relationship with clinicopathological factors and breast cancer survival. Promoter methylation of hMLH1 was more frequent in cancers with distant metastasis and in larger tumors (greater than 3 cm) whereas BRCA1 hypermethylation was higher in patients under 45 years. Furthermore, aberrant methylation of hMLH1 correlated with reduced overall survival while BRCA1 hypermethylation associated with 5-year survival. Our results indicate that BRCA1 and hMLH1 promoter methylation could be an important prognostic factor of breast cancer.
Acknowledgments
This work was supported by a Grant of the “Ministère de l’Enseignement Supérieur et de la Recherche Scientifique Tunisien”. The authors would like to thank all clinicians and pathologists at CHU Habib Bourguiba at Sfax-Tunisia, and Dr. Khemais Ben-Hadj for critical suggestions.
References
- 1.Feuer EJ, Wun L-M, Boring CC, Flanders WD, Timmel MJ, Tong T. The lifetime risk of developing breast cancer. Journal of the National Cancer Institute. 1993;85(11):892–897. doi: 10.1093/jnci/85.11.892. [DOI] [PubMed] [Google Scholar]
- 2.Mourali N, Muenz LR, Tabbane F, Belhassen S, Bahi J, Levine PH. Epidemiologic features of rapidly progressing breast cancer in Tunisia. Cancer. 1980;46(12):2741–2746. doi: 10.1002/1097-0142(19801215)46:12<2741::aid-cncr2820461234>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Ferlay J, Hamdi-Cherif M, et al. Cancer in Africa: Epidemiology and Prevention. Vol. 153. Lyon, France: IARC Scientific; 2003. Breast Cancer; pp. 262–267. [Google Scholar]
- 4.Russo J, Yang X, Hu YF, et al. Biological and molecular basis of human breast cancer. Frontiers in Bioscience. 1998;3:D944–D960. doi: 10.2741/a335. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Research. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 8.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nature Reviews Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 9.Asch BB, Barcellos-Hoff MH. Epigenetics and breast cancer. Journal of Mammary Gland Biology and Neoplasia. 2001;6(2):151–152. doi: 10.1023/a:1011306222533. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann U, Langer F, Feist H, Glockner S, Hasemeier B, Kreipe H. Quantitative assessment of promoter hypermethylation during breast cancer development. American Journal of Pathology. 2002;160(2):605–612. doi: 10.1016/S0002-9440(10)64880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21(35):5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 12.Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochemical Pharmacology. 2004;68(6):1187–1197. doi: 10.1016/j.bcp.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 13.Bae YK, Brown A, Garrett E, et al. Hypermethylation in histologically distinct classes of breast cancer. Clinical Cancer Research. 2004;10(18, part 1):5998–6005. doi: 10.1158/1078-0432.CCR-04-0667. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Lo P-K, Fackler MJ, et al. A comparative study of Korean with Caucasian breast cancer reveals frequency of methylation in multiple genes correlates with breast cancer in young, ER, PR-negative breast cancer in Korean women. Cancer Biology and Therapy. 2007;6(7):1114–1120. doi: 10.4161/cbt.6.7.4331. [DOI] [PubMed] [Google Scholar]
- 15.Hachana M, Trimeche M, Ziadi S, Amara K, Korbi S. Evidence for a role of the Simian Virus 40 in human breast carcinomas. Breast Cancer Research and Treatment. 2009;113(1):43–58. doi: 10.1007/s10549-008-9901-z. [DOI] [PubMed] [Google Scholar]
- 16.Catteau A, Harris WH, Xu C-F, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18(11):1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 17.Bianco T, Georgia CT, David CAW, John EC, Alexander D. Tumor specific distribution of BRCA1 promoter region methylation supports a pathogenic role in breast and ovarian cancer. Carcinogenesis. 2000;21(2):147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 18.Butcher DT, Rodenhiser DI. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. European Journal of Cancer. 2007;43(1):210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Murata H, Khattar NH, Kang Y, Gu L, Li G-M. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002;21(37):5696–5703. doi: 10.1038/sj.onc.1205683. [DOI] [PubMed] [Google Scholar]
- 20.Murata H, Khattar NH, Gu L, Li G-M. Roles of mismatch repair proteins hMSH2 and hMLH1 in the development of sporadic breast cancer. Cancer Letters. 2005;223(1):143–150. doi: 10.1016/j.canlet.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 21.Jascur T, Boland CR. Structure and function of the components of the human DNA mismatch repair system. International Journal of Cancer. 2006;119(9):2030–2035. doi: 10.1002/ijc.22023. [DOI] [PubMed] [Google Scholar]
- 22.Thomas DC, Umar A, Kunkel TA. Microsatellite instability and mismatch repair defects in cancer. Mutation Research. 1996;350(1):201–205. doi: 10.1016/0027-5107(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 23.Jiricny J. The multifaceted mismatch-repair system. Nature Reviews Molecular Cell Biology. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 24.Catteau A, Morris JR. BRCA1 methylation: a significant role in tumour development? Seminars in Cancer Biology. 2002;12(5):359–371. doi: 10.1016/s1044-579x(02)00056-1. [DOI] [PubMed] [Google Scholar]
- 25.Elston CW, Ellis IO, Goulging H, Pindre SE. Elston CW, Ellis IO, editors. Role of pathology in the prognosis and management of breast cancer. Systemic Pathology. (3rd edition) 1998;13:385–433. [Google Scholar]
- 26.Sobin LH, Wittekind Ch, editors. International Union Against Cancer, TNM Classification of Malignant Tumours. 5th edition. New York, NY, USA: Wiley-Liss; 1997. [Google Scholar]
- 27.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 28.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naqvi RA, Hussain A, Deo SSV, et al. Hypermethylation analysis of mismatch repair genes (hmlh1 and hmsh2) in locally advanced breast cancers in Indian women. Human Pathology. 2008;39(5):672–680. doi: 10.1016/j.humpath.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Research. 2006;8(4):1–10. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei M, Grushko TA, Dignam J, et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Research. 2005;65(23):10692–10699. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Letters. 2006;237(2):272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Duffy MJ. Estrogen receptors: role in breast cancer. Critical Reviews in Clinical Laboratory Sciences. 2006;43(4):325–347. doi: 10.1080/10408360600739218. [DOI] [PubMed] [Google Scholar]
- 34.Rusiecki JA, Holford TR, Zahm SH, Zheng T. Breast cancer risk factors according to joint estrogen receptor and progesterone receptor status. Cancer Detection and Prevention. 2005;29(5):419–426. doi: 10.1016/j.cdp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Campan M, Weisenberger DJ, Laird PW. DNA methylation profiles of female steroid hormone-driven human malignancies. Current Topics in Microbiology and Immunology. 2006;310:141–178. doi: 10.1007/3-540-31181-5_8. [DOI] [PubMed] [Google Scholar]
- 36.Widschwendter M, Siegmund KD, Müller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Research. 2004;64(11):3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 37.Rhee J, Han S-W, Oh D-Y, et al. The clinicopathologic characteristics and prognostic significance of triple-negativity in node-negative breast cancer. BMC Cancer. 2008;8(308):1–8. doi: 10.1186/1471-2407-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvin WJ, Jr., Carey LA. What is triple-negative breast cancer? European Journal of Cancer. 2008;44(18):2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 39.Xu X, Gammon MD, Zhang Y, et al. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Research and Treatment. 2009;115(2):397–404. doi: 10.1007/s10549-008-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]